Abstract
Light chain amyloidosis (AL amyloidosis) is a hematological disorder in which a clonal population of B cells expands and secretes enormous amounts of the immunoglobulin light chain protein. These light chains misfold and aggregate into amyloid fibrils, leading to organ dysfunction and death. We have studied the in vitro fibril formation kinetics of two patient-derived immunoglobulin light chain variable domain proteins, designated AL-09 and AL-103, in response to changes in solution conditions. Both proteins are members of the κI O18:O8 germline and therefore are highly similar in sequence, but they presented with different clinical phenotypes. We find that AL-09 forms fibrils more readily and more rapidly than AL-103 in vitro, mirroring the clinical phenotypes of the patients and suggesting a possible connection between the fibril kinetics of the disease protein and the disease progression.
Keywords: amyloid, light chain amyloidosis, amyloid fibril kinetics, electrostatics, immunoglobulin light chain
Introduction
Protein misfolding disorders are a major cause of morbidity and mortality [1–5]. One such disorder is light chain amyloidosis, or AL amyloidosis. In this disease, there is a mild expansion of a clonal B cell population and a subsequent production and secretion of immunoglobulin (Ig) light chains. These light chains misfold and aggregate, leading to amyloid deposition and organ malfunction. Light chain amyloidosis is typically fatal, with a median survival time of 12–40 months following diagnosis [6–8]. While the typical treatment (chemotherapy with or without autologous stem cell transplantation) targets the clonal B cell population, all evidence indicates that the pathology arises from the overproduced Ig light chain. A better understanding of the disease protein could lead to therapy that directly targets the protein misfolding.
AL amyloidosis has several unique characteristics that have drawn the attention of clinicians and researchers. AL amyloid deposits may be found in virtually every organ system (except the central nervous system), but the choice and combination of organs involved is seemingly random [9–11]. The most commonly clinically involved organs are the kidneys [12,13], heart [14], and liver [15]. While there is some correlation between germline usage and organ involvement, there is no way of predicting which organs will be affected in which patients. The differences in organ involvement lead to a wide variability in disease severity. Patients with cardiac involvement have a lower survival rate than AL amyloidosis patients as a whole, with the median survival time falling below 12 months [7]. But even within these broad categories there is a large variability in survival, with documented cardiac amyloidosis patients surviving longer than 10 years [16]. While other co-morbidities play an important role in survival, differences in the light chain sequence among different AL amyloidosis patients appear to play a key role in mediating the severity of the disease. Due to somatic hypermutation occurring during B cell maturation, each Ig light chain has a novel combination of mutations. This unique property of immunoglobulin sequences differentiates AL amyloidosis from other amyloid diseases. In AL amyloidosis, rather than having a common protein sequence with a few familial variants, each patient has a different protein sequence. While the sequences are still highly similar, particularly because a few germline sequences are highly overrepresented[14,17–19], it is possible that the few amino acid changes contribute to the diversity of phenotype seen among patients with AL amyloidosis.
We sought to determine if small but significant sequence differences might manifest as differences in the in vitro fibril formation behavior of these proteins. Towards that end, we selected two patient-derived protein samples, AL-09 and AL-103. Both proteins are derived from the κI O18:O8 germline sequence and share greater than 90% sequence identity[20], but the clinical phenotypes of these proteins differed significantly. Patient AL-09 rapidly succumbed to cardiac amyloidosis, surviving just months after diagnosis, while patient AL-103 survived for almost 3 years with prominent cardiac and skeletal muscle (tongue) involvement. Both patients had high levels of free light chains in the serum (AL-103 = 99.1mg/dL, AL-09 = 201mg/dL), were close in age, and received a standard chemotherapy regimen. We have cloned and recombinantly produced the variable domain (VL) of the dominant light chain from these patients and have analyzed the fibril formation behavior across a wide range of solution conditions using 96-well plate assays. We find that there is a correspondence between the severity of the disease and the kinetics of fibril formation for these proteins.
Methods
Protein expression and purification
Recombinant AL-09 and AL-103 VL domains were expressed in E. coli and purified as described previously[20,21]. Briefly, protein was expressed in E. coli, refolded from inclusion bodies, and purified by size exclusion chromatography with a HiLoad 16/60 Superset 75 column on an AKTA FPLC (GE Healthcare, Piscataway, NJ, USA) system. Purity was verified by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis.
Fibril Formation Kinetics
Fibril formation kinetics were followed by ThT fluorescence on a plate reader (Analyst AD, Molecular Devices, Sunnyvale, CA, USA) with an excitation wavelength of 440 nm and an emission wavelength of 480 nm. All experiments were performed in triplicate using black 96-well polystyrene plates. Plates were incubated at 37°C and shaken continuously. All reagents were obtained from Sigma-Aldrich (St Louis, MO) and were filtered prior to use. In nucleation studies, preformed aggregates were removed from the protein stock by ultracentrifugation to the sedimentation point of a 0.5S particle[22]. In elongation studies, seeds were prepared in 0.5M Na2SO4 at 50°C (AL-09) or at pH 2, 37°C (AL-103) before washing with 10mM pH 7.4 Tris-HCl and sonicating for 5 minutes. 1.0μL aliquots of fibril seeds were added to initiate the elongation reactions.
For protein concentration dependence, each well contained 10mM pH 7.4 Tris-HCl, 10μM ThT, 150mM NaCl, and the indicated concentration of protein. For pH dependence, each well contained 10μM ThT, 150mM NaCl, and 20μM protein in 10mM acetate, borate, and citrate (ABC) buffer at the indicated pH. For salt concentration dependence, each well contained 10mM pH 7.4 Tris-HCl, 10μM ThT, 20μM protein, and the indicated concentration of NaCl. t50 values represent the midpoint of the exponential phase of the reaction, the time where fibril formation is 50% complete.
Circular Dichroism Spectroscopy
Circular Dichroism (CD) spectroscopy was performed using a Jasco J-810 spectropolarimeter (Jasco, Inc., Easton, MD). Far UV-CD spectra were collected from 260–200nm at a rate of 10nm/min. Proteins were prepared at a concentration of 20μM in ABC buffer at the indicated pH. Blanks were subtracted and the raw data was converted to mean residue ellipticity (MRE) by the formula:
Transmission Electron Microscopy
Transmission electron microscopy was used to confirm the presence of fibrils. A 3-μl fibril sample was placed on a 300 mesh copper formvar/carbon grid and air-dried. The sample was negatively stained with 4% uranyl acetate, washed with distilled and deionized water, air-dried and inspected on a Philips Tecnai T12 transmission electron microscope at 80kV (FEI, Hillsboro, Oregon, USA).
Results
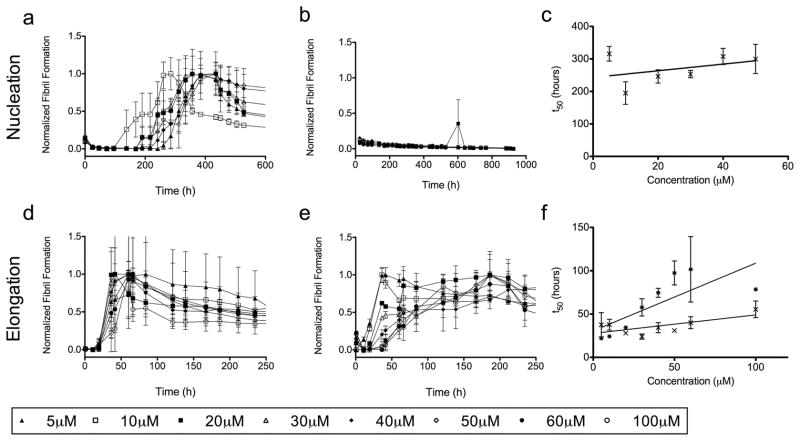
Figure 1 shows the protein concentration dependence of fibril formation. In figure 1a, AL-09 forms amyloid fibrils at all concentrations studied. A nucleated polymerization reaction should show a direct dependence on protein concentration, but in this instance there is no significant difference in the rate of fibril formation at low versus high concentrations (from 5 to 50μM). This likely indicates some competing off-pathway aggregation acting to moderate changes in the protein concentration[23]. In this scenario, the reversible off-pathway aggregates act as a reservoir of protein feeding the creation of kinetically disfavored but more energetically stable fibrils. In figure 1b, AL-103 does not form amyloid fibrils over the time course of this experiment. No ThT enhancement was observed after over 2000 hours; at that point, evaporation of solvent from the wells became problematic and the experiment was halted. Comparison of the midpoint times (t50) of the AL-09 amyloid formation reactions (figure 1c) shows that there is a weak correlation between precursor protein concentration and fibril formation within the protein concentration range tested, but the large error prevents us from drawing any further conclusions. The elongation studies (Figures 1d and 1e) show efficient seeding of fibril formation but are otherwise unremarkable. Likewise, figure 1f shows no real trends in the midpoint times for these reactions. There is a general pattern, seen throughout these experiments, of peak and decay of ThT intensity over the course of fibril formation. This loss of signal after fibril formation occurs is likely due to inner filter effects of aggregates and lateral association of fibrils as well as photobleaching of ThT.
Figure 1. Protein Concentration Dependence of Fibril Formation.
a) AL-09 amyloid fibril nucleation at 5–50μM protein. b) AL-103 amyloid fibril nucleation at 5–60μM protein. c) Comparison of the t50 values for the AL09 (✕) nucleation reactions studied in (a). d) AL-09 fibril elongation at 5–100μM protein. e) AL-103 fibril elongation at 5–100μM protein. f) Comparison of the t50 values for the AL09 (✕) and AL103 ( ) elongation reactions studied in (d) and (e), respectively.
) elongation reactions studied in (d) and (e), respectively.
Numerous studies have demonstrated the key role of pH in amyloid fibril formation with a variety of different precursor proteins including Ig light chains[24–28]. Figure 2a shows the nucleation reaction for AL-09, where amyloid fibril formation occurs rapidly at all pH values from pH 2 to 9. Due to the large variability from well to well, no statistical significance is observed for any of the reactions. This is in contrast to AL-103 (Figure 2b), where the amyloid fibril formation rate shows strong pH dependence from pH 2 to pH 6. The large variability between wells limits the ability to make quantitative comparisons, but the lag time to fibril formation at pH 6 is significantly longer than the lag time observed at lower pH values (p<0.002). It is also worth noting that even under conditions where it forms fibrils, AL-103 amyloid fibril formation occurs much more slowly than AL-09 amyloid fibril formation. Both of these observations are displayed in figure 2c, comparing the t50 for both proteins. AL-103 shows a much stronger response to changes in pH while simultaneously forming fibrils more slowly at all values studied. In figures 2d–f we see again that the elongation reactions show clear reduction in lag time without significant difference between the different conditions. Interestingly, across all four nucleation and elongation reactions for AL-09 and AL-103, high pH is inhibitory to fibril formation. AL-103 fibrils do not form at pH 7 and higher, while AL-09 fibril nucleation is inhibited at pH 10. Even more impressively, pH 10 also prevents the elongation of fibrils in a seeded reaction (Figure 2c and 2d), the only condition where we have failed to observe fibril elongation in either protein. No modifications of the protein were detected by mass spectrometry and EM was negative for fibrils (data not shown) in the pH 10 reactions. At this point it is unclear whether this effect is related to a specific event in the fibril formation pathway or a more generic perturbation of protein-protein interaction at high pH.
Figure 2. pH Dependence of Fibril Formation.
a) AL-09 amyloid fibril nucleation at pH 2–10. b) AL-103 amyloid fibril nucleation at pH 2–6. c) Comparison of the t50 values for the AL09 (✕) and AL103 ( ) nucleation reactions studied in (a) and (b), respectively. d) AL-09 fibril elongation at pH 2–10. e) AL-103 fibril elongation at pH 2–10. f) Comparison of the t50 values for the AL09 (✕) and AL103 (
) nucleation reactions studied in (a) and (b), respectively. d) AL-09 fibril elongation at pH 2–10. e) AL-103 fibril elongation at pH 2–10. f) Comparison of the t50 values for the AL09 (✕) and AL103 ( ) elongation reactions studied in (d) and (e), respectively.
) elongation reactions studied in (d) and (e), respectively.
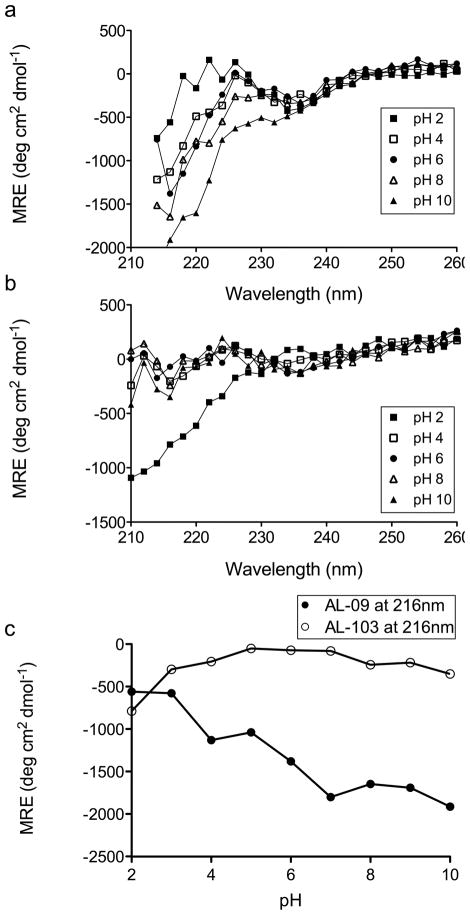
To address the possibility that denaturation of the protein is the cause of this inhibition, we used CD spectroscopy to monitor the secondary structure of these proteins over a range of pH values (Figure 3). We find that AL-09 shows a decrease in mean residue ellipticity (MRE) at 216nm with increasing pH, but this change does not correlate with a shift to a random coil structure (Figure 3a, 3c). Furthermore, the pH dependent change in MRE does not show the increase in MRE at 235nm that is associated with unfolding of the protein (Figure 3a) [21]. AL-103 shows stable β-sheet structure from pH 3 to pH 10, with clear random coil structure at pH 2 (Figure 3b, 3c). Clearly there are changes in the structure of these proteins across this range of pH, but they do not correspond to the differences in fibril formation kinetics observed in figure 2.
Figure 3. Circular Dichroism Spectroscopy of AL-09 and AL-103.
The pH dependence of AL-09 (a) and AL-103 (b) secondary structure as shown by CD spectroscopy. Every other data point is plotted for clarity. In panel (c), MRE at 216nm (β-sheet minimum) is plotted against pH.
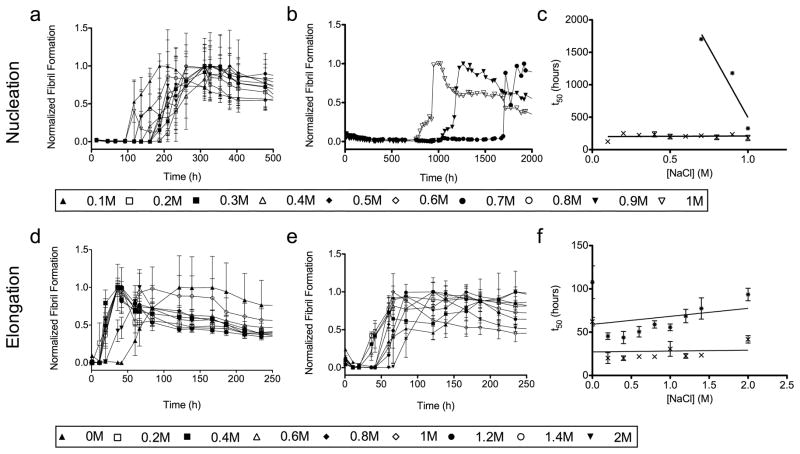
While modulating the pH will affect the charge of certain amino acid side chains, the presence of salt can have non-specific effects on all electrostatic interactions. Figure 4 shows the dependence of the fibril formation rate on NaCl concentration. Figure 4a shows that AL-09 again forms fibrils rapidly without any significant difference over a 10-fold range in salt concentration (0.1–1.0M NaCl). In figure 4b, AL-103 shows an inverse relationship between salt concentration and fibril formation, forming fibrils most rapidly at 1M NaCl, followed by 0.9 and 0.7M NaCl, though all three reaction are still slower than AL-09 fibril formation. This suggests that electrostatic interactions are critical for preventing AL-103 fibril formation. There are no error bars in figure 4b because in each case only one of the three reaction wells gave a positive result, complicating the interpretation of these results. Figure 4c shows how the rate of fibril formation for AL-103 is greatly accelerated with increasing salt concentration. In figures 4d–f, elongation studies show efficient seeding for both proteins but no significant difference in an even wider range of solution conditions than those used in the nucleation reactions (0.0M to 2.0M NaCl).
Figure 4. NaCl Concentration Dependence of Fibril Formation.
a) AL-09 amyloid fibril nucleation at 0.1–1.0M NaCl. b) AL-103 amyloid fibril nucleation at 0.7, 0.9, and 1.0M NaCl. c) Comparison of the t50 values for the AL09 (✕) and AL103 ( ) amyloid fibril nucleation reactions studied in (a) and (b), respectively. d) AL-09 amyloid fibril elongation at 0.0–2.0M NaCl. e) AL-103 fibril elongation at 0.0–2.0M NaCl. f) Comparison of the t50 values for the AL09 (✕) and AL103 (
) amyloid fibril nucleation reactions studied in (a) and (b), respectively. d) AL-09 amyloid fibril elongation at 0.0–2.0M NaCl. e) AL-103 fibril elongation at 0.0–2.0M NaCl. f) Comparison of the t50 values for the AL09 (✕) and AL103 ( ) amyloid fibril elongation reactions studied in (d) and (e), respectively.
) amyloid fibril elongation reactions studied in (d) and (e), respectively.
We utilized transmission electron microscopy to confirm the presence of amyloid fibrils. Figure 5 shows representative EM images from each of the proteins. AL-09 forms a dense network of short fibrils that is typical of in vitro AL amyloid fibril samples (Figure 5a) [21,29]. AL-103 incubated at low pH gives fibrils that are slightly thicker and longer than the AL-09 amyloid fibrils, but are still consistent with the known width and morphology of amyloid fibrils (Figure 5b). These images confirm that the increases in ThT fluorescence correspond to amyloid fibril formation.
Figure 5. Transmission Electron Microscopy.
a) AL-09 amyloid fibrils grown from 20μM protein in 0.4M NaCl, scale bar =100nm (reaction shown in figure 4a). b) AL-103 amyloid fibrils grown at pH 2, scale bar=100nm (reaction shown in figure 2b). c) Sequence alignment of AL-09 and AL-103 with the κI O18:08 germline sequence. Mutations are shown in bold.
Discussion
While no broad conclusions can be drawn from two patients given the enormous number of potential individual differences and the uncertainties surrounding the diagnosis of this rare disease, it is clear from these data that the VL protein from patient AL-09 forms fibrils much more rapidly than the VL protein from patient AL-103. While it has been proposed that amyloid fibrils are not the toxic species in protein misfolding diseases[30,31], it is reasonable to presume that mutations that favor fibril formation will also favor protein misfolding and the formation of toxic intermediate species from soluble protein. The divergent behavior of AL-09 and AL-103 is remarkable given that these proteins share >90% sequence identity (Figure 5c) and have a similar thermodynamic stability (Tm = 41.1 ±1.0°C and ΔGfolding = −3.53± 0.28 kcal/mol for AL-09; Tm = 41.6±0.5°C and ΔGfolding = −4.43±0.25 kcal/mol for AL-103)[20]. Figure 5c shows a sequence alignment of the two AL proteins with their germline sequence. AL-09 has seven mutations from the germline (S30N, N34I, K42Q, N53T, D70E, I83L, and Y87H), while AL-103 has four (N34I, D92H, iP95, and Q100P). Previous work has shown that the tyrosine in position 87 and the asparagine at position 34 are essential for maintaining a stable, soluble protein with a canonical dimer interface[32,33]. Based on this, we hypothesize that mutations stabilizing the canonical interface oppose fibril formation while those that promote this alternative dimer conformation are amyloidogenic. However, the relationship between these mutations and the arrangement of the light chain dimer is complex and must be explored further before we can say with confidence how each change relates to the fibril formation pathway.
In studying the data, several general trends emerge. While AL-09 forms fibrils quite rapidly, it shows a remarkable insensitivity to the solution conditions. From pH 2–9, a 107-fold difference in the concentration of hydrogen ions, there is no significant difference in the lag time of the reaction (figure 2a and 2c). Part of this is no doubt due to the large well-to-well variability in the data, but even if it were possible to tighten up the data considerably, there would be only minor differences between the extremes of pH. Likewise, over 10-fold ranges of salt and protein concentration there is no significant difference in fibril nucleation (figure 1c, 4c). The question then becomes whether this insensitivity is unique to this protein or a general feature of rapid fibril formation. Is AL-09 poised to form fibrils independent of its surroundings? As has been discussed, previous research has shown that AL-09 has a highly plastic dimer interface that crystallizes in a different conformation from AL-103 and other light chains[32,34]. Our hypothesis that the altered dimer interface is essential for fibril formation implies that AL-09 is much more likely to form fibrils because the somatic mutations found in this patient destabilize the canonical interface and promote the formation of the alternative conformation. AL-103 on the other hand would be better able to form fibrils under conditions that encourage the population of a conformation similar to that of AL-09. If AL-09 is poised near a conformation critical for AL amyloid fibril formation, inhibition of AL-09 fibril formation could have broad relevance to AL amyloidosis patients in general.
It is interesting to note that at approximately physiological conditions (pH 7.4, 150mM NaCl, low micromolar protein concentration), AL-103 does not form fibrils over more than 2000 hours without seeding, but does so within a week once seeds are added (Figures 1b and 1d). This implies that AL-103 fibril nucleation is an exceedingly rare event. It is impossible to know how long this patient had the disease prior to diagnosis, but it is possible that this patient had an asymptomatic over-secretion of Ig light chains for several years prior to diagnosis due to the low probability of AL-103 amyloid nucleation. The other possibility of course is that there is some cofactor present in vivo or a microenvironment that acts to accelerate fibril formation. Lowering the pH and/or increasing the concentration of salt in solution accelerate AL-103 amyloid fibril formation in vitro. Perhaps AL-103 fibrils were nucleated in the presence of locally high concentrations of hydrogen ions or electrolytes. Further study is necessary to achieve a more fine-grained understanding of these issues.
Elongation studies did not provide much separation between the two proteins since both showed rapid fibril formation across nearly all conditions. This reinforces the idea stated here and elsewhere that the nucleation step is where differences in structure and dynamics may play a critical role. The reordering of soluble protein onto a fibril template is a comparatively minor hurdle[35].
Perhaps the most intriguing data in this study is the broad inhibition of fibril formation at high pH (Figure 2). At low pH, fibril formation is rapid for both proteins, more notably so for AL-103. Other AL proteins have been shown to have partially unfolded states populated at low pH[36], but at this time we cannot say if such a condition exists for AL-09 or AL-103 based on far UV CD data collected at different pH values for both proteins (Figure 3). At pH 10, neither nucleation nor elongation reactions proceed to form amyloid fibrils, the only such condition we have observed. Furthermore, fibrils grown at low pH do not dissolve when shifted to pH 10 on a timescale of days (data not shown), indicating that stability of the fibrillar species is not the reason for the lack of elongation. Considering the possibilities of what may be occurring at pH 10, it has been noted that tyrosine residues play an important role in immunoglobulin function[37]. Given the importance of these tyrosine residues, it is plausible that its deprotonation at pH 10 disrupts key intermolecular interactions on the pathway to fibril formation, either by the loss of hydrogen bonding, π interactions, or by the introduction of a negative charge in an inappropriate position. The interactions of lysine residues may also be in flux at pH 10. We are undertaking further studies to try and understand whether this inhibition is a protein-specific effect or whether it might be a more general property of amyloid fibril formation.
In conclusion, we have shown that a protein that readily and rapidly forms amyloid fibrils corresponds to an aggressive disease phenotype, and vice versa. Further studies are needed to determine whether this is a general property of these disease proteins, but given the prominent role of the protein in this disorder we find it unlikely that the fibril formation kinetics will not have a bearing on the disease progression. Greater understanding of this relationship could help in the development of protein-targeted therapeutics that might better halt the progression of this disease.
Acknowledgments
We would like to acknowledge Dr. Roshini Abraham for the contribution of patient cDNA and members of the Ramirez-Alvarado laboratory for helpful comments on the manuscript. Funding provided by NIH GM071514 (MRA), F30DK082169 (DJM), and the generous support of patients with light chain amyloidosis and the Mayo foundation.
Abbreviations
- AL amyloidosis
Light chain amyloidosis
- Ig
immunoglobulin
- ThT
Thioflavin T
- VL
Variable domain
- t50
time at which fibril formation reaction is 50% complete
- CD
Circular Dichroism
- MRE
mean residue ellipticity
Footnotes
Declaration of Interest: The authors declare no competing financial interests.
References
- 1.Buxbaum JN. Diseases of protein conformation: what do in vitro experiments tell us about in vivo diseases? Trends Biochem Sci. 2003;28:585–592. doi: 10.1016/j.tibs.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 (Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Pepys MB. Amyloidosis. Annu Rev Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 6.Gertz MA, Kyle RA. Primary systemic amyloidosis--a diagnostic primer. Mayo Clin Proc. 1989;64:1505–1519. doi: 10.1016/s0025-6196(12)65706-1. [DOI] [PubMed] [Google Scholar]
- 7.Kyle RA, Greipp PR, O’Fallon WM. Primary systemic amyloidosis: multivariate analysis for prognostic factors in 168 cases. Blood. 1986;68:220–224. [PubMed] [Google Scholar]
- 8.Wechalekar AD, Hawkins PN, Gillmore JD. Perspectives in treatment of AL amyloidosis. Br J Haematol. 2008;140:365–377. doi: 10.1111/j.1365-2141.2007.06936.x. [DOI] [PubMed] [Google Scholar]
- 9.Gertz MA, Lacy MQ, Dispenzieri A. Amyloidosis. Hematol Oncol Clin North Am. 1999;13:1211–1233. ix. doi: 10.1016/s0889-8588(05)70122-2. [DOI] [PubMed] [Google Scholar]
- 10.Kyle RA, Linos A, Beard CM, Linke RP, Gertz MA, O’Fallon WM, Kurland LT. Incidence and natural history of primary systemic amyloidosis in Olmsted County, Minnesota, 1950 through 1989. Blood. 1992;79:1817–1822. [PubMed] [Google Scholar]
- 11.Kyle RA, Greipp PR. Amyloidosis (AL). Clinical and laboratory features in 229 cases. Mayo Clin Proc. 1983;58:665–683. [PubMed] [Google Scholar]
- 12.Gertz MA, Kyle RA. Prognostic value of urinary protein in primary systemic amyloidosis (AL) Am J Clin Pathol. 1990;94:313–317. [PubMed] [Google Scholar]
- 13.Picken MM. Immunoglobulin light and heavy chain amyloidosis AL/AH: renal pathology and differential diagnosis. Contrib Nephrol. 2007;153:135–155. doi: 10.1159/000096765. [DOI] [PubMed] [Google Scholar]
- 14.Kyle RA, Gertz MA. Primary systemic amyloidosis: clinical and laboratory features in 474 cases. Semin Hematol. 1995;32:45–59. [PubMed] [Google Scholar]
- 15.Gertz MA, Kyle RA. Hepatic amyloidosis: clinical appraisal in 77 patients. Hepatology. 1997;25:118–121. doi: 10.1002/hep.510250122. [DOI] [PubMed] [Google Scholar]
- 16.Kyle RA, Gertz MA, Greipp PR, Witzig TE, Lust JA, Lacy MQ, Therneau TM. Long-term survival (10 years or more) in 30 patients with primary amyloidosis. Blood. 1999;93:1062–1066. [PubMed] [Google Scholar]
- 17.Abraham RS, Geyer SM, Price-Troska TL, Allmer C, Kyle RA, Gertz MA, Fonseca R. Immunoglobulin light chain variable (V) region genes influence clinical presentation and outcome in light chain-associated amyloidosis (AL) Blood. 2003;101:3801–3808. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- 18.Comenzo RL, Zhang Y, Martinez C, Osman K, Herrera GA. The tropism of organ involvement in primary systemic amyloidosis: contributions of Ig V(L) germ line gene use and clonal plasma cell burden. Blood. 2001;98:714–720. doi: 10.1182/blood.v98.3.714. [DOI] [PubMed] [Google Scholar]
- 19.Prokaeva T, Spencer B, Kaut M, Ozonoff A, Doros G, Connors LH, Skinner M, Seldin DC. Soft tissue, joint, and bone manifestations of AL amyloidosis: clinical presentation, molecular features, and survival. Arthritis Rheum. 2007;56:3858–3868. doi: 10.1002/art.22959. [DOI] [PubMed] [Google Scholar]
- 20.Randles EG, Thompson JR, Martin DJ, Ramirez-Alvarado M. Structural alterations within native amyloidogenic immunoglobulin light chains. J Mol Biol. 2009;389:199–210. doi: 10.1016/j.jmb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin RW, De Stigter JK, Sikkink LA, Baden EM, Ramirez-Alvarado M. The effects of sodium sulfate, glycosaminoglycans, and Congo red on the structure, stability, and amyloid formation of an immunoglobulin light-chain protein. Protein Sci. 2006;15:1710–1722. doi: 10.1110/ps.051997606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zagorski MG, Yang J, Shao H, Ma K, Zeng H, Hong A. Methodological and chemical factors affecting amyloid beta peptide amyloidogenicity. Methods Enzymol. 1999;309:189–204. doi: 10.1016/s0076-6879(99)09015-1. [DOI] [PubMed] [Google Scholar]
- 23.Powers E, Powers D. Mechanisms of Protein Fibril Formation: Nucleated Polymerization with Competing Off-Pathway Aggregation. Biophysical Journal. 2007;94:379–391. doi: 10.1529/biophysj.107.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ionescu-Zanetti C, Khurana R, Gillespie JR, Petrick JS, Trabachino LC, Minert LJ, Carter SA, Fink AL. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc Natl Acad Sci U S A. 1999;96:13175–13179. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiti F, Bucciantini M, Capanni C, Taddei N, Dobson CM, Stefani M. Solution conditions can promote formation of either amyloid protofilaments or mature fibrils from the HypF N-terminal domain. Protein Sci. 2001;10:2541–2547. doi: 10.1110/ps.10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frare E, Mossuto MF, de Laureto PP, Tolin S, Menzer L, Dumoulin M, Dobson CM, Fontana A. Characterization of oligomeric species on the aggregation pathway of human lysozyme. J Mol Biol. 2009;387:17–27. doi: 10.1016/j.jmb.2009.01.049. [DOI] [PubMed] [Google Scholar]
- 27.Sasahara K, Yagi H, Sakai M, Naiki H, Goto Y. Amyloid nucleation triggered by agitation of beta2-microglobulin under acidic and neutral pH conditions. Biochemistry. 2008;47:2650–2660. doi: 10.1021/bi701968g. [DOI] [PubMed] [Google Scholar]
- 28.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci U S A. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Z, Hu D, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46:3521–3531. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 30.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 31.Hartley DM, Walsh DM, Ye CP, Diehl T, Vasquez S, Vassilev PM, Teplow DB, Selkoe DJ. Protofibrillar intermediates of amyloid beta-protein induce acute electrophysiological changes and progressive neurotoxicity in cortical neurons. J Neurosci. 1999;19:8876–8884. doi: 10.1523/JNEUROSCI.19-20-08876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baden EM, Randles EG, Aboagye AK, Thompson JR, Ramirez-Alvarado M. Structural insights into the role of mutations in amyloidogenesis. J Biol Chem. 2008;283:30950–30956. doi: 10.1074/jbc.M804822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson FC, Baden EM, Owen BA, Volkman BF, Ramirez-Alvarado M. A single mutation promotes amyloidogenicity through a highly promiscuous dimer interface. Structure. 2010;18:563–570. doi: 10.1016/j.str.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. Altered dimer interface decreases stability in an amyloidogenic protein. J Biol Chem. 2008;283:15853–15860. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc Chem Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 36.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 37.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]