Abstract
Galactose-α1,3-galactose(αGal) epitopes, the synthesis of which requires the enzyme product of α1,3-galactosyltransferase (α1,3GT), are sugar chains on the cell surface of most mammalian species. Notable exceptions are higher primates including Old World monkeys, apes, and humans. The αGal-negative species as well as mice with deletion of the α1,3GT gene produce abundant anti-αGal antibodies. The evolutionary loss of αGal epitopes has been attributed to point mutations in the coding region of the gene. Because no transcripts could be found in the higher primate species with Northern blot analysis, a potential alternative explanation has been loss of upstream regulation of the gene. Here, we have demonstrated that the rhesus promoter is functional. More importantly, a variety of full-length transcripts were detected with sensitive PCR-based methods in the tissues of rhesus monkeys, orangutans, and humans. Five crucial mutations were delineated in the coding region of the human and rhesus and three in the orangutan, any one of which could be responsible for inactivation of the α1,3GT gene. Two of the mutations were shared by all three higher primates. These findings, which elucidate the molecular basis for the evolutionary loss of αGal expression, may have implications in medical research.
Most mammals express the cell surface carbohydrate epitope galactose-α1,3-galactose (αGal),1 with notable exceptions that include Old World monkeys, apes, and humans (1). With the loss of the αGal epitopes, the synthesis of which is dependent on enzyme product of the α1,3-galactosyltransferase(α1,3GT) gene, higher primates produce anti-αGal antibodies (2) that are responsible for the hyperacute rejection of organs transplanted from αGal-positive donors (3).
In 1989, Joziasse et al. (4) reported the sequence of full-length cDNA clone of the bovine α1,3GT gene and demonstrated the presence of this gene in the DNA of human cell lines. Using an 804-bp fragment derived from bovine full-length cDNA as a probe, they also detected mRNA transcripts in bovine and marmoset (New World monkey) but not in human or African green monkey cell lines (4). The absence of α1,3GT mRNA has since been widely viewed as a feature of all Old World monkeys, apes, and humans (5, 6). The molecular basis for the inactivation of the α1,3GT gene in the αGal-negative species has been attributed to a mutation(s) localized to a partial sequence of exon 9 (4, 5, 7).
Stimulated by the current interest in producing transgenic pigs for clinical use as tissue and organ xenograft donors, the cDNA for the α1,3GT gene of the αGal-positive pig was isolated (8), and the coding regions were characterized (9–12). Further, the full genomic organization of the porcine α1,3GT, gene including its upstream regulatory region, was completed (12). A CpG island (i.e. a C connected by a 3′–5′ phosphodiester bond to a G) characteristic of housekeeping genes was observed around exon 1 of the pig α1,3GT gene (12). It is of interest that a CpG island was not present in the mouse gene (13–15), suggesting that the upstream regulatory regions might be species-specific even though the enzyme function of the α1,3GT gene product was identical (15). If so, it would be conceivable that the loss of αGal expression in higher primates could be due to mutation in the upstream regulatory region (16).
As a further step in a phylogenetic comparison of the αGal-positive pig with the higher primates, we employed a PCR-based cloning strategy to avoid the potential artifacts of extrapolating from partial sequences and cross-species hybridization. After obtaining the full coding region of the αGal-positive marmoset and cebus α1,3GT gene, we identified the full coding region of the gene in the αGal-negative rhesus monkey, orangutan, and human, as well as the genomic organization and upstream regulatory region in the rhesus and human. In all three αGal-negative primate species, we found that the α1,3GT gene produces a variety of transcripts that were confirmed in the rhesus samples with Northern blot analysis. The transcripts contained crucial mutations in addition to the one(s) described previously in exon 9 of the rhesus (61, orangutan (6), and human (5, 6). Taken together, our findings suggest substantial revisions of the characterization of the α1,3GT gene that have implications for the molecular study of primate evolution and for the planning of xenotransplantation initiatives.
EXPERIMENTAL PROCEDURES
Genomic Organization of Human, Orangutan, and Rhesus Monkey α1,3GT Genes
After high molecular genomic DNA was isolated from the PBMC of multiple healthy adult male humans, two rhesus monkeys, and an orangutan, libraries for the three species were constructed using the Universal GenomeWalker™ Library Kit (CLONTECH, Palo Alto, CA). The initial gene-specific and nested primer pairs were designed from the partial exon 9 sequences of the α1,3GT gene available in the GenBank™ (Table I). Gene-specific and nested primer pairs for other regions were designed from the exonic sequences obtained from RACE or RT-PCR experiments (not shown).
Table I. Sequences of oligonucleotides used as PCR primers.
| Human |
| H9a: 5′-CTGAATGGGTGTTCCTCCAGAAATGGCT-3′ |
| H9b: 5′-TGGAAGACCTGGTCCACATCCATGCAG-3 |
| H9m: 5′-GAAGAGGTGGCAAGACATCAGCATGATGC-3′ |
| H9n: 5′-CTGCATGGATGTGGACCAGGTCTTCCA-3′ |
| Orangutan |
| U9a: 5′-TAGGCCAGTCGGTGGCTCAGCTACAGG-3′ |
| U9b: 5′-TGGCGGTACAAGGC AGATCCCGATGAC-3′ |
| U9m: 5′-GTGATGTTGAGAACCTGAATGGGTGTTG-3′ |
| U9n: 5′-CTGGCCAAATGGAATATATGCTGCTGACTC-3′ |
| Rhesus |
| R9a: 5′-GAGGTCAAGCCAGAGAAGAGGTGGCAACA-3′ |
| R9b: 5′-GACTTCCTCTTCTGCATGGATGTAGACCAG-3′ |
| R9m: 5′-ATGTTGAGAACCTGAATGGGTGTTCCTCC-3′ |
| R9n: 5′-CTGGCCAAATGGAATGCATGCTGCTGACTC-3′ |
5′- or 3′-RACE Analyses
Using Trizol™ reagent (Invitrogen) according to the manufacturer’s instructions, total RNA was extracted from rhesus PBMC and spleen samples, from human PBMC and placenta (provided in the CLONTECH Marathon™ amplification kit), and from orangutan PBMC. After treatment with DNase I (Ambion Inc., Austin, TX), poly(A)+ RNA was separated from total RNA using the Dynabeads™ mRNA purification Kit (Dynal, Oslo, Norway) in accordance with the protocol provided. Using the primers listed in Table I, the 5′- and 3′-RACE procedures were performed with the Marathon™ cDNA amplification kit. The thermal cycling conditions on a PerkinElmer GeneAmp System 9600 or 9700 thermocycler and TaKaRa LA Taq (Takara Shuzo, Shiga, Japan) enzyme were used for PCR procedures.
RT-PCR
Using total RNA derived from two rhesus monkeys, first-strand cDNA templates for RT-PCR were generated with the Super-Script Preamplification™ system (Invitrogen) according to the manufacturer’s protocol. Based on the sequences obtained from the RACE experiments, primers were designed and RT-PCR was performed. Comparable experiments were performed with the orangutan and human-derived specimens. The primers are listed in Table I.
Subcloning and Sequencing of Amplified Products
PCR products amplified in 5′- or 3′-RACE, GenomeWalker™-PCR, and RT-PCR were subcloned into the pCR II vector provided with the Original TA Cloning™ Kit (Invitrogen, Carlsbad, CA). Automated fluorescent sequencing of cloned inserts was performed using an ABI 377 Automated DNA Sequence Analyzer (Applied Biosystems, Inc., Foster City, CA).
Northern Blot Analysis
To confirm the results of RT-PCR in the rhesus species with NorthernMaxTM-GlyNorthern™ blots system (Ambion), total RNA was extracted from rhesus spleen tissue using Trizol™ as described above. Using the protocol provided with the kit, ~10 μg of total RNA from each species was blotted to the BrightStar-Plus™ membrane. The BrightStar™ Psoralen-biotin nonisotopic labeling kit was employed for the biotinylated DNA probe (a 744-bp fragment of the rhesus α1,3GT exon 9).
Luciferase Assay for Promoter Activity
The Dual Luciferase™ reporter assay (Promega, Madison, WI) was used to evaluate the promoter activity. For this purpose, a fragment of 1431 bp of the 5′-flanking region of rhesus exon 1 was inserted into the multicloning site of the pGL3-Basic vector (termed pGL3-RA), which was co-transfected with pRL-TK vector for normalization of transfection efficiency into pig aortic endothelial cells using LipofectAMINE (Invitrogen). Relative luciferase activity of the pGL3-RA was measured by luminometer and compared with that of the pig promoter (12).
RESULTS
Genomic Organization of the Rhesus and Human α1,3GT Gene
The full-length genomic organization determined by the combined results of GenomeWalker-PCR, RACE, and RT-PCR consisted of nine exons and eight introns (Fig. 1), corresponding to the previously delineated porcine and mouse genomic structure (8–14). However, some cryptic exons or aberrant splicing sites were observed as described below. The total rhesus and human gene size is calculated to be more than 50 kb.
Fig. 1. Genomic organization on a kb scale of the α1,3GT gene of the human (above horizontal line) and rhesus (below horizontal line).
The numbered boxes indicate the approximate location of exons within the locus of the two species but do not accurately reflect the length of each exon. The translation start and stop codons are located in exons 4 and 9. The horizontal thick bar above exon 9 denotes the GenBank™ data for human (J05421, 794 bp) contributed by Larsen et al. (5), and the short thick bar below exon 9 denotes the data for rhesus (M73306, 371 bp) contributed by Galili and Swanson (6). Shown below the gene locus representation is a contig of overlapping subgenomic clones derived from GenomeWalker™ libraries (horizontal arrows) or from long PCR experiments (horizontal lines).
5′-RACE, 3′-RACE, and Northern Blot Analysis of the α1,3GT Gene
5′ - and 3′-RACE experiments revealed sequences of the αGal-negative species that were highly homologous with the α1,3GT gene of αGal-positive species. 3′-RACE demonstrated the presence of poly(A) (adenylation) signal and poly(A) tail. Based on these sequences, we performed RT-PCR using a pair of primers that spanned several exons. Multiple bands were seen in human (Fig. 2A, a) and rhesus (Fig. 2A, c), whereas only one major band was seen in the orangutan (Fig. 2A, b). The results of any exon of a given species were identical with all three PCR methods (i.e. RACE, RT-PCR, and GW-PCR) (Fig. 2A). Moreover, the presence of transcripts for the rhesus α1,3GT gene was confirmed with Northern blot hybridization (Fig. 2A, d). We concluded that the sequences identified with all of the detection methods were derived from authentic α1,3GT mRNA in the respective αGal-negative species.
Fig. 2.

A, electrophoresis of PCR (a–c) and Northern blot hybridization (d). Lane 1, electrophoresis results of RT-PCR in the human, orangutan, and rhesus cDNA libraries, respectively, using a pair of primers spanning the coding region. Lane M, DNA size marker ladder (Invitrogen, 1-kb DNA size ladder). d, Northern blot of total RNA from bovine (lane 1) or rhesus spleen tissue (lane 2) hybridized with a probe derived from rhesus exon 9 fragment. As expected, with in-species versus cross-species hybridization, gene expression was lower in the bovine than in the monkey spleen cells. B, schematic splicing variations and cryptic exons of human (a), orangutan (b), and rhesus (c) α1,3GT gene. The closed boxes denote exons in the respective species. The open boxes with arrows denote cryptic exons that were not observed in the αGal-positive species. Exon 6Rc in rhesus has exon 6, intron 6, and exon 7 (a retained intron).
Cryptic Exons
Cryptic exons were found in both the rhesus and human α1,3GT gene, e.g. between exons 2 and 4, designated 3H in human and 3R in rhesus (Fig. 2B, a and c). Exons 3H and 3R showed no homology with each other, and neither exon had any homology with the previously described mouse exon 3 (13). A cryptic human exon with two size variants, both of which had poly(A) tails, was found in 18 of 22 humans between exons 7 and 8 (Fig. 2B, a) and was designated 7H. At the end of the 7Ha variant, a donor splicing site was found that spliced to exon 9 (Fig. 2B, a). In rhesus, cryptic exons designated 6Ra and 6Rb were found between exons 6 and 7, and in addition, a third variant was found (6Rc) that included a retained intron (Fig. 2B, c).
Aberrant Splicing Sites
In the 5′-untranslated region, rhesus exon 2 is 112 bp, whereas human exon 2 is 269 bp (Fig. 3). Comparative analysis revealed that the first half (110 bp, termed 2A) of the human exon 2 is highly homologous to rhesus exon 2, and the last half (159 bp, termed 2B) is similar to the rhesus intron 2 that extends from exon 2. Although the first half (138 bp, termed 8Ra) of the rhesus exon 8 (327 bp) showed high homology with the porcine exon 8 (138 bp), the remaining half (189 bp, termed 8Rb) resembled the porcine intron 8 sequences extending from exon 8 (Figs. 2B, c, and 3). A single nucleotide deletion of G was found only in rhesus at the border between the region 8Ra and 8Rb (Fig. 3, position [d]), resulting in utilization of another downstream splicing site.
Fig. 3. Comparison of exon-intron boundaries of the human (H), orangutan (Ou), and rhesus (R) α1,3GT gene.
Capital or lowercase letters indicate the nucleotides of the exon or intron in each species, respectively. The numbers in parentheses indicate exon size in bp. The start codon in exon 4 and the authentic stop codon in exon 9 are underlined.
Alternative mRNA Splicing of the Human and Rhesus α1,3GT Gene
A variety of alternative splicing patterns were demonstrated in the RACE and RT-PCR experiments (Fig. 2, A and B). In the coding region, the predominant pattern in human consisted of exons 4 through 9. Because of the cryptic exons and aberrant splicing sites in the human and rhesus genes, the alternative splicing patterns were far more varied in these αGal-negative species than in αGal-positive murine or porcine species (8–13).
Exon-Intron Splice Junctions of Human, Orangutan, and Rhesus α1,3GT Gene
A comparative sequence analysis of splice junctions was performed on rhesus, orangutan, and human genomic DNA (Fig. 3). The results were further compared with the porcine species sequence (GenBank™ accession numbers AF221510–AF221517) (not shown), revealing high homology among the three species. As shown in Fig. 3, the pyrimidine richness in the intron 7-exon 8 boundary of both the rhesus and human was less than the pyrimidine content of the intron 3–exon 4 boundary, explaining the frequency of splicing of exon 8 in the 2 species.
Regulatory Region and 5′-Untransplated Region of the Rhesus α1,3GT Gene
GenomeWalker-PCR starting at exon 1 revealed that the rhesus and human upstream regulatory region contains several SP1/GC-box, AP2, and other consensus motifs in the absence of a TATA- or CAAT-box. The findings were similar to those in the pig gene (12). In both the rhesus and human, as well as the pig, the region comprises a CpG island. In the upstream region between −340 and −470, the high degree of homology between the rhesus and pig, but not between the human and pig, gene is noteworthy (Fig. 4). The transcription initiation site has been identified in the pig (12). When the transcription initiation site is identified in the human and rhesus gene, the question may be clarified as to whether there are differences in gene regulation between the human and rhesus.
Fig. 4. Exon 1 vicinity of α1,3GT gene of rhesus (R) compared with pig (P).
The rhesus exon 1 and its potential regulatory region (capital letters) are aligned with the corresponding porcine exon 1 sequence (12). The lowercase letters denote intron 1. An asterisk indicates that the nucleotide is identical between both species.
Rhesus α1,3GT Gene Promoter Activity
The intensity of expression promoted expressed by the pGL3-RA construct was 18-fold greater than the pGL3-Basic control, and that of pig was 81-fold higher (Fig. 5).
Fig. 5.
Luciferase assay for promoter activity of the rhesus α1,3GT gene
3′-Untranslated Region of the Rhesus α1,3GT Gene
The nucleotide divergence of the rhesus exon 9 sequence from that of the orangutan and human was found starting 1273 bp down-stream from the 5′-end of exon 9 and spanning 420 bp (data not shown). A GenBank™ BLAST search revealed that this divergence had a striking homology with a retrovirus long terminal repeat sequence present in the baboon (GenBank™ accession number NC-000863; Ref. 17).
Homology in the Coding Region
We compared our results of the human, orangutan, and rhesus sequences to our αGal-positive marmoset and cebus New World monkey sequences (deposited in the GenBank™). Our marmoset sequence also was compared with an independently submitted sequence (18). Although the overall sequence from the start codon in exon 4 to the stop codon in exon 9 in marmoset is 1131 bp, the corresponding sequence of the human (from exon 4 to exon 9 through exon 8) or rhesus (from exon 4 to exon 9 through exon 8Ra) was 1128 or 1129 bp, respectively. Homology between human, orangutan, or rhesus to marmoset is 94.3, 94.7, or 92.4%, respectively.
Comparison of Predicted Amino Acid Sequences
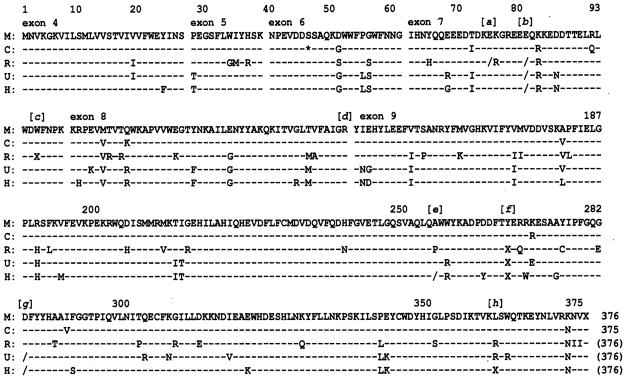
There were no frameshift mutations in the rhesus, orangutan, or human exons 4–6. However, five crucial mutations that might account for loss of the α1,3GT expression were identified in the human and rhesus, and three were detected in the orangutan. It is noteworthy that the mutations at the position [b] and [f] were shared by all three species.
In the rhesus, at position [a] and [b] (Fig. 7), corresponding to the deduced amino acid positions 75 and 81, respectively, in marmoset, a single nucleotide deletion resulted in a frameshift. At position 96 (position [c]), a single nucleotide substitution resulted in a premature stop codon. As described under “Aberrant Splicing Sites,” a single nucleotide deletion was detected at position [d] of the exon 8 splicing junction. At position 268 [f], a single nucleotide substitution also resulted in a premature stop codon.
Fig. 7. Comparison of crucial point mutations of the α1,3GT gene in primates.
The numbered open boxes (top) are exons in which crucial mutations in the primates are shown with the corresponding nucleotide sequence. No mutations were observed in αGal-positive primates. Crucial mutations were observed in exons 7 and 9 of the three higher primate species examined (lower boxes). Note that the mutations at position [b] and [f] (lower closed boxes with thick lines) are shared by rhesus, orangutan, and human. (F/S), frameshift; (Stop), stop codon.
In the orangutan, a single nucleotide deletion was observed at position [b] and [g]. A premature stop codon was observed at position [f].
In human, a single nucleotide deletion was observed at the position [b], [e] and [g]. A premature stop codon was observed at position [f] and [h].
Three critical mutations were observed within exon 7, leading to the predicted nonfunctional α1,3GT expression in all frames (Figs. 6 and 7). At position [a] or [b] in the rhesus, corresponding to the deduced amino acid at positions 75 or 81, respectively, in marmoset, a single nucleotide deletion, G, from the triplet GAA (Glu) was identified, resulting in a frameshift. At position 96 (position [c]), a single nucleotide substitution resulted in a premature stop codon (from TGG (Trp) to TGA). At position 268 ([f], a single nucleotide substitution resulted in an additional premature stop codon (TAT (Tyr) to TAG).
Fig. 6. Comparisons of deduced amino acid sequences of the α1,3GT gene in marmoset (M), cebus (C), rhesus (R), orangutan (O), and human (H).
Nucleotide sequences are shown from the start codon to the stop codon of the marmoset monkey cDNA in one-letter abbreviations. Dashed horizontal lines indicate that each nucleotide is identical to the corresponding nucleotide of the marmoset base-line sequence. The single asterisk in cebus denotes a triplet nucleotide deletion not present in the marmoset. A single nucleotide deletion, depicted by a slash (/), was observed at positions [a], [b], [e], and [g]. Premature termination, depicted by an X, was observed at positions [c], [f], and [h].
A single nucleotide deletion at position [g] was observed in both human and orangutan but not in rhesus. The premature stop codon at position [h] was observed only in human. Thus, five crucial mutations that might account for loss of α1,3GT expression were determined in human and rhesus and three in orangutan. It is noteworthy that the mutations at position [b] and [f] were shared by the rhesus monkey, ape, and human.
DISCUSSION
The αGal epitopes are chemically related to the ABO blood type antigens. In the absence of expression of A or B epitopes or of αGal epitopes, antibodies develop against the respective antigens. Phylogenetic data on the emergence of the ABO antigen system (19) and especially molecular studies on ABO transferase (20) have been valuable in determining higher primate evolution. In contrast, little comparable information is known about the molecular basis of inactivation of the gene expressing the enzyme α1,3GT upon which synthesis of the αGal epitope depends.
The αGal system in lower mammals was not discovered until the 1980s (1, 2). Evidence of the presence of α1,3GT gene homologues in some αGal-negative higher primates was published in 1989–1991 by three pioneer groups. Joziasse et al. (7) described an 89-bp sequence (HGT-10) in human cells that was similar to a fragment of the murine and bovine exon 4. Larsen et al. (5) characterized a 703-bp genomic sequence in human tissue with a high degree of homology to exon 9 of the mouse α1,3GT gene. Galili and Swanson (6) reported a 371-bp sequence in rhesus cells that resembled part of mouse and bovine exon 9.
All three groups concluded that point mutations in exon 9 at some time in the evolutionary past of the higher primates could be the cause of the loss of αGal epitope expression. We confirmed the point mutations that had been described earlier in exon 9 of the rhesus (6), orangutan (6), and human α1,3SGT gene (5, 6), and demonstrated four additional point mutations in exons 7 and 8. The crucial mutations in the different species that result in frameshift and premature termination of translation include a deletion mutation at position [b] in exon 7 and a premature termination at position [f] in exon 9 that are shared by all three of these higher primates.
Because no α1,3GT transcripts could be found in the early investigations of the African green monkey (COS) and human cell lines (4), the possibility could not be dismissed that inactivation of the gene in the higher primates is due to mutations in the upstream regulatory regions of the α1,3GT gene (16). This potential explanation for loss of the αGal epitope was eliminated in our study using sensitive PCR methods for detection of mRNA transcripts that were not available at the time of the seminal investigations (4–7). A variety of transcripts with more than 10 different splicing patterns of the α1,3GT gene were observed in the full coding region in all rhesus, orangutan, and human tissues examined. Their presence was confirmed with Northern blot of total RNA derived from rhesus spleen.
Joziasse et al. (7, 16) estimated that inactivation of the α1,3GT gene occurred between 40 and 25 millions years ago, following the divergence between higher primates (catarrhines) and the New World monkey but prior to the further divergence of the individual higher primates. The hypothesis was based on their discovery of an intron-less processed cDNA homologue (termed HGT-2) in the human genome (7, 16). In contrast, Galili and Swanson (6) postulated that inactivation occurred at a much later time and independently in Old World monkeys, apes, and humans (6). Our finding of crucial point mutations shared by the rhesus, orangutan, and human at positions [b] and [f] of exons 7 and 9, respectively, appears to be more consistent with Joziasse’s hypothesis (16) that the α1,3GT gene inactivation occurred in a common ancestor than with the suggestion by Galili and Swanson (6) that it occurred independently in the three different lineages. However, the time and the process of the inactivation remain to be determined (6, 7). The high homology of the α1,3GT gene in the αGal-negative species to the shared sequence of the αGal-positive marmoset and cebus (Fig. 7) suggests that the inactivation may have occurred more recently than previously thought.
The development of preformed antibodies directed against a wide range of microorganisms or macroparasites that express αGal-like substance has been the conventional explanation for the evolution of the αGal-negative state in higher primates (1, 2). The possibility cannot yet be excluded, however, that some other survival advantage drove the gene inactivation and that the protection from such infecting agents afforded by the “natural” anti-αGal antibodies was fortuitous. Whatever the reason, antibodies against the αGal epitope are responsible for the immediate (hyperacute) rejection by αGal-negative recipients of tissues and organs from αGal-positive donors, precluding successful clinical xenotransplantation from αGal-positive animals (3). Detailed information about the α1,3GT gene may help in mapping strategies for transgenic modification of the αGal-positive species.
If higher primates were to spontaneously recover the expression of αGal epitopes, as has been described in normal breast and MCF7 human breast carcinoma cell lines (21), the potential for the development of αGal antibody-mediated autoimmune disease is implicit. Galili and co-workers (22) observed the presence of αGal epitopes in thyroid cells of a human with autoimmune Grave’s disease, but no transcripts could be detected in these cells with Northern blot. A search for α1,3GT mRNA transcripts in tissues from such “autoimmune suspect” patients may be fruitful with more sensitive PCR technology used in our study.
Acknowledgments
We thank Rickquel Tripp for technical assistance and Terry Mangan for manuscript preparation.
Footnotes
This work was supported by National Institutes of Health Grants DK 29961, R01 AI/DK 38899, AI 40329, DK 49615, and R01 DK 54232 and by Juvenile Diabetes Foundation Grant 4-1999-807.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s)AF384428 (marmoset α1,3GT), AY034181 (cebus α1,3GT), AY026225-AY026237 (rhesus α1,3GT), AF456457 (orangutan α1,3GT), and AF378121-AF378123 (human α1,3GT).
The abbreviations used are: αGal, galactose-α1,3-galactose; α1,3GT, α1,3-galactosyltransferase; RACE, rapid amplification of cDNA ends; PBMC, peripheral blood mononuclear cells; RT-PCR, reverse transcriptase PCR.
References
- 1.Galili U, Shohet SB, Kobrin E, Stults CLM, Macher BA. J Biol Chem. 1988;263:17755–17762. [PubMed] [Google Scholar]
- 2.Galili U, Clark MR, Shohet SB, Buehler J, Macher BA. Proc Natl Acad Sci U S A. 1987;84:1369–1373. doi: 10.1073/pnas.84.5.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper DKC, Koren E, Oliol R. Lancet. 1993;342:682–683. doi: 10.1016/0140-6736(93)91791-j. [DOI] [PubMed] [Google Scholar]
- 4.Joziasse DH, Shaper JH, Van den Eijnden DH, Van Tunen AJ, Shaper NL. J Biol Chem. 1989;264:14290–14297. [PubMed] [Google Scholar]
- 5.Larsen RD, Rivera-Marrero CA, Ernst LK, Cummings RD, Lowe JB. J Biol Chem. 1990;265:7055–7061. [PubMed] [Google Scholar]
- 6.Galili U, Swanson K. Proc Natl Acad Sci U S A. 1991;88:7401–7404. doi: 10.1073/pnas.88.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joziasse DH, Shaper JH, Jabs EW, Shaper NL. J Biol Chem. 1991;266:6991–6998. [PubMed] [Google Scholar]
- 8.Sandrin MS, Dabkowski PL, Henning MM, Mouhtouris E, McKenzie IFC. Xenotransplant. 1994;1:81–88. [Google Scholar]
- 9.Strahan KM, Gu F, Andersson L, Gustafsson K. Immunogenetics. 1995;41:101–105. doi: 10.1007/BF00182319. [DOI] [PubMed] [Google Scholar]
- 10.Vanhove B, Goret H, Soulillou JF, Pourcel C. Biochim Biophys Acta. 1997;1356:1–11. doi: 10.1016/s0167-4889(96)00151-6. [DOI] [PubMed] [Google Scholar]
- 11.Katayama A, Ogawa H, Kadomatsu K, Kurosawa N, Kobayashi T, Kaneda N, Uchimura K, Yokoyama I, Muramatsu T, Takagi H. J Glycoconj. 1998;15:583–589. doi: 10.1023/a:1006963809894. [DOI] [PubMed] [Google Scholar]
- 12.Koike C, Friday RP, Nakashima I, Luppi P, Fung JJ, Rao AS, Starzl TE, Trucco M. Transplantation. 2000;70:1275–1283. doi: 10.1097/00007890-200011150-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joziasse DH, Shaper NL, Kim D, Van den Eijnden DH, Shaper JH. J Biol Chem. 1992;267:5534–5541. [PubMed] [Google Scholar]
- 14.Larsen RB, Rajan VP, Ruff MM, Kukowska-Latallo J, Cummings RD, Lowe JB. Proc Natl Acad Sci U S A. 1989;86:8227–8231. doi: 10.1073/pnas.86.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike C, Friday RP, Fung JJ, Starzl TE, Trucco M. Transplant Proc. 2001;33:710–711. doi: 10.1016/s0041-1345(00)02216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joziasse DH, Oriol R. Biochem Biophys Acta. 1999;1455:403–418. doi: 10.1016/s0925-4439(99)00056-3. [DOI] [PubMed] [Google Scholar]
- 17.Mang R, Goudsmit J, van der Kuyl AC. J Virol. 1999;73:7021–7026. doi: 10.1128/jvi.73.8.7021-7026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henion TR, Macher BA, Anaraki F, Galili U. Glycobiology. 1994;4:193–201. doi: 10.1093/glycob/4.2.193. [DOI] [PubMed] [Google Scholar]
- 19.Oliol R, Le Pendu J, Mollicone R. Vox Sang. 1986;51:161–171. doi: 10.1111/j.1423-0410.1986.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 21.Petryniak J, Varani J, Ervin PR, Goldstein IJ. Cancer Lett. 1991;60:59–65. doi: 10.1016/0304-3835(91)90049-n. [DOI] [PubMed] [Google Scholar]
- 22.Etienne-Decerf J, Winand R, Galili U. Autoimmunity. 1991;10:81–87. doi: 10.3109/08916939109004811. [DOI] [PubMed] [Google Scholar]