Abstract
In systemic lupus erythematosus (SLE), T helper cells exhibit increased and prolonged expression of cell-surface CD40 ligand (CD154), spontaneously overproduce interleukin-10 (IL-10), but underproduce interferon-gamma (IFN-γ). We tested the hypothesis that the imbalance of these gene products reflects skewed expression of CD154, IL-10, and IFN-γ genes. Here, we demonstrate that the histone deacetylase inhibitor, trichostatin A, significantly down-regulated CD154 and IL-10 and up-regulated IFN-γ gene expression in SLE T cells. This reversal corrected the aberrant expression of these gene products, thereby enhancing IFN-γ production and inhibiting IL-10 and CD154 expression. That trichostatin A can simultaneously reverse the skewed expression of multiple genes implicated in the immunopathogenesis of SLE suggests that this pharmacologic agent may be a candidate for the treatment of this autoimmune disease.
The hallmark of systemic lupus erythematosus (SLE) is dysregulated production of autoantibodies that ultimately leads to irreversible, immune complex-mediated end-organ failure (1, 2). This dysregulated state seems to stem from a functional imbalance of T lymphocyte subsets, the fundamental disorder of cellular immunity in SLE (3). Characterized by a diametric increase in CD4+ T helper (Th) function and decrement in CD8+ T cytotoxic/suppressor activity, the outcome of this imbalance of regulatory activity is Th-driven polyclonal B cell production of both natural antibodies and pathogenic autoantibodies.
Within the Th subpopulation, there is also an imbalance between Th1 and Th2 cytokine production in SLE. Activated Th2 cells overproduce interleukin 6 (IL-6), IL-10, and tumor necrosis-α, whereas Th1 cells underproduce IL-2, interferon-γ (IFN-γ), and transforming growth factor-β (4–8). Moreover, activated Th cells overexpress cell-surface CD40 ligand (CD154) that persists over an extended time interval compared with activated normal T cells (9, 10). The ongoing interaction between CD154 on T cells and CD40 on B cells, in the presence of high levels of IL-6 and IL-10 and in the absence of effective regulatory CD8 T cytotoxic/suppressor cells, persistently activates B cells, resulting in high-output Ig production. This altered homeostasis ultimately leads to polyclonal hypergammaglobulinemia.
One mechanism that might account for this skewed expression of CD154, IL-10, and IFN-γ in SLE T cells is altered chromatin structure, in part as a result of an imbalance of histone acetylation. The histone proteins, H2A, H2B, H3, and H4, possess globular carboxyl (C)-terminal and highly basic amino (N)-terminal tail domains. The C-terminal domain forms the core of the nucleosome, whereas the tail domain is located outside of the core particle and is accessible to posttranslational covalent modifications, especially acetylation and methylation (11). Reversible acetylation of the ɛ-amino group of lysine by histone acetylases and histone deacetylases (HDACs) is a dynamic process that plays a role in regulation of chromatin structure (12). Lysine acetylation neutralizes histone proteins, whereas deacetylation restores positively charged lysines. Following deacetylation, interactions between positively charged lysines and negative charges in the DNA phosphodiester backbone stabilize the DNA/histone complex, restricting nucleosome mobility on the DNA and hindering accessibility of the promoter to the transcription machinery (13). DNA methylation seems to further enhance this process by promoting histone deacetylation (14). By contrast, acetylation seems to result in dissociation of DNA–histone interactions in the nucleosome, enhancing accessibility of the promoter to transcription factors and RNA polymerase, thereby promoting transcriptional initiation (15). Parenthetically, histone acetylases and HDACs can also acetylate/deacetylate non-histone proteins, including p53, which can also regulate gene transcription (16). Thus, if deficient acetylation exists and results in skewed gene expression of CD154, IL-10, and IFN-γ in SLE T cells, then augmenting acetylation might reverse this disorder.
Inhibition of HDAC activity can increase histone acetylation by preventing lysine deacetylation. In both yeast and mammalian species, HDACs exist in multiprotein complexes that are composed of transcriptional co-repressors (16, 17) and are recruited to regions of the genome by DNA-binding factors, such as the methylated DNA-binding protein MeCP2 (18, 19). There, these complexes seem to promote transcriptional repression (16, 20). In eukaryotic cells, however, the outcome of histone acetylation and deacetylation on chromatin remodeling and, ultimately, gene transcription seems to be gene-specific. Histone acetylation results in up-regulation [p21Cip1/waf1, p27Kip1, HIV-1 (21–23)], repression [c-myc, IL-8 (22, 24)], or no change [glyceraldehyde-3-phosphate dehydrogenase (GAPDH), γ-actin (22–24)] in the transcription of these genes. In fact, expression of only 1–2% of genes seems to be modified by histone hyperacetylation (22, 23). We reasoned, therefore, that if altered expression of CD154, IL-10, and IFN-γ is associated with relative under-acetylation, then increasing histone acetylation by inhibiting HDAC activity might correct the disorder.
To investigate the idea that aberrant expression of CD154, IL-10, and IFN-γ in SLE T cells reflects skewed gene expression because of altered acetylation of these genes, we tested the capacity of trichostatin A (TSA) to reverse this disorder. TSA is a specific, reversible inhibitor of HDACs in vitro and in vivo that is active at nanomolar concentrations (25). Crystallographic analyses suggest that the hydroxamic acid moiety binds to the zinc in the tubular pocket, a portion of the active catalytic site formed by a tubular pocket, a zinc-binding site, and two asparagine–histidine charge-relay systems (26). This agent and a structurally related compound, e.g., suberoylanilide hydroxamic acid, have been used effectively and without toxicity in mouse models (27, 28). Here, we demonstrate that TSA significantly down-regulated CD154 and IL-10 and up-regulated IFN-γ gene expression in SLE T cells. This reversal corrected the aberrant expression of these gene products, thereby enhancing IFN-γ production and inhibiting IL-10 and CD154 expression. That TSA can simultaneously affect expression of multiple genes involved in the immunopathogenesis of SLE warrants further study of the potential efficacy of this agent as a candidate for the treatment of this autoimmune disease.
Materials and Methods
Study Subjects.
Nine adult subjects with an established diagnosis of SLE were randomly recruited from our Lupus Clinic. All SLE subjects fulfilled ≥4 ARA criteria for the classification of SLE (29). Of these, eight were female, five were Caucasian, and four were African-American. The mean (±SEM) age of these subjects was 39 ± 4.1 yr, and the mean systemic lupus erythematosus disease activity index was 4.2 ± 1. Two subjects were being treated with ≤10 mg/day prednisone; none were being treated with or had taken immunosuppressive agents within 6 months. Corticosteroid therapy was held for 24 h before venipuncture. Age, gender, and racially matched normal controls were also studied. All subjects signed an informed consent form approved by the Wake Forest University Baptist Medical Center Institutional Review Board before participation in this study.
Isolation of Blood Mononuclear Cells.
Peripheral blood mononuclear cells (PBMCs) were isolated from samples obtained by venipuncture or leukopheresis; enrichment of CD3+ T cells was performed as previously detailed (30). T cell preparations were a mean (±SEM) 96% ± 1.2 CD3+ as determined by flow cytometry.
Cell Culture Procedure and TSA Treatment.
Freshly isolated PBMCs or enriched T cells were cultured as previously described (30, 31). Briefly, 1 × 106 cells/ml were cultured in RPMI-1640 media supplemented with 25 mM Hepes/10% heat-inactivated FCS/100 IU penicillin/100 μg/ml streptomycin/2 mM L-glutamine for varying intervals to 72 h. In experiments using TSA, cells were preincubated with the inhibitor for 18 h and were then subsequently activated. Because SLE T cells respond suboptimally to activation by antigens, mitogens, or anti-CD3/anti-CD28 mAbs because of defects of signal transduction (32, 33), cellular activation was induced by 20 ng/ml phorbol 12-myristate 13-acetate (PMA) and 0.5 μM ionomycin (IO) for varying intervals. Activation of cells by PMA + IO bypasses the defective proximal signaling cascade, stimulating protein kinase C and releasing intracellular Ca2+. The combination of PMA + IO enhances activation of SLE T cells but does not necessarily normalize the response. Cell culture supernatants were harvested, aliquoted, and immediately frozen at −70°C at specific times throughout the course of experiments.
Measurement of Cell-Surface Expression and Cytokine Secretion.
For quantification of the proportion of T cells expressing cell-surface CD3 and CD154 by flow cytometry, 1 × 106 viable T cells were stained with saturating concentrations of FITC-anti-CD3 and phycoerythrin-(PE)-anti-CD154 mAbs (Caltag, South San Francisco, CA), fixed in 1% paraformaldehyde, and extensively washed with physiologic buffered saline. The percentage of PE-CD154+ cells was determined by quantifying the number of cells with fluorescence above background levels when cells were stained with PE-IgG alone. Other cell-surface markers were quantified by flow cytometry as detailed (34). Secretion of IL-10 and IFN-γ into culture supernatants was quantified by ELISA (R & D Systems).
Isolation of RNA and Semiquantitative Reverse Transcriptase–PCR.
Total RNA from untreated and TSA-treated cells was isolated by using TRIZOL reagent (Life Technologies, Grand Island, NY). Three micrograms of total RNA was converted to cDNA by standard methods by using reverse transcriptase (Life Technologies, Gaithersburg, MD) and random primers (Promega). For semiquantitative PCR, 5 μl of cDNA was amplified by PCR by using the following specific primers: CD154: 5′-GAATCCTCAAATTGCGGCAC-3′ and 5′-CAGAAGGTGACTTGGCATAG-3′; GAPDH: 5′-GGTGAAGGTCGGAGTCAACG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′; IL-10: 5′-TTGCCTGGTCCTCCTGACTG-3′ and 5′-GATGTCTGGGTCTTGGTTCT-3′; IFN-γ: 5′-ATGAAATATACAAGTTATATCTTGGCTTT-3′ and 5′-GATGCTCTTCGACCTCGAAACAGCAT-3′. The reaction mixtures were subjected to 30 cycles of denaturation (94°C, 1 min) and annealing for 1 min at 53°C (CD154), 40°C (GAPDH), and 55°C (IL-10 and IFN-γ). Extension was for 2 min at 72°C; a final extension of 7 min at 72°C was performed by using a DNA thermal cycler (Perkin-Elmer 2400). PCR amplifications were carried out in duplicate. PCR products were separated by 2% PAGE, and band intensities were quantified by using an Alpha Imager 2000 (Alpha Innotech, San Leandro, CA). Independent amplification of the control gene, GAPDH, was used to correct for differences in efficiency of RNA isolation and reverse transcription. The final normalized results were calculated by dividing the relative transcript levels of the test genes by the relative amounts of GAPDH transcripts.
Statistical Analysis.
Statistical significance (P ≤ 0.05) was calculated by the paired Student's t test, Mann–Whitney U rank-sum test, or ANOVA (SigmaStat, Jandel, San Rafael, CA). Mean values (±SEM) are used throughout the text.
Results
TSA Decreases Expression of CD154 mRNA and CD154 Protein.
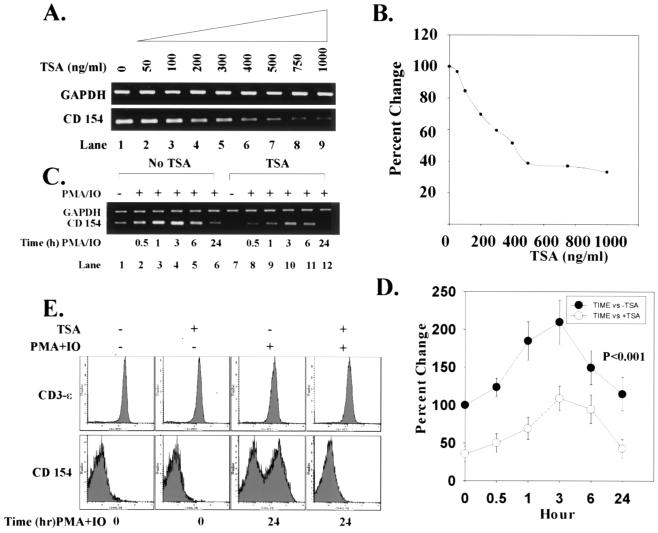
To determine whether TSA can down-regulate CD154 mRNA production, SLE T cells were treated with increasing concentrations of TSA over 18 h. Fig. 1 A and B demonstrate that TSA maximally inhibited CD154 transcript by 60% but did not modify GAPDH mRNA expression. When SLE T cells were activated with PMA + IO, CD154 mRNA content increased 100%, peaked at 3 h, and waned thereafter (Fig. 1 C and D). Under these conditions, however, GAPDH mRNA remained stable, demonstrating that cellular activation does not modify this gene's expression. By contrast, when T cells were preincubated with TSA for 18 h and then activated by PMA + IO over intervals to 24 h, up-regulation of CD154 transcript was significantly reduced throughout the entire time course compared with cells not exposed to TSA (Fig. 1 C and D; P < 0.001). Thus, in SLE T cells, TSA reduces the degree of up-regulation of CD154 transcript expression.
Figure 1.
Down-regulation of CD154 transcript levels and protein expression by TSA. (A) Increasing concentrations of TSA (0–1000 ng/ml) progressively decrease levels of CD154 mRNA relative to GAPDH mRNA in SLE T cells. (B) Based on densitometric scanning of the gel in A, this graph depicts the percent change of CD154 mRNA quantity with increasing concentrations of TSA over 24 h. GAPDH mRNA expression is stable and unchanged in the presence of TSA. These data are representative of independent experiments using T cells from three SLE subjects. (C) T cells were incubated in the absence or presence of 1,000 ng/ml TSA over 18 h. T cells were then stimulated with 20 ng/ml PMA + 0.5 μM IO for intervals to 24 h. CD154 mRNA levels relative to GAPDH mRNA levels are shown. (D) This graph depicts the percent change in CD154 mRNA amount over time in the absence (●) or presence (○) of TSA. (E) SLE T cells were cultured in the absence or presence of 1,000 ng/ml TSA for 18 h and subsequently activated with 20 ng/ml PMA + 0.5 μM IO for 24 h. Flow cytometric analysis of CD154 and CD3-ɛ expression on SLE T cells is compared. The abscissa denotes the number of cells and ordinate the intensity of the cell fluorescence signal. These data are representative of independent experiments performed with T cells from nine SLE and normal control subjects, respectively. Data from normal T cells are comparable to SLE T cells and, therefore, are not shown. Statistical analyses were performed by paired Student's t test or one-way ANOVA.
In agreement with previous work (9, 10), we found that an increased proportion of SLE and normal control T cells expresses cell-surface CD154 when stimulated with PMA + IO. To determine whether TSA-dependent down-regulation of CD154 mRNA reduces surface expression of CD154, SLE T cells were treated for 18 h with TSA and the proportion of CD154+ cells quantified by flow cytometry. Compared with untreated cells, TSA significantly reduced cell-surface CD154+ expression over 24 h (P = 0.029) (Fig. 1E and Table 1). From dose titration experiments, the optimal dose of TSA to maximally reduce cell-surface expression of CD154 was 1000 ng/ml (data not shown). By contrast, the decrease in the proportion of CD154+ T cells from normal subjects was not statistically different from untreated cells (Table 1). Interestingly, activation of both SLE and normal T cells with PMA + IO over 24 h induced a new population of CD154+ cells that expressed a comparable proportion of the cells with the receptor (Fig. 1E and Table 1). However, CD154 expression was completely inhibited when cells were pretreated with TSA before activation (SLE, P = 0.005; normal, P < 0.001) (Fig. 1E and Table 1). By contrast, cell-surface CD3-ɛ expression remained stable under these varying conditions, indicating that the effect of TSA on CD154 surface expression is not generalized (Fig. 1E). In sum, these experiments reveal that TSA down-regulates both CD154 mRNA levels and protein expression, but not GAPDH mRNA levels or CD3-ɛ cell-surface expression, in SLE and control T cells.
Table 1.
Effect of TSA on CD154 surface expression, IL10 production, and IFN-γ secretion in SLE and control T cells
| 24-h culture | CD154+ T cells
(%) Mean ± SEM
|
IL-10 (pg/ml) Mean ±
SEM
|
IFN-γ (pg/ml) Mean ± SEM
|
|||
|---|---|---|---|---|---|---|
| Normal | SLE | Normal | SLE | Normal | SLE | |
| Medium | 4.1 ± 1.4 | 5.0 ± 1.2 | 0 | 242.4 ± 144.2 | 0 | 0 |
| TSA (1000 ng/ml) | 2.5 ± 0.8 | 2.1 ± 0.7 | 0 | 0 | 70.8 ± 27.4 | 0 |
| PMA (20 ng/ml) + IO (0.5 μM) | 26.7 ± 4.0 | 24 ± 3.2 | 0 | 65.9 ± 49.3 | 2862.2 ± 1258.8 | 23.8 ± 13.9 |
| TSA (1000 ng/ml) + PMA (20 ng/ml) + IO (0.5 μM) | 5.6 ± 1.6 | 12.3 ± 3.2 | 0 | 0 | 2384.0 ± 970.9 | 291.4 ± 143.4 |
| P value | ||||||
| Medium, TSA | P = N.S. | P = 0.029 | P = 0.008 | |||
| PMA + IO, TSA + PMA + IO | P < 0.001 | P = 0.005 | P = 0.299 | P = 0.011 | ||
TSA Decreases Expression of IL-10 mRNA and IL-10 Protein Secretion.
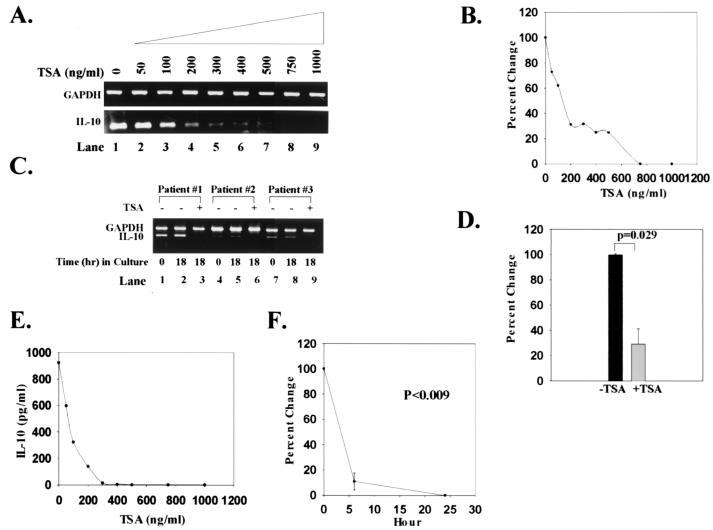
T cells from SLE subjects produce markedly increased amounts of IL-10, resulting in high serum levels of the cytokine (35, 36). To determine whether TSA could down-regulate IL-10, we carried out a dose-response analysis. Like CD154, increasing concentrations of TSA progressively decreased IL-10 transcript expression (Fig. 2 A and B). No detectable IL-10 mRNA was identified at TSA concentrations of 700–800 ng/ml (Fig. 2 A and B). A similar analysis in T cells from normal controls could not be performed because these cells do not express IL-10 mRNA. Stimulation of T cells with PMA + IO did not increase the amount of IL-10 transcript or IL-10 secretion in either SLE or normal controls (Table 1). As shown in Fig. 2C, IL-10 transcripts were present in freshly isolated T cells (0 h; lanes 1, 4, 7) and remained stable relative to GAPDH transcripts after culturing cells for 18 h in the absence of mitogens, antigens, or cytokines (lanes 2, 5, 8). However, when SLE T cells were cultured in the presence of TSA for 18 h, no detectable IL-10 transcripts were identified (lanes 3, 6, 9). When IL-10 transcripts from SLE subjects were quantified relative to GAPDH transcripts, TSA decreased levels of IL-10 mRNA by 71% (Fig. 2D; P = 0.029). Thus, TSA significantly diminished IL-10 transcript expression in SLE T cells over 24 h.
Figure 2.
Down-regulation of IL-10 transcript levels and protein expression by TSA. (A) Increasing concentrations of TSA (0–1,000 ng/ml) progressively decrease the amount of IL-10 mRNA relative to GAPDH mRNA. (B) Based on densitometric scanning of the gel in A, this graph quantifies the percent change in IL-10 transcript levels with increasing concentrations of TSA over 24 h. These data are representative of nine SLE subjects studied. T cells from normal controls do not express detectable IL-10 mRNA. (C) IL-10 and GAPDH transcripts from T cells of three SLE subjects are shown. Transcripts from freshly isolated T cells are shown in lanes 1, 4, and 7. Transcripts from T cells cultured for 18 h in the absence or presence of 1,000 ng/ml TSA are shown in lanes 2, 5, and 8 and 3, 6, and 9, respectively. These data are representative of nine SLE subjects studied. (D) Based on densitometric scanning of gels, this graph shows the percent change in amount of IL-10 mRNA from SLE T cells cultured in the absence or presence of 1,000 ng/ml TSA. (E) This graph illustrates the inhibition of IL-10 secretion by increasing concentrations of TSA over 24 h. (F) This graph depicts the percent change of IL-10 secretion over time. IL-10 protein secretion was undetectable in supernatants from normal T cells. Statistical analysis was performed by paired Student's t test.
That culturing SLE T cells with TSA produced such a marked decrement in IL-10 transcript expression prompted us to quantify IL-10 in culture supernatants. During 24 h in culture, normal PBMCs produced no IL-10; by contrast, SLE PBMCs secreted a mean of 242.4 pg/ml of IL-10 (Table 1). These results verify previous analyses demonstrating that SLE PBMCs spontaneously produce high concentrations of IL-10 (37). Treatment of PBMCs from SLE subjects over 18 h with increasing concentrations of TSA resulted in a dose-dependent inhibition of IL-10 protein secretion that was maximal at 300 ng/ml of the inhibitor (Fig. 2E). Within 6 h, TSA inhibited IL-10 secretion by 90%; at 24 h, there were undetectable levels of IL-10 in culture supernatants (P = 0.008) (Fig. 2F and Table 1). Thus, like CD154, TSA is able to block expression of IL-10 transcript, abolishing IL-10 production by SLE T cells.
TSA Increases Expression of IFN-γ mRNA and IFN-γ Protein Secretion in Response to Mitogen.
Low production of IFN-γ by SLE T cells (7, 35, 36) may reflect down-regulation of gene expression (36). To establish whether TSA can augment IFN-γ expression, SLE T cells were treated for 18 h in the absence or presence of TSA. When T cells were activated with PMA + IO in the absence of TSA, peak IFN-γ transcript expression increased 13-fold at 1 h over basal levels relative to GAPDH transcript but waned thereafter. By contrast, activation of T cells in the presence of TSA induced a peak 37-fold increase in IFN-γ mRNA levels at 6 h over untreated cells relative to GAPDH (Fig. 3 A and B; P = 0.031). Thus, TSA up-regulates expression of IFN-γ transcripts in SLE T cells, yielding both a significantly increased and prolonged production of the transcript.
Figure 3.
Up-regulation of IFN-γ transcript levels by TSA. (A) SLE T cells were incubated in the absence or presence of 1,000 ng/ml TSA over 18 h. T cells were then stimulated with 20 ng/ml PMA + 0.5 μM IO for intervals to 24 h. IFN-γ mRNA levels relative to GAPDH are shown. (B) Based on densitometric scanning of the gel in A, this graphs depicts the fold increase of IFN-γ mRNA in cells cultured in the absence (●) or presence (○) of 1,000 ng/ml TSA during intervals to 24 h. (C) T cells were cultured in the absence or presence of 1,000 ng/ml TSA for 24 h and then stimulated with 20 ng/ml PMA + 0.5 μM IO for 24 h. This graph shows the fold increase of IFN-γ protein secretion. Statistical analyses were performed by paired Student's t test or one-way ANOVA.
This strong up-regulation of IFN-γ transcript was reflected in significantly increased secretion of IFN-γ protein by 24 h. In the absence of stimulation, neither SLE nor control PBMCs produced measurable IFN-γ over 24 h (Table 1). When SLE PBMCs were activated with PMA + IO for 24 h, the mean IFN-γ secretion was 23.8 pg/ml (Table 1). By contrast, similar activation of normal PBMC stimulated cells to produce 120-fold more IFN-γ production under the same conditions (Table 1). These data confirm that SLE PBMC activated in vitro produce significantly less IFN-γ than normal cells (7). However, activation of PBMCs in the presence of TSA further enhanced IFN-γ output by >12-fold (P = 0.011), whereas there was actually a small decrease in IFN-γ production by control PBMC (Fig. 3C and Table 1). This reduction of IFN-γ production by treatment of normal PBMC with TSA has been previously observed (38). Taken together, these results demonstrate that TSA rapidly up-regulates and stabilizes IFN-γ transcript levels in SLE PBMC, resulting in enhanced IFN-γ production in response to cellular activation.
Discussion
A fundamental disorder of the cellular immune response in SLE is dysregulated expression of CD154, IL-10, and IFN-γ. In contrast to normal T cells, freshly isolated SLE T cells display a higher epitope density of cell-surface CD154 (9, 10) and spontaneously produce increased amounts of IL-10 (35, 39). Moreover, in vitro activated SLE T cells secrete significantly less IFN-γ than control T cells (7, 35). Here, we demonstrate that the specific HDAC inhibitor, TSA, significantly down-regulated CD154 and IL-10 and up-regulated IFN-γ transcript expression in SLE T cells. TSA maximally inhibited CD154 and IL-10 mRNA transcripts by 60% and 71%, respectively, whereas TSA up-regulated IFN-γ mRNA content in SLE T cells by 24-fold in response to mitogen. That TSA could simultaneously reverse the aberrant expression of these genes suggested that expression of their products might be corrected.
Indeed, reversal of skewed gene expression corrected the aberrant production of these gene products. When freshly isolated SLE T cells were cultured in the presence of TSA, cell-surface expression of CD154 was significantly reduced compared with controls. Moreover, TSA completely inhibited the induction of new CD154+ T cell subpopulations in both SLE and controls in response to cellular activation. Strikingly, TSA completely inhibited IL-10 secretion and augmented IFN-γ production by 12-fold. Of interest is that the capacity of TSA to normalize the expression of these gene products did not necessarily require a return of gene transcripts to levels observed in controls. Apparently, down-regulation, but not inhibition, of CD154 transcript is sufficient to correct receptor expression and to prevent its appearance on a new subpopulation of T cells following cellular activation.
TSA is a prototype first generation pharmacologic inhibitor capable of modifying the expression of multiple genes concomitantly. Indeed, suberoylanilide hydroxamic acid, a synthetic analogue that inhibits HDAC activity at micromolar concentrations, is currently in phase I clinical trials for certain cancers (40, 41). By modulating multiple genes simultaneously, the efficacy of TSA is likely to be superior to single or even multiple biologic agents. Because it down-regulates CD154 and IL-10, TSA could have a salutary effect on disease progression in vivo. CD154–CD40 interactions regulate diverse pathways of the immune system (42). For example, this interaction (i) influences T cell priming and T cell-mediated effector functions; (ii) up-regulates costimulatory molecules; and (iii) mediates the immunopathogenesis of organ-specific autoimmune diseases (e.g., multiple sclerosis), chronic inflammation (e.g., atherosclerosis or pulmonary fibrosis), and organ transplant rejection. The capacity of TSA to down-regulate CD154 expression would provide an effective means to ameliorate the inflammatory process by interfering with the CD40–CD154 axis. Similarly, up-regulation of IL-10 is associated with various autoimmune diseases and allograft rejection. The dramatic down-regulation of IL-10 secretion by TSA also supports its potential use as a therapeutic agent in these disorders. Finally, the inability to mount a protective immune response to pathogens is the leading cause of mortality in subjects with SLE. Up-regulation of IFN-γ by TSA in response to T cell activation could potentially augment immune defenses by stimulating bactericidal activity of phagocytes, natural killer cell activity, antigen presentation, as well as integrating cellular interactions (43). Thus, our demonstration that TSA corrects defective cytokine production and altered CD154 expression suggests that this agent may be a potential candidate for treatment of SLE.
Further investigation of the capacity of TSA to modify gene expression of CD154, IL-10, and IFN-γ in other autoimmune diseases is certainly warranted. Moreover, the role of HDACs in the immunopathogenesis and the heritability of SLE also remain to be established.
Acknowledgments
We thank members of the Kammer Laboratory for ongoing informative and critical discussions and Drs. G. Gilkeson, A. Perl, and G. Tsokos for critical review of this manuscript. N.M. is a postdoctoral fellow of the Arthritis Foundation. D.R.B. was a predoctoral fellow of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant RO1 AR39501, the Lupus Foundation of America, General Clinical Research Center of the Wake Forest University School of Medicine Grant MO1 RR07122, and National Cancer Institute Grant 5P30 CA12197.
Abbreviations
- SLE
systemic lupus erythematosus
- TSA
trichostatin A
- Th
T helper
- HDAC
histone deacetylases
- PBMC
peripheral blood mononuclear cells
- PMA
phorbol 12-myristate 13-acetate
- IO
ionomycin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kammer G M, Tsokos G C, editors. Lupus: Molecular and Cellular Pathogenesis. Totowa, NJ: Humana; 1999. pp. 1–708. [Google Scholar]
- 2.Wener M H. In: Principles of Molecular Rheumatology. Tsokos G C, editor. Totowa, NJ: Humana; 2000. pp. 127–144. [Google Scholar]
- 3.Horwitz D A, Stohl W, Gray J D. In: Dubois' Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 155–194. [Google Scholar]
- 4.Linker-Israeli M, Deans R J, Wallace D J, Prehn J, Ozeri-Chen T, Klinenberg J R. J Immunol. 1991;147:117–123. [PubMed] [Google Scholar]
- 5.Richaud-Patin Y, Alcocer-Varela J, Llorente L. Rev Invest Clin. 1995;47:267–272. [PubMed] [Google Scholar]
- 6.Alcocer-Varela J, Alarcon-Segovia D. J Clin Invest. 1982;69:1388–1392. doi: 10.1172/JCI110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsokos G C, Boumpas D T, Smith P L, Djeu J Y, Balow J E, Rook A H. Arthritis Rheum. 1986;29:1210–1215. doi: 10.1002/art.1780291005. [DOI] [PubMed] [Google Scholar]
- 8.Ohtsuka K, Gray J D, Stimmler M M, Toro B, Horwitz D A. J Immunol. 1998;160:2539–2545. [PubMed] [Google Scholar]
- 9.Koshy M, Berger D, Crow M K. J Clin Invest. 1996;98:826–837. doi: 10.1172/JCI118855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai-Mehta A, Lu L J, Ramsey-Goldman R, Datta S K. J Clin Invest. 1996;97:2063–2073. doi: 10.1172/JCI118643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer V A, Davie J R. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 12.Kornberg R D, Lorch Y. Curr Opin Gen Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 13.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne A W. Nature (London) 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 15.Lee D, Hayes J, Pruss D, Wolffe A P. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 16.Ng H H, Bird A. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 17.Cress W D, Seto E. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Razin A. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng H H, Bird A. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 20.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 21.Gray S G, Yakovleva T, Hartmann W, Tally M, Bakalkin G, Ekstrom T J. Exp Cell Res. 1999;253:618–628. doi: 10.1006/excr.1999.4661. [DOI] [PubMed] [Google Scholar]
- 22.Van Lint C, Emiliani S, Verdin E. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 23.Richon V M, Sandhoff T W, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. . (First published August 22, 2000; 10.1073/pnas.180316197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang N, Katz J P, Martin D R, Wu G D. Cytokine. 1997;9:27–36. doi: 10.1006/cyto.1996.0132. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 26.Finnin M S, Donigian J R, Cohen A, Richon V M, Rifkind R A, Marks P A, Breslow R, Pavletich N P. Nature (London) 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi I, Miyaji H, Yoshida T, Sato S, Mizukami T. J Antibiot. 1996;49:453–457. doi: 10.7164/antibiotics.49.453. [DOI] [PubMed] [Google Scholar]
- 28.Butler L M, Agus D B, Scher H I, Higgins B, Rose A, Cordon-Cardo C, Thaler H T, Rifkind R A, Marks P A, Richon V M. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 29.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F, Schaller J G, Talal N, Winchester R J. Arthritis Rheum. 1982;15:1271–1275. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 30.Mishra N, Khan I U, Tsokos G C, Kammer G M. J Immunol. 2000;165:2830–2840. doi: 10.4049/jimmunol.165.5.2830. [DOI] [PubMed] [Google Scholar]
- 31.Laxminarayana D, Khan I U, Mishra N, Olorenshaw I, Taskén K, Kammer G M. J Immunol. 1999;162:5639–5648. [PubMed] [Google Scholar]
- 32.Dayal A K, Kammer G M. Arthritis Rheum. 1996;39:23–33. doi: 10.1002/art.1780390104. [DOI] [PubMed] [Google Scholar]
- 33.Liossis S N C, Ding X Z, Dennis G J, Tsokos G C. J Clin Invest. 1998;101:1448–1457. doi: 10.1172/JCI1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laxminarayana D, Kammer G M. J Immunol. 1996;156:497–506. [PubMed] [Google Scholar]
- 35.Hagiwara E, Gourley M F, Lee S, Klinman D M. Arthritis Rheum. 1996;39:379–385. doi: 10.1002/art.1780390305. [DOI] [PubMed] [Google Scholar]
- 36.Handwerger B S, Luzina I G, da Silva L, Storrer C E, Via C S. In: Lupus: Molecular and Cellular Pathogenesis. Kammer G M, Tsokos G C, editors. Totowa, NJ: Humana; 1999. pp. 321–340. [Google Scholar]
- 37.Viallard J F, Pellegrin J L, Ranchin V, Schaeverbeke T, Dehais J, Longy-Boursier M, Ragnaud J M, Leng B, Moreau J F. Clin Exp Immunol. 1999;115:189–195. doi: 10.1046/j.1365-2249.1999.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dangond F, Gullans S R. Biochem Biophys Res Commun. 1998;247:833–837. doi: 10.1006/bbrc.1998.8891. [DOI] [PubMed] [Google Scholar]
- 39.Llorente L, Zou W, Levy Y, Richaud-Patin Y, Wijdenes J, Alcocer-Varela J, Morel-Fourrier B, Brouet J C, Alarcon-Segovia D, Galanaud P, et al. J Exp Med. 1995;181:839–844. doi: 10.1084/jem.181.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marks P A, Richon V M, Rifkind R A. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 42.Grewal I S, Flavell R A. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 43.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]