Abstract
Reliable model systems are needed to elucidate the role cancer stem cells (CSCs) play in pediatric brain tumor drug resistance. The majority of studies to date have focused on clinically distinct adult tumors and restricted tumor types. Here, the CSC component of 7 newly established primary pediatric cell lines (2 ependymomas, 2 medulloblastomas, 2 gliomas, and a CNS primitive neuroectodermal tumor) was thoroughly characterized. Comparison of DNA copy number with the original corresponding tumor demonstrated that genomic changes present in the original tumor, typical of that particular tumor type, were retained in culture. In each case, the CSC component was approximately 3–4-fold enriched in neurosphere culture compared with monolayer culture, and a higher capacity for multilineage differentiation was observed for neurosphere-derived cells. DNA content profiles of neurosphere-derived cells expressing the CSC marker nestin demonstrated the presence of cells in all phases of the cell cycle, indicating that not all CSCs are quiescent. Furthermore, neurosphere-derived cells demonstrated an increased resistance to etoposide compared with monolayer-derived cells, having lower initial DNA damage, potentially due to a combination of increased drug extrusion by ATP-binding cassette multidrug transporters and enhanced rates of DNA repair. Finally, orthotopic xenograft models reflecting the tumor of origin were established from these cell lines. In summary, these cell lines and the approach taken provide a robust model system that can be used to develop our understanding of the biology of CSCs in pediatric brain tumors and other cancer types and to preclinically test therapeutic agents.
Keywords: cancer stem cells, drug resistance, etoposide, pediatric
Brain tumors are the leading cause of death in children who develop solid tumors. The incidence rate is approximately 30 per million,1 with an overall 10-year period survival estimate of 59%.2 Diagnosis is based on clinical history and radiological imaging but relies heavily on histological conformation.3 Tumors are characterized into 7 main groups depending on the location of the tumor, histopathological features, and immunohistochemistry. These features largely determine grade, which in turn loosely reflects outcome.4 Although there have been some improvements in the outcome for children with brain tumors,5 the prognosis for children with high grade or diffuse intrinsic pontine gliomas, relapsed medulloblastoma, and ependymoma is extremely poor.6,7 In order to achieve the necessary delicate balance between improved treatment and avoidance of damage to the developing brain, we need a better understanding of the underlying biology of these tumors.
Several studies have demonstrated the existence of a highly tumorigenic subpopulation of cells in brain tumors.8–18 Although there is a debate regarding naming these cells (cancer “stem” or “initiating” or “propagating” cells) and regarding their frequency within brain tumors, studies have clearly shown an association between this subgroup and (i) the expression of stem cell markers such as CD133, Sox2, and nestin, (ii) the ability to self-renew as shown by secondary neurosphere formation, (iii) the ability to undergo limited multipotent differentiation, and (iv) successful serial implantation of xenografts.13,17,19 Importantly, significantly higher expression of CD133 has been reported in recurrent glioblastoma multiforme (GBM) tissue than in respective newly diagnosed tumors,20 and expression has been associated with a poor prognosis.18 Furthermore, CD133-positive cells have been shown to be more resistant to radiation and standard chemotherapy compared with CD133-negative cells.8,20,21 Therefore, cancer stem cells (CSCs) could be the source of tumor resistance and subsequent recurrence. However, such a hypothesis requires a thorough understanding of the cellular mechanisms that sustain CSC growth and resistance in brain tumors.
Generating cell lines from pediatric brain tumors is difficult and often is limited by the size of the tumor tissue available and the ability of dissociated cells to grow under cell culture conditions. This has resulted in a dearth of appropriate model cell lines in which to investigate prognostic targets and develop potential therapies. For several tumor types, such as ependymoma, there are no “commercially” available cell lines. Others are more readily available, but their relationship with the tumor of origin has not been studied, and neither has their retention of CSCs and degree of tumorigenicity. In this paper, CSCs from 7 newly established primary pediatric cell lines were characterized. The percentages of CSCs in all cell lines and the growth characteristics of neurospheres enriched with CSCs were determined. The potential for differentiation was assessed, and xenograft models were used to establish tumorigenicity. Furthermore, the resistance of neurosphere-derived cells to etoposide, an important chemotherapeutic agent, was determined, and mechanisms that may contribute to this resistance were investigated.
Material and Methods
Cell Culture
Brain tumor cell lines were derived at the Children's Brain Tumour Research Centre (CBTRC) (nMED1, nMED2, nEPN1, nEPN2, nOLIG1, and ncPNET1) and at the University of Birmingham (bGB1) as approved by the Local Research Ethics Committee. The tumors of origin were graded and classified by a pathologist (J.L.) according to WHO criteria (Supplementary Material, Table S1 and Fig. S1). Blood clots and vessels were removed from tumor specimens, and samples were washed with Hank's balanced salt solution (HBSS) and finely minced. Birmingham samples were then incubated in Dulbecco's modified Eagle's medium (DMEM)/F12/l-glutamine medium with 15 mM Hepes (Invitrogen) and 15% fetal bovine serum (FBS; Invitrogen). Samples derived at the CBTRC were further incubated with 1× enzyme solution22 in HBSS (0.2 mg/mL DNase type 1 [2000 U/mg], Worthington; 0.083 mg/mL neutral protease [dispase; 1 U/mg], Worthington; 0.4 mg/mL collagenase type 1a [125 U/mg], Worthington; 0.1 mg/mL hyaluronidase, 10 mL Worthington; HBSS [calcium magnesium free], Invitrogen). Samples were rotated at 37°C at 70 rpm for 60 minutes and passed through a 40-µm cell strainer, and dissociated cells were centrifuged at 800 rpm (180×g) for 5 minutes at room temperature, resuspended in 6 mL of tumor media, in a 25-cm2 tissue culture flask. Monolayers were grown in tumor medium: DMEM/l-glutamine (Sigma) supplemented with 15% FBS harvested using trypsin and EDTA (Invitrogen) and split (1:20) every 3–4 days into fresh medium. NTERA-2, PFSK-1, U87, and SNB19 cell lines were obtained from American Type Culture Collection and grown as recommended. To generate neurospheres, cells grown as monolayers were washed with HBSS (Sigma) dissociated with trypsin and resuspended into the serum-free stem cell medium: DMEM high glucose (Sigma) and Ham's F-12 solution (Invitrogen) (70/30%), 2% B27 (Invitrogen), 5 ng/mL heparin (Sigma), supplemented with 20 ng/mL human recombinant epidermal growth factor (hrEGF; Invitrogen), and 20 ng/mL human basic recombinant fibroblast growth factor (bFGF; BD Bioscience). To generate secondary neurospheres, primary neurospheres were dissociated into single cells using accutase (PAA Laboratories) or mechanically, passed through a 40-µm strainer and reseeded at a minimum dilution of 1:2 and a maximum dilution of 1:4. All neurospheres used in this study were passaged at least twice as neurospheres before assessment. All cell lines were maintained in standard humidified incubators at 5% CO2.
Differentiation of Tumor Cell Lines
Monolayer or neurosphere-derived single cells were seeded on chamber slides at a concentration of either 1 × 104 or 1 × 102 cells/mL and fixed after 1 or 10 days, respectively. Cells were plated in DMEM high glucose (Sigma) supplemented with 3% fetal calf serum in the absence of hrEGF and bFGF and with or without platelet-derived growth factor alpha (PDGF-α) (10 ng/mL; Sigma). Cells were fixed with 0.4% paraformaldehyde (PFA) for 30 minutes at room temperature and washed in HBSS, then slides were stored at 4°C under HBSS. To test significance, paired t-tests were applied. For staining protocols, see Supplementary Methods.
Neurosphere Immunofluorescence Preparation
For cryostat sectioning, neurospheres were allowed to settle for 30 minutes at room temperature, fixed with 0.4% PFA, and resuspended in 30% sucrose/HBSS. Neurosphere pellets were embedded in optimal cutting temperature compound (BDH Laboratory Supplies, 361603E) and sectioned (Leica CM 1850) at a thickness of 10 µm onto 3-aminopropyltriethoxysilane (VWR International)–coated slides. Slides were stored at −20°C. To cytospin, 50 µL of neurosphere suspensions were loaded into each well and samples centrifuged at 1000 rpm for 2 minutes using the a Cell-Prep cytospin (Centurion Scientific, model 4050). Cells were fixed in 0.4% PFA and processed immediately for immunofluorescence. For staining protocols, see Supplementary Methods.
Single-Nucleotide Polymorphism Array and Data Analysis
Freshly harvested monolayer and neurosphere cells or frozen tumor material were lysed in a buffer (50 mM Tris, pH 8.0, 100 mM EDTA, 1% SDS, 100 mM NaCl, and proteinase K 4 mg/mL), homogenized for 1 minute, and incubated on a thermomixer (Eppendorf) at 37°C, 450 rpm overnight. Standard phenol–chloroform extraction was then carried out. All array experiments were performed by Almac Diagnostics, using the Affymetrix GeneChip® Human Mapping 500 K Array Set. Row data were produced using the Affymetrix software GCOS and GTYPE, and copy number alterations were identified using Copy Number Analyser for GeneChip.23 DNA from each cell line was compared with tumor and where possible (nEPN1 and bGB1) with constitutional DNA. Annotated output files were then imported into the Spotfire Decision Site (www.spotfire.tibco.com) to allow visualization and comparison.
Microsomal Membrane Extraction
Frozen cell pellets were thawed and resuspended in lysis buffer (250 mM sucrose, 10 mM Tris, pH 7.4, 0.2 mM CaCl2, 1× protease inhibitor cocktail [Roche, 10946900]) and homogenized with a 1-mL syringe (25 gage needle). Homogenized cells were incubated on ice for 20 minutes and centrifuged at 300×g, 4°C for 3 minutes. The supernatant was then ultracentrifuged at 100 000×g, 4°C for 1 hour (Optima Max Ultracentrifuge 130 000 rpm, Beckman Coulter). The microsomal membrane pellets were resuspended in cold phosphate buffered saline (PBS). Samples were added to buffer (0.76% Tris, pH 6.8, 15% glycerol, 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 0.05% bromophenol blue, pH 6.8) and incubated at 37°C for 45 minutes prior to SDS–polyacrylamide gel electrophoresis (PAGE).
Clonogenic Assay
Single-cell suspensions were incubated in DMEM only (Sigma) for 30 minutes at 37°C in 5% CO2 to recover membrane integrity. Approximately 5 × 105 cells were added to 5 mL of dimethyl sulfoxide with etoposide at the test concentration (6.25, 12.5, 25, 50, and 100 µM) or to 5 mL of media with 0.1% dimethyl sulfoxide (Sigma) and incubated for 2 hours at 37°C in 5% CO2. Treated cells were then plated in 2 independent 6-well plates (all monolayers at 300 cells/well; neurospheres for nMED1, 500 cells/well; nEPN1, 1000 cells/well; bGB1 and nOLIG1, 2000 cells/well) and supplemented with 3 mL/well of neurosphere or tumor medium. At day 4, neurospheres were replenished with hrEGF and bFGF. After 8 days, monolayers were fixed with 4% PFA and stained with crystal violet, and colonies were counted. For neurospheres, 1 mL of 4% PFA was added, images of each well were obtained (Nikon SMZ1500), and neurospheres ≥100 µm were counted using Adobe Photoshop vCS3. The survival fraction was the number of colonies in the test well as a percentage of the control. To test significance, the generalized linear model was applied using Statistical Package for the Social Sciences (SPSS).
In Vivo Growth of Cell Lines
Cells from semiconfluent monolayers were harvested with 0.025% EDTA and resuspended for in vivo administration at 6 × 105 cells in 5 μL of PBS and injected via a small burr-hole 2 mm anterior to the Bregma and 2 mm lateral of the midline of the skull. The clinical condition of the mice was regularly monitored for any signs of adverse effect for up to 30 days. Pretermination mice were injected intraperitoneally with 150 mg/kg bromodeoxyuridine and 60 mg/kg pimonidazole. Brains were excised, fixed in formalin, and paraffin embedded. The project was run under Home Office project PPL 40/2962, which was awarded in November 2006 (Watson) following local ethical approval and adhered to the guidelines of the UK Coordinating Committee for Cancer Research.
Comet Assay
The comet assay was based on previously described protocols.24,25 Monolayers and neurospheres were dissociated and treated with etoposide at 100 μm for 2 hours and harvested immediately after treatment or 2 and 4 hours after drug removal. A total of 100 randomly selected individual cells per gel from 2 separate slides were analyzed to give a representative result for the population of cells. The percentage of DNA in the comet tail was selected to assess DNA damage, and the mean and SEM were determined. Comet image capture and analysis utilized Komet software (Version 5.5, Andor Technology) and an epifluorescence microscope (Olympus BH2) fitted with an excitation filter of 515–535 nm and a barrier filter of 590 nm, at a magnification of ×400. To test significant differences between early time points of monolayers and neurospheres, an analysis of variance test was applied using SPSS. To test significant differences in the rate of DNA damage over time between monolayers and neurospheres, the generalized linear model was applied using SPSS.
Results and Discussion
Primary Pediatric Brain Tumor Cell Lines Are Stable and Can Form Neurospheres
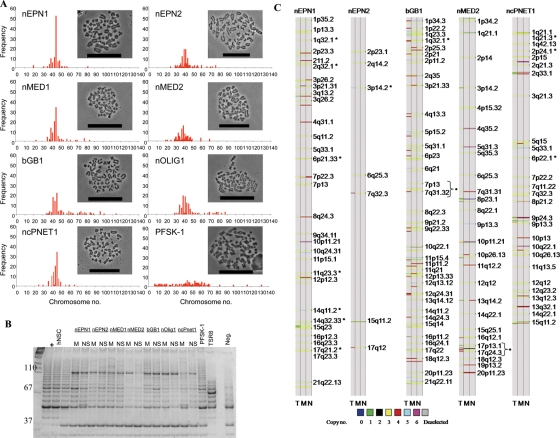
To date, we have successfully established 35 primary pediatric cell lines. Seven of these were selected for a detailed characterization, namely 2 ependymomas (nEPN1 recurrent and nEPN2 primary), 2 medulloblastomas (nMED1 primary and nMED2 recurrent), 1 glioblastoma (bGB1 primary), 1 oligodendroglioma (nOLIG1 primary), and 1 CNS primitive neuroectodermal tumor (ncPNET1 primary). The clinical history for all of the original tumors was obtained (Table 1), and histology was reviewed using standard structural stains and immunohistochemistry hematoxylin/eosin sections showing histological features of each tumor (Supplementary Material, Fig. S1). Monolayers from all cell lines were successfully passaged for more than 60 generations. Neurospheres were serially passaged for up to 11 generations, could be readily derived even at late passage from our primary monolayers, and had retained the relative CSC component (Supplementary Material, Fig. S2), indicating that the conditions used maintain CSCs. In contrast, it was not possible to derive neurospheres from the commercially available sPNET cell line (PFSK-1). Metaphase spreads of each of the cell lines confirmed a near diploid chromosome number in contrast to PFSK-1 (Fig. 1A, Supplementary Methods), suggesting that they are more chromosomally stable.
Table 1.
Clinical details of patients from whom the cell lines were derived
| Cell line | Age (mo) | Diagnosis | WHO grade | P/R | Primary tumor site | Surgical resection status | Treatment | Follow up (mo) | Outcome | Metastatic status |
|---|---|---|---|---|---|---|---|---|---|---|
| nMED1 | 41 | Medulloblastoma | IV | P | Cerebellum | Partial | High-dose cyclophosphamide. UK infant medulloblastoma protocol posterior fossa RT | 8 | Deceased | M2 at relapse |
| nMED2 | P:127 | Medulloblastoma | IV | R | Metastatic to supratentorial compartment (frontal bilateral) | Partial resection (T2) | Chemotherapy and standard craniospinal RT. POG 3021 | 42 | Deceased | M0 |
| bGB1 | 43 | “Giant cell” GBM | IV | P | Cerebrum frontal lobe | Partial | Cisplatin − temozolomide × 6 courses (partial response) repeat surgery − radiotherapy (focal) | 52 | Alive | M0 |
| nOLIG1 | 78 | Oligodendroglioma | II | P | Cerebrum right fronto temporo-parietal | Partial | PCV − progression high-dose Carboplatin + Etoposide. Repeat surgery − Focal radiotherapy (54 Gy) | 24 | Alive | M0 |
| ncPNET1 | 61 | sPNET | IV | P | Cerebrum left frontal | Partial | Headstart chemotherapy. High-dose Carboplatinum + Thiotepa. Local relapse 3 February 2006. Repeat surgery. Craniospinal radiotherapy. Further local relapse 4 mo post-RT | 21 | Deceased | M0 |
| nEPN1 | P:162 3rd R:268 | Ependymoma | II | R | Cerebrum (right parietal) | Complete | Repeat surgery at relapse − (GTR) Focal radiotherapy and chemotherapy. Repeat surgery 18 mo later | 111 | Deceased | M0 |
| nEPN2 | 41 | Ependymoma | II | P | Fourth ventricle | Macroscopic resection | UKCCSG Infant Ependymoma Study | 20 | Alive | M0 |
GBM, glioblastoma multiforme; GTR, gross total resection; P, primary; PCV, procarbazine/CCNU (lomustine)/vincristine; R, recurrent; RT, radiotherapy; UKCCSG, UK Coordinating Committee for Cancer Research.
Fig. 1.
Primary cell lines have retained tumor-specific chromosomal abnormalities and have active telomerase. (A) Chromosome number ranges for our cell lines and PFSK-1. 100 cells were counted for each cell line. Bars represent 50 µm. (B) Telomerase products are shown as incremental 6 bp ladder bands starting from 50 bp. The positive control cell lines used were hNSCs, adult glioma cells (U87), and ncPNET1 cells (PFSK-1), all of which have high telomerase activity. Telomerase activity in monolayers (M) and neurospheres (NS) derived from nEPN1, nEPN2, nMED1, nMED2, bGB1, nOLIG1, and ncPNET1 cell lines was consistently present in 3 individual experiments (a representative figure is shown). Additional controls were the telomerase positive cells (+), TSR8 PCR control template, and the lysis buffer only sample (Neg.). (C) DNA copy number analysis for nEPN1, nEPN2, bGB1, nMED2, ncPNET1, and the corresponding patient's tumors. DNA was extracted from tumor sample (T), monolayers (M), and neurospheres (N). *Aberrant regions commonly seen in that classification of tumor.
In comparison with human neural stem cells (hNSCs), the adult GBM U87 line, and PFSK-1, all of our cell lines demonstrated a high level of telomerase processivity under both growth conditions using the telomere repeat amplification protocol assay (Fig. 1B, Supplementary Methods). Human neural stem cells retain high levels of telomerase activity, whereas commercial cell lines (U87 and PFSK-1) that have been cultured for long periods of time are believed to have reactivated telomerase. Whether our cells have retained or reactivated telomerase was not ascertained.
Chromosomal Abnormalities Present in the Original Tumor Are Retained in Culture
To ensure that the growth conditions used in this study enrich for aberrant cancer cells, single-nucleotide polymorphism (SNP; Affymetrix 500 K arrays) analysis was performed on 5 of the primary pediatric cell lines grown as monolayers or neurospheres (nEPN1, nEPN2, bGB1, nMED2, and ncPNET1). DNA copy number was compared with that detected in primary tumor samples from which they were derived. Aberrant regions in patient tumors were retained in primary cell lines under both culture conditions (Fig. 1C). For the recurrent nEPN1, chromosome copy number gains in regions 1q32.1, 2q32.1, 6p21.33, 11q23.3, 14q11.2, and 17q21.2 have been reported previously.26 The primary ependymoma nEPN2 cells retained gains in regions 2p23.1, 2q14.2, 3p14.2, 6q25.3, 7q32.3, and 17q12, and loss of 15q11.2. Amplification of 3p14.2 has also been reported in ependymoma.26 Copy number aberrations of bGB1 SNP in region 1q32.1 and changes in chromosome 7 have been detected in other glioblastoma tumors.27 For the nMED2 cell line, gain of 1p34.2 and loss of 8p23.1 have recently been reported in medulloblastoma.28 Aberrations of ncPNET1 included 1q21.3, 2p24.1, and 6p22.1, which are also gained regions commonly seen in this tumor type (S.M., unpublished data).
Enrichment of the tumor initiating population and associated genomic alterations in the cultured cells was accompanied by a corresponding decrease in other cell types harboring genomic alterations that are likely to be less important for tumor initiation and maintenance. These cells are most likely to be those that vary in aneuploidy and chromosome stability.29,30
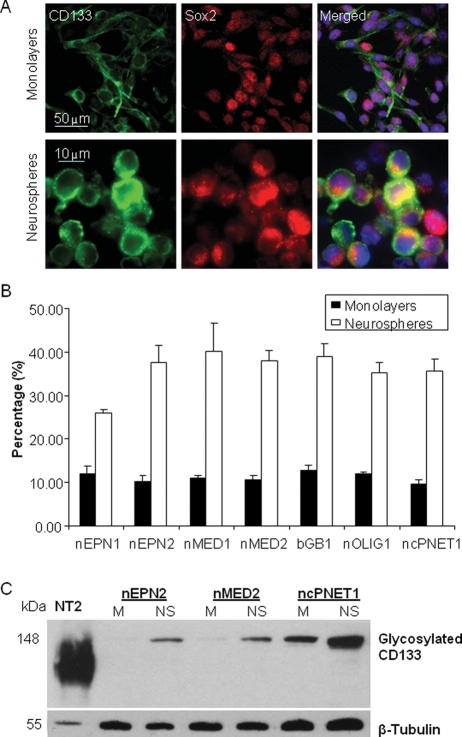
Cells Expressing Stem Cell Markers Are Enriched in Neurosphere Compared with Monolayer Culture
To confirm the presence of CSCs in our cell lines, the percentage of cells coexpressing Sox2 and CD133 was determined using immunofluorescence (Fig. 2 and Supplementary Material, Fig. S2). When grown as neurospheres, costained cells appeared small and round compared with those grown in monolayers (Fig. 2A). A 3–4-fold increase in the percentage of CD133 and Sox2 costained cells was detected in neurospheres (mean 36.3 ± 1.8%) compared with monolayers (mean 10.9 ± 0.5%; P = 1 × 10−5), thus confirming enrichment of CSCs in cultured neurospheres (Fig. 2B).
Fig. 2.
Growing cells as neurospheres enriches for cells expressing membranous CD133 and nuclear/cytosolic Sox2. (A) Monolayers were plated on chamber slides and neurospheres were cryosectioned, and each was stained to detect CD133 (green) and Sox2 (red). Nuclei are stained with DAPI. (B) The percentage of cells costained with CD133 and Sox2. The data represent 3 independent experiments and error bars show the standard error of the mean (SEM). (C) Glycosylated CD133 (140 kDa) expression was detected in membrane fractions prepared from cell lines grown as neurospheres or monolayers. β-Tubulin was used as a loading control. This is a representative blot of 3 independent experiments.
To confirm that neurosphere cultures are enriched for cells expressing membranous CD133, membrane fractions were resolved on SDS-PAGE (Fig. 2C). Significantly, a differential band of approximately 140 kDa, representing glycosylated CD133, was detected at a higher intensity in neurosphere membranes compared with monolayers. In contrast, CD133 protein levels overall did not change between monolayers and neurospheres (Supplementary Material, Fig. S3), confirming that only membranous glycosylated CD133 is a biomarker for CSCs.31
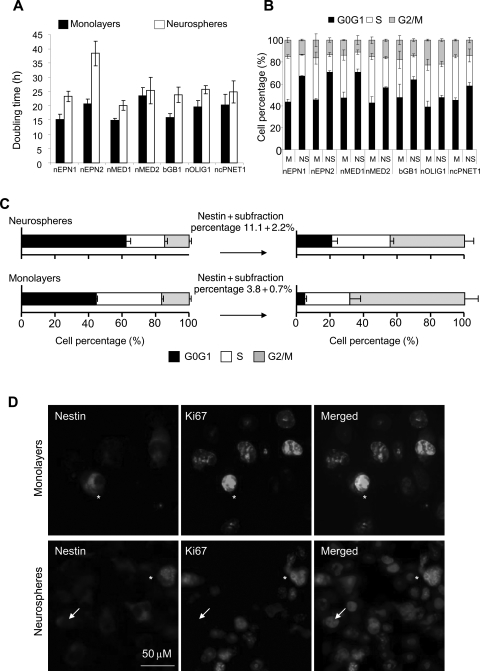
Cells Grown as Neurospheres Have an Altered Cell Cycle Profile
Many chemotherapeutic agents specifically target cycling cells; therefore, understanding the nature of the cell cycle of CSCs is vital. Currently, it is unclear whether CSCs are inherently quiescent31,32 or whether they can alter their cell cycles in a niche-responsive manner.33–35 To investigate this, cell cycle dynamics were studied. The doubling time for cells grown as neurospheres was significantly higher than for their corresponding monolayers (25.9 vs 18.6 hours; P = .008; Fig. 3A). Cell cycle profiles for cell lines were analyzed (Fig. 3B). Growing cells as neurospheres resulted in an increase in the percentage of cells in G0G1 (44% vs 64%; P = .0003). This was accompanied by a significant decrease in cells in S-phase (39% vs 23%; P = .0003) and G2/M (16% vs 15 %; P = .0434).
Fig. 3.
Cells grown as neurospheres have an altered cell cycle profile. (A) The doubling times of monolayer and neurosphere cells grown from all cell lines. (B) Cell cycle profiles of monolayer and neurosphere cells. M, monolayers; N, neurospheres. The data represent 3 independent experiments, and error bars show the SEM. (C) Cell cycle profiles of nestin-expressing cells in monolayers and neurospheres. Cells of equivalent passages were used. These data are representative of 2 independent repeats for each of the cell lines. (D) Representative images of monolayer or neurosphere cells expressing nestin (red) that are either ki67 (green) positive (*) or ki67 negative (→).
To investigate whether the observed 20% increase in G0G1 in neurospheres was due to the enrichment of quiescent CSCs, the cell cycle profile of the nestin-positive CSC component was analyzed. Similar to CD133 immunocytochemistry, a 3-fold increase in nestin-positive cells was detected by flow cytometry in neurospheres compared with monolayers (11.1 ± 2.2 and 3.8 ± 0.7, respectively, P = .0043; Fig. 3C). Nestin-positive cells from neurospheres were detected in all phases of the cell cycle (21% G0G1, 35% S, and 45% G2M; Fig. 3C and Supplementary Material, Fig. S4B). Therefore, nestin-positive cells, representing the CSC component, do not solely account for the accumulation seen in G0G1 of neurosphere cultures. Consistent with these data, immunofluorescent analysis of the nEPN1 and nEPN2 cell lines demonstrated that approximately one-third of nestin-positive cells within neurospheres remained Ki67 positive (nestin+Ki67+ 30.5 ± 9.9 and 25.3 ± 10.4, respectively; Fig. 3D). In contrast, for monolayer-derived nestin-positive cells, the majority of cells (68%) were detected in G2M and almost 100% of cells coexpressed the proliferation marker Ki67, consistent with the previous observations of CD133-positive monolayer cells.33 Since the enrichment of nestin-positive cells in neurospheres coincides with an increase in the G0G1 fraction, it is possible that some nestin-positive cells utilize cellular quiescence in order to preserve their own replicative capacity. The caveat for these data is that nestin positivity may not account for all the CSCs present.36
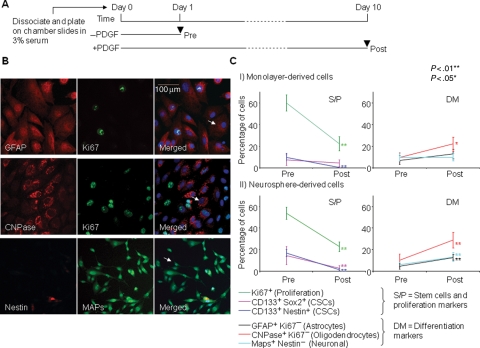
Cells Derived from Neurospheres Have a Higher Capacity for Multilineage Differentiation
An essential property that links brain tumor CSCs and tumor heterogeneity is their capacity, albeit limited, for multilineage differentiation.37 Differentiation of all 7 cell lines in PDGF-α in combination with 3% serum was assessed using double immunofluorescence and cell morphology (Fig. 4). After differentiation, a significantly lower proportion of cells were Ki67 positive (monolayers P = 3e−6, neurospheres P = 8e−5), suggesting a decrease in proliferation. The percentage of cells costaining for stem cell markers had also decreased (monolayers CD133+nestin+ P = .002, neurospheres CD133+nestin+ P= .007, and CD133+Sox2+ P= .004). For monolayers, a significant proportion of cells had differentiated into oligodendrocytes (P= .012), whereas the ability of these cells to differentiate into astrocytes (P= .105) and neurons (P= .733) did not reach significance, suggesting that this was suppressed. However, neurosphere-derived cells were able to differentiate down all 3 lineages (astrocytes, P= .049; oligodendrocytes, P= .001; and neurons, P= .005), indicating that neurospheres enriched with brain CSCs have a higher capacity to undergo multipotent differentiation. Therefore, using PDGF-α resulted in trilineage differentiation of neurosphere-derived, but not monolayer-derived, cells, indicating that neurospheres may be more multipotent. However, in keeping with their tumorigenic potential, the degree of differentiation was limited, as a large proportion of cells continued to divide and many costained with glial fibrillary acidic protein (GFAP) and microtubule-associated proteins (data not shown).
Fig. 4.
Neurosphere-derived cells undergo multipotent differentiation. (A) Schematic diagram of the protocol. (B) Differentiated astrocyte cells stained with GFAP (red) are negative for Ki67 (green) (first column), oligodendrocytes stained with CNPase (red) are negative for Ki67 (green) (second column), and neuronal cells stained with microtubule-associated proteins (MAPs; green) positivity are nestin (red) negative (third column). Arrows on merged images (bottom row) indicate the examples of counted differentiated cells. (C) The percentages of cells counted pre- and postdifferentiation in all cell lines grown as (I) monolayers or (II) neurospheres. CSCs refer to CSCs. Each value plotted represents the mean and SEM. derived from all 7 cell lines, with 3 independent experiments completed for each cell line. Significance values were determined using the paired t-test comparing values for pre- and postdifferentiation. **P < .01 and *P < .05.
Orthotopic Xenografts Result in Clinically Relevant Mouse Models
All of our cell lines rapidly form neurospheres when cultured under appropriate conditions (Supplementary Material, Figs. S1 and S5A), which was not always the case for commercially available cell lines. Similarly, our cell lines were able to rapidly form subcutaneous xenografts (Supplementary Material, Fig. S5B), prompting the suggestion that the rate of neurosphere formation could be an indicative factor for tumor growth potential and that the tumor forming cells within the newly derived lines are different from those present in the long-term established cell lines.13,38
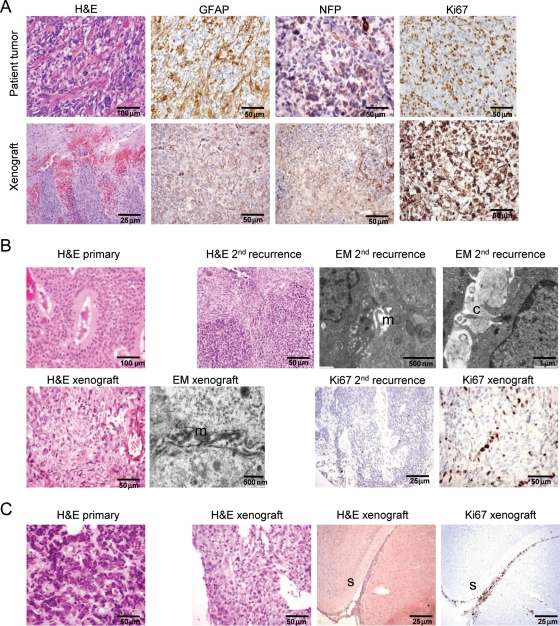
In the light of this enhanced tumorigenicity, orthotopic xenografts of each of the cell lines into immune-compromised mice were then assessed, and it was revealed that these were capable of producing a tumor that reflected the gross immunohistological characteristics of their tumor of origin (Fig. 5). Every injection resulted in a clinically relevant animal model (n= 4 for each tumor, except for n= 3 for nEPN1, where complications resulted from surgery in 1 case).
Fig. 5.
Comparison of xenografts in mouse brain to original patient tumors. (A) Upper panel shows patient tumor from which bGB1 was derived: hematoxylin and eosin (H&E) shows a pleomorphic cellular glial tumor. GFAP and neurofilament protein (NFP) expression are present in a proportion of tumor cells. Ki-67 staining shows a high proliferation rate. Lower panel shows the mouse xenograft tumor derived from bGB1 transplanted cells. H&E shows a cellular glial tumor-infiltrating mouse brain. GFAP expression and NFP expression are seen in a proportion of cells, similar to that in the original tumor. Ki67 staining shows a high proliferation index. (B) Upper panel shows the original patient tumor, which demonstrated cellular and epithelial patterns on H&E. The recurrent tumor, from which nEPN1 was derived 8 years later, shows a spindle cell and cellular pattern with a less well defined tubular pattern. Ultrastructural analysis electron microscopy [EM] confirmed desmosomal junctions with spaces lined by microvilli (m) and cilia (c). In the lower panel, H&E of the mouse xenograft derived from nEPN1 shows a glial-pattern tumor with no obvious ependymal pattern, resembling the spindle cell component seen in the patient sample. EM revealed no desmosomal junctions but did show spaces lined by microvilli (m). The Ki67 labeling index was low in both the recurrent patient sample and the mouse xenograft, consistent with a grade II tumor. (C) H&E of the patient sample shows an embryonal tumor corresponding to a classical medulloblastoma. This was a recurrence that had metastasized in the subarachnoid space to the supratentorial compartment. The transplanted tumor showed features of a primitive lesion with a high apoptotic rate. The tumor spread in the subarachnoid space (s) and had a very high Ki67 proliferation index that was similar to that seen in the original patient sample.
The original tumor from which bGB1 was derived was described as a giant cell GBM based on the presence of anaplasia and multinuclear giant cells. Immunostaining of the xenograft, however, revealed that in addition to GFAP, it was also positive for neurofilament protein (Fig. 5A). Re-examination of the original tumor demonstrated that it was also positive for both glial and neuronal markers (Fig. 5A), suggesting that the diagnosis should be revised to malignant glioneuronal tumor (MGNT). MGNTs have been linked to better survival in children,39 which could explain why this patient is still alive 7 years after diagnosis, even though the tumor was only partially resected. MGNTs have also been linked to metastasis, and a clear invasive front was seen in the orthotopic xenografts. Immunostaining of nOLIG1 confirmed the formation of a glial lineage tumor.
nEPN1 was derived from the second tumor relapse. The original resection of tumor 8 years before and first recurrence 2 years after had shown an epithelial-pattern ependymoma with prominent tubules, grade II. The second recurrence in 2001 from which the xenograft was derived showed cellular ependymoma with areas having a preserved but less well-defined tubular pattern together with areas of the spindle-cell pattern. The histology of the xenograft did not show any distinct pattern and resembled the spindle-cell component of the tumor of origin. Electron microscopy had confirmed ependymal differentiation with desmosomal junctions in the tumor of origin with lumina showing cilia and microvilli. Ultrastructural examination of the xenograft did not show cilia but revealed microvillar surfaces. Ki67 staining (20%–30%; grade II) in the xenograft was similar to the tumor of origin (Fig. 5B). Immunostaining of the nEPN2 xenograft was negative for glial and neuronal markers. Although derived from an original tumor that had clear ependymal features, the xenograft is less well differentiated having lost classical morphological features of ependymoma.
ncPNET1 was derived from a highly aggressive grade IV tumor classed as a supratentorial PNET. The resultant xenografts revealed a primitive tumor with evidence of a high proliferation index and an invasive growth pattern. The 2 medulloblastoma tumors (nMED1 and nMED2) both formed tumors that were positive for neuronal markers and had a high proliferation index, in keeping with their classification as grade IV medulloblastomas. nMED2 was derived from a recurrent medulloblastoma that had metastasized to the supratentorial compartment. Perhaps reflecting a preserved capacity to spread in this manner, clear leptomeningeal spread was seen in the orthotopic xenografts (Fig. 5C).
Neurosphere-Derived Cells Are Significantly More Resistant to Etoposide Compared with Monolayers
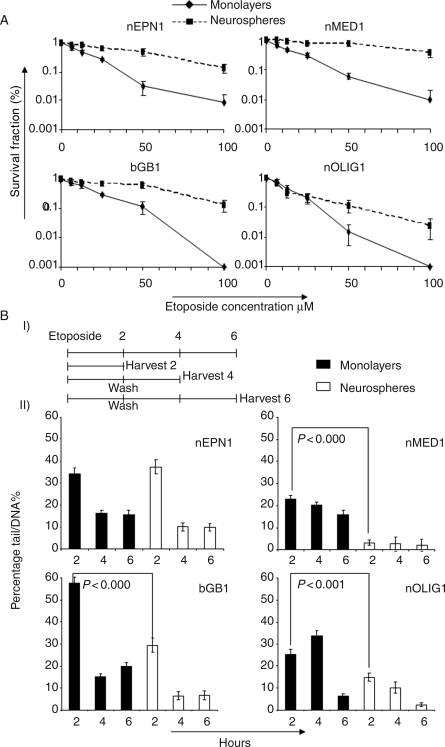
To investigate the drug resistance potential of our cell lines, the clonogenic survival of monolayers and neurospheres to the topoisomerase II poison etoposide was tested. Apart from nOLIG1, neurosphere-derived cells were significantly more resistant to etoposide compared with monolayers (nEPN1 P < .001, bGB1 P < .001, and nMEDl P < .001; Fig. 6A). The half maximal inhibitory concentration values for these cell lines were significantly higher for neurospheres compared with monolayers (50 compared with 12.5 μM for nEPN1, 60 compared with 20 μM for bGB1, and 80 compared with 12 μM for nMEDl). To detect the DNA damage after exposure to 100-μM etoposide, the alkaline comet assay was used (Fig. 6B). Apart from nEPN1, monolayers from all 3 cell lines had an increase in the initial DNA damage after 2 hours compared with neurospheres, with means of 35.3 and 15.8, respectively (P= .03; nEPN1: monolayer 34.4 ± 2.5 and neurospheres 37.5 ± 3.2; nMED1: monolayer 23.1 ± 1.6 and neurospheres 3.2 ± 1.2; bGB1: monolayer 57.8 ± 2.7 and neurospheres 29.3 ± 3.2; nOLIG1: monolayer 25.2 ± 2.3 and neurospheres 14.8 ± 1.7). In addition, the level of DNA damage in nEPN1, bGB1, and nOLIG1 is reduced faster over time in neurospheres compared with monolayers, suggesting that DNA repair is more efficient in neurospheres (nEPN1 P < .001, bGB1 P < .001, and nOLIG1 P < .001). Thus, 2 possible mechanisms are potentially active: (i) the reduction in the initial DNA damage may be due to the efflux of etoposide via ABC multidrug transporters, thus preventing damage (nMED1, bGB1, and nOLIG1); and (ii) the ability of neurospheres to repair their DNA may be due to enhanced DNA repair mechanisms (nEPN1, bGB1, and nOLIG1).
Fig. 6.
Neurospheres have an increased resistance to etoposide compared with monolayers. (A) Clonogenic survival of singularized neurosphere and monolayer cells, from nEPN1, bGB1, nOLIG1, and nMED1, 8 days after 2–3 hours of treatment with etoposide. Points represent the mean of 3 independent experiments with SEM (B) (I) Schedule of treatment for the Comet assay. (II) Comet assays showing DNA tail %. nEPN1, bGB1, and nOLIG1 neurospheres have lower early DNA damage and are faster at DNA repair than monolayers. Representative graphs of 3 independent experiments are shown.
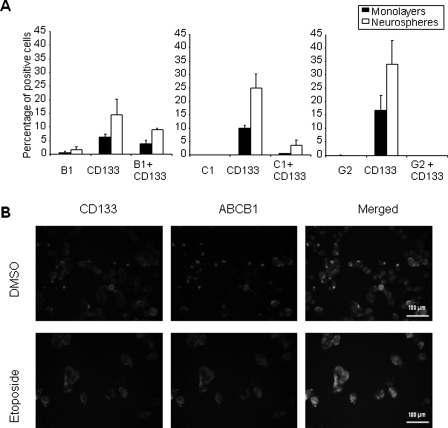
To investigate the role of ATP-binding cassette (ABC) multidrug transporters, cells derived from nMED1 neurospheres, which had demonstrated background levels of DNA damage after exposure to etoposide for 2 hours, were costained for CD133 with ABCB1 or ABCC1 or ABCG2 (Fig. 6BII). Coexpression analysis of CD133 and the 3 main multidrug transporters (ABCB1, ABCC1, and ABCG2), all of which are known to transport etoposide, revealed that ABCB1 and ABCC1 were enriched in CSCs (Fig. 7A). Twice as many CD133 cells expressed ABCB1 (8%) than ABCC1, and immunofluorescent analysis of surviving clones indicated that this expression was further increased by etoposide treatment (Fig. 7B). This supports the hypothesis that ABCB1 may be responsible for etoposide efflux.
Fig. 7.
The medulloblastoma cell line nMED1 shows increased coexpression of CD133 and ABCB1 following etoposide treatment. (A) Endogenous expression levels of CD133 and ABC transporters ABCB1, ABCC1, and ABCG2 as detected by flow cytometry. Standard error and the mean of 3 experiments are shown. (B) CD133 and ABCB1 are coexpressed at a higher level in cells after exposure to etoposide treatment than in vehicle (DMSO)-treated cells. Scale bars represent 50 µm.
Conclusion
Establishing the current hypothesis that only a subpopulation of cells is responsible for driving tumor progression, and relapse is of crucial importance as proof, will have a major impact on the development of new therapeutic regimes for brain tumors.14,19,37,40 In the first part of this study, 7 newly derived pediatric brain tumor cell lines, representative of a broad range of tumor types, were fully characterized. Neurospheres were readily derived from our primary cultures, indicating the presence of CSCs. Comparison with the original tumor demonstrated that these lines not only retained genomic changes present in the original tumor, but also harbored alterations typical of their particular tumor type. These cell lines have retained the CSC component, are able to undergo limited multilineage differentiation, and can rapidly form tumors in vivo. Importantly, comparing neurosphere with monolayer culture demonstrates a dynamic system to model CSC and bulk tumor response to treatments. Our ability to continuously derive CSC-enriched neurospheres from standard monolayer cultures means that we can assay therapeutic response in a robust system that is true to the tumor of origin but also representative of that tumor type. We have demonstrated this in our investigations of DNA damage repair and drug transporter expression. Thus, these cell lines can be used to understand the biology of CSCs in pediatric brain tumors and to screen new therapeutic agents that could assist in developing antitumor therapy based on tumor biology.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online.
Funding
CBTRC and Samantha Dickson Brain Tumor Trust project grant (SDBTT 17/38 to B.C., I.D.K., and R.G.G.). The support of the Emily Hughes Hallet fund and Wellcome Trust (077212/Z/05/Z) enabled microscopy and ultracentrifugation, respectively. W.P. was funded by a Royal Thai Government Studentship.
Acknowledgments
The authors would like to thank Trevor Grey for electron micrographs and Dr Mark Edmonds-Jones for statistical support.
Conflict of interest statement. None declared.
References
- 1.Blaney SM, Kun LE, Hunter J, et al. Tumours of the central nervous system. In: Pizzo PA, Poplack DG, editors. Principles and Practice of Pediatric Oncology. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 786–864. [Google Scholar]
- 2.Arndt V, Lacour B, Steliarova-Foucher E, et al. Up-to-date monitoring of childhood cancer long-term survival in Europe: tumours of the sympathetic nervous system, retinoblastoma, renal and bone tumours, and soft tissue sarcomas. Ann Oncol. 2007;18:1722–1733. doi: 10.1093/annonc/mdm189. doi:10.1093/annonc/mdm189. [DOI] [PubMed] [Google Scholar]
- 3.Abdullah S, Qaddoumi I, Bouffet E. Advances in the management of pediatric central nervous system tumors. Ann N Y Acad Sci. 2008;1138:22–31. doi: 10.1196/annals.1414.005. doi:10.1196/annals.1414.005. [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merchant TE, Fouladi M. Ependymoma: new therapeutic approaches including radiation and chemotherapy. J Neurooncol. 2005;75:287–299. doi: 10.1007/s11060-005-6753-9. doi:10.1007/s11060-005-6753-9. [DOI] [PubMed] [Google Scholar]
- 6.Howell L, Mensah A, Brennan B, Makin G. Detection of recurrence in childhood solid tumors. Cancer. 2005;103:1274–1279. doi: 10.1002/cncr.20896. doi:10.1002/cncr.20896. [DOI] [PubMed] [Google Scholar]
- 7.Messahel B, Ashley S, Saran F, et al. Relapsed intracranial ependymoma in children in the UK: patterns of relapse, survival and therapeutic outcome. Eur J Cancer. 2009;45:1815–1823. doi: 10.1016/j.ejca.2009.03.018. doi:10.1016/j.ejca.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. doi:10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 10.Gilbertson RJ, Hill DA, Hernan R, et al. ERBB1 is amplified and overexpressed in high-grade diffusely infiltrative pediatric brain stem glioma. Clin Cancer Res. 2003;9:3620–3624. [PubMed] [Google Scholar]
- 11.Harris MA, Yang H, Low BE, et al. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68:10051–10059. doi: 10.1158/0008-5472.CAN-08-0786. doi:10.1158/0008-5472.CAN-08-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. doi:10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. doi:10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. doi:10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Tunici P, Bissola L, Lualdi E, et al. Genetic alterations and in vivo tumorigenicity of neurospheres derived from an adult glioblastoma. Mol Cancer. 2004;3:25. doi: 10.1186/1476-4598-3-25. doi:10.1186/1476-4598-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. doi:10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 19.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. doi:10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 20.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. doi:10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133- cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys. 2007;67:1–5. doi: 10.1016/j.ijrobp.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 22.Ali-Osman F. Culture of human normal brain and malignant brain tumors for cellular, molecular, and pharmacological studies. In: Jones GE, editor. Methods in Molecular Medicine: Human Cell Culture Protocols. Totowa, NJ: Humana Press Inc.; 1996. pp. 63–80. [DOI] [PubMed] [Google Scholar]
- 23.Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. doi:10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 24.Godard T, Deslandes E, Sichel F, Poul JM, Gauduchon P. Detection of topoisomerase inhibitor-induced DNA strand breaks and apoptosis by the alkaline comet assay. Mutat Res. 2002;520:47–56. doi: 10.1016/s1383-5718(02)00174-2. [DOI] [PubMed] [Google Scholar]
- 25.Moneef MA, Sherwood BT, Bowman KJ, et al. Measurements using the alkaline comet assay predict bladder cancer cell radiosensitivity. Br J Cancer. 2003;89:2271–2276. doi: 10.1038/sj.bjc.6601333. doi:10.1038/sj.bjc.6601333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. doi:10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 27.TCGA. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. doi:10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubuc AM, Northcott PA, Mack S, Witt H, Pfister S, Taylor MD. The genetics of pediatric brain tumors. Curr Neurol Neurosci Rep. 2010;10:215–223. doi: 10.1007/s11910-010-0103-9. doi:10.1007/s11910-010-0103-9. [DOI] [PubMed] [Google Scholar]
- 30.Weaver BA, Cleveland DW. The role of aneuploidy in promoting and suppressing tumors. J Cell Biol. 2009;185:935–937. doi: 10.1083/jcb.200905098. doi:10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lardon J, Corbeil D, Huttner WB, Ling Z, Bouwens L. Stem cell marker prominin-1/AC133 is expressed in duct cells of the adult human pancreas. Pancreas. 2008;36:e1–e6. doi: 10.1097/mpa.0b013e318149f2dc. doi:10.1097/mpa.0b013e318149f2dc. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. doi:10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 33.Jaksch M, Munera J, Bajpai R, Terskikh A, Oshima RG. Cell cycle-dependent variation of a CD133 epitope in human embryonic stem cell, colon cancer, and melanoma cell lines. Cancer Res. 2008;68:7882–7886. doi: 10.1158/0008-5472.CAN-08-0723. doi:10.1158/0008-5472.CAN-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llaguno SA, Chen J, Kwon CH, Parada LF. Neural and cancer stem cells in tumor suppressor mouse models of malignant astrocytoma. Cold Spring Harb Symp Quant Biol. 2008;73:421–426. doi: 10.1101/sqb.2008.73.005. [DOI] [PubMed] [Google Scholar]
- 35.Viale A, De Franco F, Orleth A, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. doi:10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 36.Piccirillo SG, Combi R, Cajola L, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. doi:10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 37.Sakariassen PO, Immervoll H, Chekenya M. Cancer stem cells as mediators of treatment resistance in brain tumors: status and controversies. Neoplasia. 2007;9:882–892. doi: 10.1593/neo.07658. doi:10.1593/neo.07658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serakinci N, Erzik C. Road for understanding cancer stem cells: model cell lines. Regen Med. 2007;2:957–965. doi: 10.2217/17460751.2.6.957. doi:10.2217/17460751.2.6.957. [DOI] [PubMed] [Google Scholar]
- 39.Varlet P, Soni D, Miquel C, et al. New variants of malignant glioneuronal tumors: a clinicopathological study of 40 cases. Neurosurgery. 2004;55:1377–1391. doi: 10.1227/01.neu.0000143033.36582.40. discussion 1391–1392 doi:10.1227/01.NEU.0000143033.36582.40. [DOI] [PubMed] [Google Scholar]
- 40.Dirks PB. Brain tumor stem cells: bringing order to the chaos of brain cancer. J Clin Oncol. 2008;26:2916–2924. doi: 10.1200/JCO.2008.17.6792. doi:10.1200/JCO.2008.17.6792. [DOI] [PubMed] [Google Scholar]