Abstract
The majority of meningiomas are probably benign but a number of tumors display considerable histological and/or clinical aggressivity, sometimes with unexpectedly high recurrence rates after radical removal. Understanding the potential behavior of these tumors in individual patients is critical for rational therapeutic decision-making. This study aimed to identify gene expression profiles and candidate markers associated with original and recurrent meningiomas. Unsupervised hierarchical clustering of the samples confirmed 2 main groups of meningiomas with distinct clinical behaviors. The gene expression profiling study identified genes and pathways potentially associated with meningioma recurrence, revealing an overall lower level of gene expression. The differential gene expression profiling analyses of original and recurrent meningiomas identified 425 known genes and expressed sequence tags related to meningioma recurrence, with SFRP1 (8p12), TMEM30B (14q23), and CTGF (6q23) showing the most disparate expression. Most of the differentially expressed genes were located at 1p, 6q, and 14q and were underexpressed in recurrences. Loss of such chromosomal regions has previously been associated with a higher risk of meningioma recurrence or malignant progression. Thus, at these locations, we propose the existence of novel candidate genes that could be involved in meningioma recurrence. In addition, the overexpression of genes of histone cluster 1 (6p) in recurrent meningiomas is reported here for the first time. Finally, the altered genes related to meningioma recurrence are involved in pathways such as Notch, TGFβ, and Wnt, as described previously, and in other pathways such as cell cycle, oxidative phosphorylation, PPAR, and PDGF, not related before to meningioma recurrence.
Keywords: meningioma, recurrence, expression profiling, candidate genes
Meningiomas are the most common primary brain tumors, accounting for over 34% of all tumors and having an incidence of 6.17 per 100 000 person-years, as reported in a recent population-based study.1 Many small meningiomas go unnoticed during life and are found incidentally in up to 1.4% of people in autopsy series.2
The World Health Organization (WHO) classification of tumors of the nervous system distinguishes between grade I (benign), grade II (atypical), and grade III (anaplastic or malignant) meningiomas.3 About 90% of all meningiomas are slowly growing benign tumors of WHO grade I. Atypical meningiomas constitute about 6%–8% of cases, although using more current definitions, it has been reported in up to 20%. These WHO grade II meningiomas are histologically defined by increased mitotic activity (4 or more mitoses per 10 high-power microscopic fields) and/or at least 3 of the following criteria: increased cellularity, high nucleus/cytoplasm ratio, prominent nucleoli, uninterrupted patternless, or sheet-like growth and necrosis. Approximately 2%–3% of all meningiomas show histological features of frank malignancy, including a high level of mitotic activity (20 or more mitoses per 10 high-power microscopic fields) and/or a histological appearance similar to sarcoma, carcinoma, or melanoma.3
Tumor recurrence is the major clinical complication in meningiomas, occurring in 10%–15% and 25%–37% of patients undergoing curative surgery after 5- and 10-year follow-up periods, respectively.4 Most meningiomas can be totally resected and many studies have highlighted the extent of resection as a powerful predictor of recurrence.4–7 However, among other disease features, such as patient age, tumor location, extent of tumor resection, and proliferation-associated markers, tumor histopathology has long been known to be a powerful independent prognostic factor of recurrence-free survival.4,8 Accordingly, although only a small proportion of all histologically benign WHO grade I meningiomas relapse, most recurrences of atypical and anaplastic tumors happen in the early years following complete surgical resection.4,5 Despite this, in absolute numbers, the majority of the relapses correspond to histologically benign meningiomas, even after apparently radical removal.4 Therefore, the prediction of relapse occurrence in meningiomas during the first few years following diagnostic surgery still remains a major challenge.
Recent studies have described genetic alterations involved in the progression of meningiomas.9 Other authors have studied recurrences using fluorescence in situ hybridization (FISH),4 but to date, very few studies of the genetic alterations involved in the recurrence of meningioma have been published. Likewise, little is known about the changes at the transcript level that are associated with meningioma relapse. To understand the molecular basis of meningioma recurrence better, we performed microarray-based expression profiling of meningiomas of different malignancy grades, including relapses.
The purpose of this study was to identify the novel candidate genes and pathways associated with meningioma recurrence as well as gene expression patterns that might be helpful for distinguishing meningiomas with a greater probability of recurrence.
Materials and Methods
Patients and Tissue Samples
Samples were collected between 1999 and 2008 from 3 Spanish hospitals: Virgen de la Salud Hospital (Toledo), Xeral-Cies Hospital Complex (Vigo), and Clinic Hospital (Barcelona). The investigation was approved by the Institutional Review Board of the Virgen de la Salud Hospital.
All samples were reviewed by means of tissue sections stained with hematoxylin and eosin (H&E) to verify tumor viability and confirm the diagnosis according to WHO guidelines3 by 4 of the authors (M.M., C.F., T.R., and G.P.-B.).
A total of 112 tumor samples (76 benign WHO grade I, 31 atypical WHO grade II, and 5 anaplastic WHO grade III) from 104 patients were included in this study. Sixteen of these patients relapsed after an average of 48.36 months (range: 17.77–85.43 months) following surgery. Criteria for confirming a recurrent tumor were applied in patients with radically resected tumors (Simpson grade 1, 2, and 3) and who had no evidence of residual tumor in postoperative-enhanced MRIs at least 3 months after surgery, and who presented a clearly growing enhanced mass in control-enhanced MRIs.6 Thirty-two patients were male and 72 were female; the mean age at operation was 60 years (range: 10–88 years). Detailed clinical information on the patients is summarized in Table 1 and Supplementary Table S1.
Table 1.
Summary of clinical information of patients
| WHO grade I | WHO grade II | WHO grade III | |
|---|---|---|---|
| Mean age (range; yrs) | 59.2 (16–88) | 64.88 (32–87) | 47 (10–69) |
| Females/males | 51/22 | 20/7 | 1/3 |
| Location | |||
| Convexity | 36 | 17 | 3 |
| Skull base | 31 | 7 | 2 |
| Falx | 3 | 3 | – |
| Intraventricular | 1 | – | – |
| Spinal | 2 | – | – |
| Gross total resection (%) | 56 (77%) | 23 (85%) | 3 (75%) |
| Relapsed original tumors | 7 | 6 | 3 |
| RFS (mos) | 53.78 | 47.35 | 31.97 |
| Malignant progression | 0 | 1 | 0 |
| Total | 73 | 27 | 4 |
Abbreviation: RFS, Recurrence-free survival.
To carry out the expression profiling study, 44 cases with available frozen tissue and RNA of sufficient quality (see below) were used, including 36 cases of original meningiomas (21 benign WHO grade I, 12 atypical WHO grade II, and 3 anaplastic WHO grade III meningiomas) and 8 recurrences (2 benign WHO grade I, 5 atypical WHO grade II, and 1 anaplastic WHO grade III meningiomas). The quantitative reverse transcription–PCR (qRT–PCR) analyses involved 47 meningiomas, most of which (40 samples) were used for the molecular profiling analysis. Tissue microarrays (TMAs) were constructed using 105 samples obtained from the paraffin-embedded tissue blocks, including 74 benign WHO grade I, 28 atypical grade II, and 3 anaplastic grade III cases. Nine of these samples were tumors collected at recurrence (Supplementary Table S1). For TMAs, samples of normal brain, normal meningothelial tissue, 1 tonsil, and 1 mesothelioma were used as controls.
RNAs from normal meningothelial tissues were used as controls: 1 RNA was obtained commercially from Clontech; and 3 other RNAs were extracted from normal meninges of adult autopsies of patients who had died from diseases unrelated to cancer.
Microarray Expression Assays
The oligomicroarrays used in this study (Agilent Technologies) contained 44 290 DNA clones covering the whole-human genome.
Total RNA was extracted from the frozen tissue using Trizol (Invitrogen), following the manufacturer's protocol. RNA quality was checked by electrophoresis and quantified with NanoDrop-1000 (NanoDrop Technologies Inc.). Fluorescent complementary RNA (cRNA) synthesis was carried out from 1 µg of total RNA from each sample using the Low RNA Input Linear Amplification Kit (Agilent Technologies). Complementary DNA (cDNA) was synthesized using MMLV-RT enzyme, and cRNA was synthesized using T7 RNA polymerase, which simultaneously incorporates cyanine 3- or cyanine 5-labeled CTP. A universal human reference RNA (Stratagene), composed of total RNA from 10 human cell lines, was labeled with cyanine 3 and tumoral or normal meningothelial tissue samples were labeled with cyanine 5. Amplified and labeled samples were purified with Qiagen's RNeasy mini-spin columns and quantified with NanoDrop-1000. Hybridization was carried out overnight at 65°C using 0.75 µg of labeled cDNA. Slides were washed with Gene Expression Wash Buffer (Agilent Technologies) following the manufacturer's instructions. After washing and drying, slides were scanned using an Axon GenePix 4100A microarray scanner (Axon Instruments Inc.). Images were analyzed with the GenePix Pro 6.0 program (Axon Instruments). Data sets for spots not recognized by the software were excluded from further consideration.
Microarray Data Analysis
Data were normalized by the print-tip loess method and background subtraction with the Diagnosis and Normalization Array Data (DNMAD) tool (Gene Expression Pattern Analysis Suite [GEPAS], version 4.0, CIPF; http://gepas.bioinfo.cipf.es/). Because of the presence in the microarray of probe replicates, and since each gene is represented by more than 1 probe, replicates of the probes representing the same gene were averaged. Then, as some of the samples did not have valid expression data, only those genes with expression data in more than 70% of the samples were used. Preprocessing was performed using the GEPAS. After normalization and preprocessing the data, 13 591 genes for each patient sample were used for subsequent analysis.
Unsupervised hierarchical cluster was carried out to classify samples normalized with respect to the median expression of tumoral samples using the UPGMA algorithm method and assuming Euclidean distances. Initially, this analysis included the 13 591 probe set obtained from the normalization analysis, but it was also done by filtering the data to limit the probe set to those that had an SD >1 and >0.5, resulting in probe sets of 1563 and 11 096 genes, respectively.
We used t-tests to identify genes differentially expressed between groups. Genes shown to have significantly different relative expression in the 2 groups, with a false discovery rate (FDR) of <0.20, were selected for further analysis.
Pathways related to the genes differentially expressed in the original and the recurrent meningiomas were identified using Get Set Enrichment Analysis (GSEA), version 2.0 (Broad Institute). Given an a priori set of genes, S, the goal of GSEA is to determine whether the members of S are randomly distributed throughout a ranked list of genes obtained from microarray analyses, or if they are preferentially found at the top or the bottom of the list.10 The gene set of the molecular pathways was obtained from the Molecular Signatures Database (MsigDB; http://www.broadinstitute.org/gsea/msigdb). Gene sets used for GSEA analyses are shown in Supplementary Table S2.
Immunohistochemistry
TMAs were constructed using formalin-fixed, paraffin-embedded archival tissue blocks as described previously.11 H&E-stained full sections from each donor block were used as the morphological selections of the representative areas of each tumor.
For immunohistochemical staining, 4-μm-thick paraffin-embedded tissue sections were subsequently dewaxed, rehydrated, and subjected to antigen retrieval using EnVision FLEX Target Retrieval solution at pH 6 or pH 10, depending on the antibody, and heated at 95°C. The slides were cooled and treated with EnVision FLEX Peroxidase-Blocking Reagent solution (Dako) for 5 minutes. Sections were then immunostained with monoclonal antibody by the Dako EnVision FLEX/HRP Technique (Dako), counterstained with hematoxylin, and mounted. The primary antibodies used were: LMO4 (LifeSpan BioSciences) and HIST1H1C (Sigma-Aldrich). For statistical analyses of data from LMO4 and HIST1H1C, tumors were classified as positive (nuclear staining in >5% of cells) or negative (nuclear staining in ≤5% of cells).
Real-Time qRT–PCR Analysis
To verify the selected microarray data, the expression levels of 4 genes (TMEM30B, SFRP1, CTGF, and TGFBR3) were determined using the ABI PRISM 7500 Real-Time PCR System (Applied Biosystems). Primers, probes of the targets, and internal controls (GAPDH and TBP) were purchased from Applied Biosystems (SFRP1, Hs00610060_m1; TMEM30B, Hs01089489_s1; CTGF, Hs00170014_m1; TGFBR3, Hs0114253_m1; GAPDH, 4333764T; and TBP, Hs00427620_m1).
RNA was reverse transcribed by adding 1 μg of total RNA, 100 pmol random primers, 10 mM deoxyribonucleoside triphosphate, 40 U RNAsin, and 200 U Superscript II reverse transcriptase (Invitrogen), in a volume of 22 μL. A singleplex reaction mixture with the corresponding Taqman probes was prepared according to the manufacturer's recommendations. All determinations were performed in triplicate, and the expression levels for each gene were normalized with respect to the endogenous controls TBP and GAPDH. We used duplicated samples from a pool of 4 different non-neoplastic meningothelial tissues as reference tissues. Results were normalized and analyzed by the comparative Ct method 2, performed with the Sequence-Detector 7500 system program (version 1.2.3f2; Applied Biosystems).
Fluorescence In Situ Hybridization
FISH assays were performed on paraffin-embedded tumor TMAs using the LSI 1p36/1q25 dual color probe set purchased from Vysis Inc., as described previously.12,13 Fluorescence signals were scored and for each sample, approximately 130 well-defined nuclei (mean: 134 cells; range: 102–226 cells) were analyzed. Tumors were considered to have 1p loss when an imbalance of 1p/1q (1 vs 2 signals) was identified in more than 25% of tumor cells.
Statistical Analyses
Statistical analyses were carried out with SPSS v17.0 (SPSS). The chi-squared test was used to assess the statistical significance of the differences observed between groups. Survival curves were plotted according to the Kaplan–Meier method, and the log-rank test was used to establish the statistical significance of the differences between curves. Differences of recurrence-free survival based on 1p loss were analyzed. The threshold for a statistical significance in all cases was taken to be P < .05.
Results
Clustering of Samples
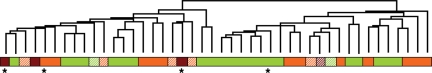
Unsupervised hierarchical clustering of all 44 samples analyzed for expression profiling, using the expression values of the 13 591 filtered genes and several clustering methods, classified the tumors into 2 main groups. A representative hierarchical cluster of the samples is shown in Fig. 1. Although the relationship between the samples varied slightly with the clustering method used, 2 basic groupings always emerged. There was a more aggressive group, which consisted of most recurrences (5 samples: 1 and 4 WHO grade I and II, respectively), all 3 anaplastic tumors, 9 WHO grade I tumors, and 5 WHO grade II meningiomas. Remarkably, 3 meningioma samples from this aggressive group correspond to those collected at the diagnosis of tumors that relapsed later. In addition, of the 9 benign meningiomas included in this aggressive group, 5 tumors had high proliferation indices, 1p loss, or tumor regrowth. The other group consisted of most of the WHO grade I (12) and II (7) tumors, whereas only 3 recurrences and 1 original meningioma that had relapsed were included.
Fig. 1.
Unsupervised hierarchical cluster analysis of 44 meningiomas corresponding to all WHO grades and including 8 recurrences. Meningiomas of WHO grade I, II, and III are shown in green, orange and dark red, respectively. Recurrences are indicated with striped bars. Asterisks denote samples collected at diagnosis from original tumors that relapsed.
Expression Profiles of Recurrent Meningiomas
In order to address the question of recurrence in meningiomas, we intended to compare matched samples of recurrent tumors collected at diagnosis and at recurrence. However, as no frozen tissue was available from these matched tumors, we compared the expression profile of 4 samples collected at diagnosis from original tumors that recurred later (1, 1, and 2 WHO grades I, II, and III, respectively), and 8 samples collected at recurrence from different recurrent tumors (2, 5, and 1 WHO grades I, II, and III, respectively). Strikingly, no differentially expressed genes (DEGs) were identified (FDR ≥ 0.9). Thus, expression profiles of original tumors that recurred later and the recurrent tumors were highly similar; and although there are few samples included in this statistical analysis, these results suggest that there are no differences at the gene expression level between the sample at diagnosis of the tumor that recurs later and the sample at recurrence.
Differential Expression Analysis in Original Meningiomas vs Recurrences
In order to search for differences in gene expression between the original and the recurrent meningiomas independently of the malignancy grade, we compared 33 original (samples at diagnosis) and 7 recurrent (samples at recurrence) meningiomas of WHO grades I and II, thus excluding any WHO grade III tumors. However, meningiomas are slow-growing tumors, so the possibility of the recurrence of tumors considered as original cannot be entirely excluded and therefore should be taken into account. This t-test analysis identified 425 genes and expressed sequence tags (ESTs) that were significantly differentially expressed (FDR < 0.20) in original and recurrent tumors. Among these DEGs, 127 had a more than 2-fold difference in expression with 81.9% (104 of 127) and 18.1% (23 of 127) under- and overexpressed, respectively, in recurrences relative to original tumors. The complete list of DEGs is shown in Supplementary Table S3. The 3 genes with the greatest differences of expression (SFRP1, TMEM30B, and CTGF) were downregulated in recurrences compared with original meningiomas. These genes are related to the Wnt signaling pathway (SFRP1), cell cycle (TMEM30B), and cell growth (CTGF).
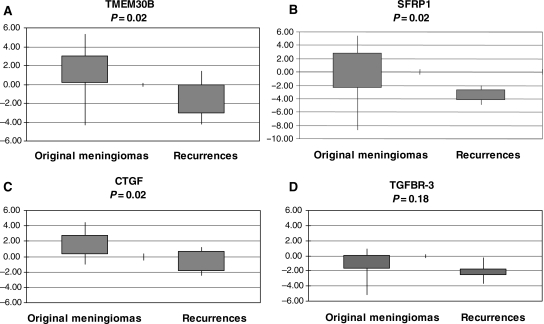
Four DEGs between original and recurrent meningiomas that were downregulated in recurrences were selected for validation by a real-time PCR: SFRP1, TMEM30B, CTGF, and TGFBR3, the first 3 of which had the greatest differences of expression. These assays confirmed the microarray expression data for SFRP1, TMEM30B, and CTGF, showing significantly lower mRNA levels in recurrences than in original meningiomas (P < .05). In addition, lower levels of TGFBR3 expression in recurrent compared with original meningiomas were observed, but the differences in the means were not statistically significant (P = .18; Fig. 2).
Fig. 2.
Validation of selected genes differentially expressed in original and recurrent meningioma by qRT–PCR analysis. (A) TMEM30B, (B) SFRP1, (C) CTGF, and (D) TGFBR3. Mean expression values related to the internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or TATA box binding protein (TBP) and to the mean of nontumoral meningothelial tissue pools are plotted. Box plots indicate the expression values of the 25th and 75th percentiles, and the extremes of the vertical lines represent the maximum and the minimum log2 expression. P < .05 for all genes except for TGFBR3.
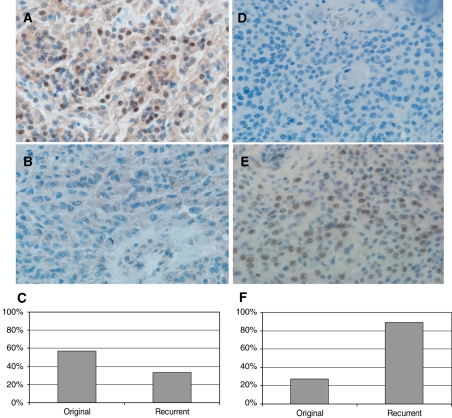
We also used immunohistochemical assays to validate expression data of 2 other genes, 1 underexpressed (LMO4) and the other overexpressed (HIST1H1C), and in recurrent meningiomas relative to original tumors. These analyses revealed an absence of expression of LMO4 in 32% (28 of 87) and 50% (4 of 8) of the original and recurrent samples, respectively (Fig. 3). As the immunohistochemical expression of HIST1H1C was focal, only those data obtained from complete sections were considered. Accordingly, 27% (3 of 11) and 89% (8 of 9) of original and recurrent tumors, respectively, presented positive HIST1H1C staining (Fig. 3).
Fig. 3.
Immunohistochemistry results of 2 genes differentially expressed in original and recurrent tumors. Photomicrograph of meningioma samples with (A) positive and (B) negative expression of LMO4 (original magnification, ×400). (C) Percentage of cases with LMO4-positive expression. Photomicrograph of tumor tissue with (D) negative and (E) positive expression of HIST1H1C (original magnification, ×400). (F) Percentage of cases with HIST1H1C-positive expression.
Deregulated Pathways Distinguishing Original from Recurrent Meningiomas
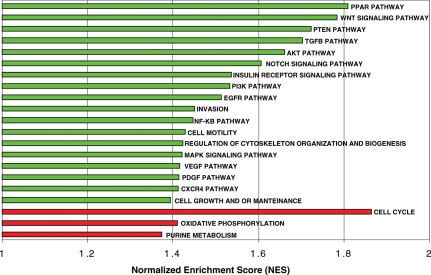
GSEA was performed to discover the molecular pathways involved in the development of meningioma recurrence. It detected 21 significant gene sets (FDR < 0.15) enriched in recurrent meningiomas compared with original tumors. Three of these were upregulated and 18 were downregulated in recurrent tumors (Fig. 4).
Fig. 4.
Molecularly altered pathways related to meningioma recurrence ordered by the normalized enrichment score. This value is obtained by normalizing the enrichment score that reflects the degree to which a set of genes, S, is over-represented at the extremes (top or bottom) of an entire ranked list, L. Molecular pathways under- or overexpressed in green or red, respectively, in recurrences.
Among the enriched molecular pathways downregulated in recurrences, Notch, TGFβ, Wnt, and insulin receptor signaling pathways have previously been associated with meningioma progression.9,14–16 Importantly, new gene sets were found to be downregulated in recurrences relative to original tumors: cell growth and maintenance, cell motility, regulation of actin cytoskeleton, and the PTEN, EGFR, AKT, and PPAR signaling pathways. Remarkably, only cell cycle, purine metabolism, and oxidative phosphorylation were upregulated in meningioma recurrence.
Chromosomal Locations of Recurrence-Associated DEGs
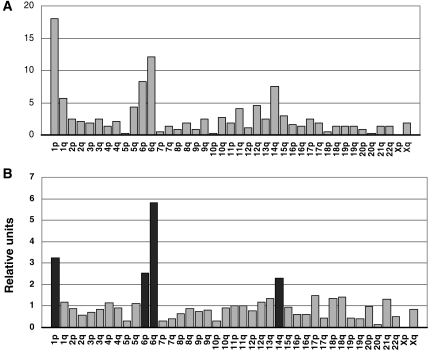
Table 2 shows the most DEGs located in the most important chromosomal locations. Analysis of the chromosomal location of the genes differentially expressed in original and recurrent tumors revealed that the most frequent locations were 1p (18.0%), 6q (12.1%), 6p (8.3%), 14q (7.5%), and 1q (5.6%) (Fig. 5A; Table 3). The locations of the DEGs were then compared with those of all the genes represented in the oligomicroarray. The percentage of DEGs localized on chromosomes 1p, 6q, 6p, and 14q was significantly higher than the percentage of all the analyzed probes of the microarray at these locations (P < .001; Fig. 5B; Table 3). Examining this in greater detail, analysis of the under- and overexpressed genes in recurrent tumors revealed that most of the genes located in 1p (67 of 67), 6q (44 of 45), and 14q (28 of 28) had a lower level of expression in recurrences than in original tumors, whereas most of the genes located in 1q (16 of 21) and 6p (22 of 31) were more strongly expressed (Table 3; Supplementary Fig. S1).
Table 2.
DEGs in original and recurrent meningiomas mapping at frequent significant chromosomal locations
| Gene symbol | Gene description | R vs O* | R** | O** |
|---|---|---|---|---|
| 1p | ||||
| PPAP2B | Phosphatidic acid phosphatase type 2B | −3.6 | −3.3 | −1.4 |
| LMO4 | LIM domain only 4 | −3.6 | n | 1.8 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | −3.0 | −1.8 | n |
| EXTL2 | Exostoses (multiple)-like 2 | −2.5 | n | 0.9 |
| F3 | Coagulation factor III | −2.5 | −0.8 | 0.6 |
| DDAH1 | Dimethylarginine dimethylaminohydrolase 1 | −2.5 | −1.4 | n |
| PGM1 | Phosphoglucomutase 1 | −2.4 | −0.8 | n |
| DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | −2.4 | −1.3 | n |
| DHRS3 | Dehydrogenase/reductase member 3 | −2.3 | −1 | n |
| BCAR3 | Breast cancer antiestrogen resistance 3 | −2.3 | −1.3 | n |
| TMEM50A | Transmembrane protein 50A | −2.3 | −1.7 | n |
| TGFBR3 | Transforming growth factor, β receptor III | −2.1 | −1.9 | −0.8 |
| PNRC2 | Proline-rich nuclear receptor coactivator 2 | −2.1 | −1.2 | n |
| CMPK | Cytidylate kinase | −2.0 | −1.4 | n |
| MKNK1 | MAP kinase interacting serine/threonine kinase 1 | −2.0 | −1.2 | n |
| DPYD | Dihydropyrimidine dehydrogenase | −2.0 | −1.7 | −0.7 |
| MAN1C1 | Mannosidase, alpha, class 1C, member 1 | −2.0 | −0.8 | n |
| LPHN2 | Latrophilin 2 | −1.9 | −1.5 | n |
| SFRS4 | Splicing factor, arginine/serine-rich 4 | −1.9 | −1.8 | −0.8 |
| RPS8 | Ribosomal protein S8 | −1.9 | −1.2 | n |
| IPP | Intracisternal A particle-promoted polypeptide | −1.8 | n | 0.6 |
| HMGN2 | High-mobility group nucleosomal binding domain 2 | −1.8 | −1.7 | −0.9 |
| GSTM1 | Glutathione S-transferase M1 | −1.8 | n | 0.6 |
| GADD45A | Growth arrest and DNA-damage-inducible, alpha | −1.8 | −1.3 | n |
| CYP4X1 | Cytochrome P450, family 4, subfamily X, polypeptide 1 | 2.4 | 0.8 | 2.1 |
| 6p | ||||
| HIST1H1C | Histone cluster 1, H1c | 5.1 | 2.1 | n |
| PRPH2 | Peripherin 2 | 3.2 | n | 1.7 |
| HIST1H1E | Histone cluster 1, H1e | 3.0 | 1.2 | n |
| HIST1H1D | Histone cluster 1, H1d | 2.5 | 1.5 | n |
| HIST1H2AD | Histone cluster 1, H2ad | 2.2 | n | −1 |
| HIST1H2BH | Histone cluster 1, H2bh | 2.2 | 1.1 | n |
| HIST1H2BO | Histone cluster 1, H2bo | 2.1 | 1.1 | n |
| HIST1H2BN | Histone cluster 1, H2bn | 2.0 | 1 | n |
| HIST1H2BF | Histone cluster 1, H2bf | 2.0 | 1 | n |
| HIST1H2AE | Histone cluster 1, H2ae | 2.0 | n | −0.6 |
| HIST1H2BI | Histone cluster 1, H2bi | 2.0 | 0.9 | n |
| HIST1H2BL | Histone cluster 1, H2bl | 2.0 | 1 | n |
| HIST1H2BC | Histone cluster 1, H2bc | 1.9 | 1.3 | n |
| HIST1H2BE | Histone cluster 1, H2be | 1.9 | 0.8 | n |
| HIST1H2BG | Histone cluster 1, H2bg | 1.8 | 1.2 | n |
| HIST1H2BD | Histone cluster 1, H2bd | 1.8 | 1.5 | 0.7 |
| HIST1H2BM | Histone cluster 1, H2bm | 1.8 | 1 | n |
| GMDS | GDP-mannose 4,6-dehydratase | −1.9 | n | 0.9 |
| RAB23 | RAB23, member RAS oncogene family | −2.2 | n | 0.9 |
| HSPA1A | Heat shock 70 kDa protein 1A | −2.8 | −1 | n |
| 6q | ||||
| CTGF | Connective tissue growth factor | −5.5 | −2.3 | n |
| PRSS35 | Protease, serine, 35 | −3.6 | n | 1.8 |
| MAP3K5 | Mitogen-activated protein kinase 5 | −2.6 | −2.4 | −1.1 |
| RNF146 | Ring finger protein 146 | −2.4 | −1.5 | n |
| EPB41L2 | Erythrocyte membrane protein band 4.1-like 2 | −2.3 | −1.1 | n |
| PNRC1 | Proline-rich nuclear receptor coactivator 1 | −2.2 | −1.2 | n |
| SERINC1 | Serine incorporator 1 | −2.2 | −1.7 | −0.6 |
| AKAP7 | A kinase (PRKA) anchor protein 7 | −2.1 | −1.5 | n |
| SESN1 | Sestrin 1 | −2.1 | −1.5 | n |
| CNKSR3 | CNKSR family member 3 | −1.9 | −0.6 | n |
| SNX3 | Sorting nexin 3 | −1.9 | −1.7 | −0.7 |
| TSPYL4 | TSPY-like 4 | −1.9 | −1.1 | n |
| FOXO3A | Forkhead box O3A | −1.9 | n | 0.6 |
| CD164 | CD164 molecule, sialomucin | −1.9 | −0.9 | n |
| DLL1 | Delta-like 1 | −1.8 | −2.3 | −1.5 |
| ME1 | Malic enzyme 1, NADP(+)-dependent, cytosolic | −1.8 | n | 0.6 |
| CCDC28A | Coiled-coil domain containing 28A | −1.8 | −0.7 | n |
| 14q | ||||
| TMEM30B | Transmembrane protein 30B | −5.8 | −2.3 | n |
| SMOC1 | SPARC-related modular calcium binding 1 | −3.0 | −0.6 | 1 |
| LTBP2 | Latent transforming growth factor β binding protein 2 | −2.9 | −1.6 | n |
| KLHDC2 | Kelch domain containing 2 | −2.1 | −0.8 | n |
| SIPA1L1 | Signal-induced proliferation-associated 1 like 1 | −1.9 | −0.8 | n |
| PRKCH | Protein kinase C, eta | −1.8 | −1.9 | −1 |
| TRMT5 | TRM5 tRNA methyltransferase 5 homolog | −1.8 | −1.6 | −0.8 |
| ALDH6A1 | Aldehyde dehydrogenase 6 family, member A1 | 2.2 | n | 1.2 |
Negative and positive values indicate the lower and the higher levels of expression, respectively, in recurrences vs original tumors, recurrences vs nontumoral tissue, or original tumors vs nontumoral tissue. n, expression data similar to that of nontumoral tissue.
*Differences of expression between recurrent (R) and original (O) tumors.
**Log2 mean expression values of recurrent (R) and original (O) meningiomas normalized to nontumoral meningothelial tissues.
Fig. 5.
(A) Diagram of the frequency of locations of genes differentially expressed in original (O) and recurrent (R) meningiomas. (B) The locations of the genes differentially expressed (O vs R) relative to the locations of the genes in the microarray. Black bars indicate the significant and most frequent locations.
Table 3.
Most frequent chromosomal locations of genes differentially expressed in original and recurrent tumors
| Cytoband | Genes in the microarray (%) | DEGs O vs R |
||
|---|---|---|---|---|
| Total (%) | Lower level of expression in R | Higher level of expression in R | ||
| 1p** | 750 (5.56) | 67 (18.01) | 67 | 0 |
| 6q** | 281 (2.08) | 45 (12.10) | 44 | 1 |
| 6p** | 444 (3.29) | 31 (8.33) | 9 | 22 |
| 14q** | 438 (3.25) | 28 (7.53) | 28 | 0 |
| 1q* | 635 (4.67) | 21 (5.65) | 5 | 16 |
O, original meningioma; R, recurrences. Significant comparisons with the location of the genes included in the microarray are shown. Only 423 genes with known location.
**P < .001; *P < .05.
Comparison of the expression data in recurrences with nontumoral tissue showed overall downregulation of the DEGs, especially concentrated on 1p (79%, 53 of 67), 6q (80%, 36 of 45), and 14q (75%, 21 of 28). Conversely, upregulation of genes was concentrated in the 1q (62%, 13 of 21) and 6p (61%, 19 of 31) regions.
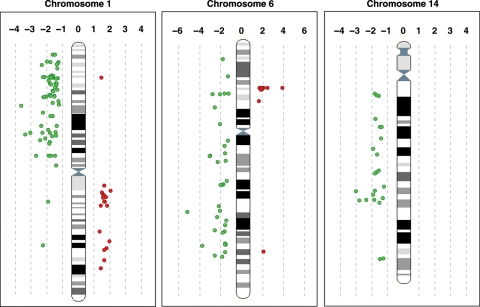
Detailed analysis of the location of the genes differentially expressed in original and recurrent meningiomas revealed them to be distributed homogeneously along the 1p, 1q, 6q, and 14q chromosome arms. However, almost all the 6p genes with stronger expression in recurrences than in original tumors concentrated on 6p22.1, the location of histone cluster 1 (Fig. 6).
Fig. 6.
The location of the genes differentially expressed in original and recurrent meningiomas on chromosomes 1, 6, and 14, plotted according to their map position. Genes with lower (green) and higher (red) levels of expression in recurrences than in original tumors are shown on the left and the right, respectively, of the chromosome ideogram.
Analysis of 1p Loss
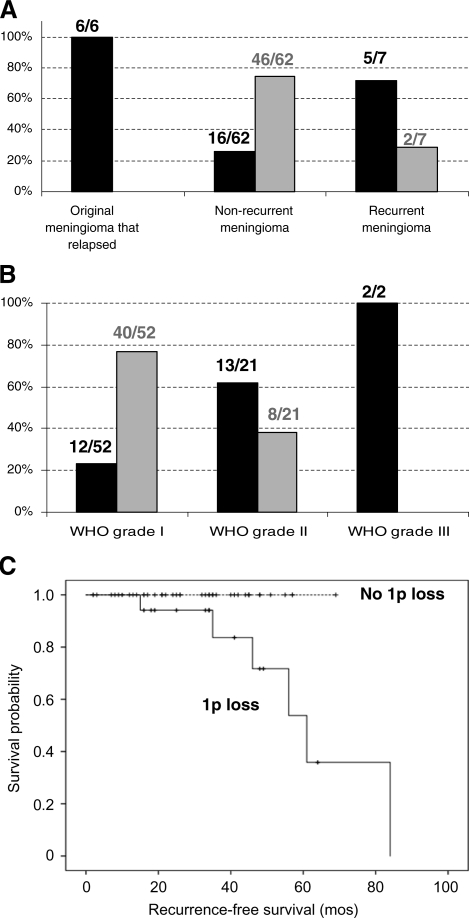
As 1p was the most frequent location of the DEGs (18%), and since all 1p genes showed a lower level of expression in recurrences compared with original tumors, we tested whether underexpression of the genes located in 1p was a consequence of the chromosomal loss of 1p in meningioma recurrence by conducting FISH assays of chromosome 1p on paraffin-embedded tissue sections of the tumors included in the TMA. These assays revealed a loss of chromosome 1p in 5 out of 7 of the recurrent meningiomas (71.4%; Fig. 7). Furthermore, all the original meningiomas that had relapsed (6 of 6) presented 1p loss, whereas only 25.8% (16 of 62) of the meningiomas that had not relapsed showed the loss of this chromosome.
Fig. 7.
(A) Frequency of 1p loss in original meningioma that relapsed, nonrecurrent meningioma, and recurrences. The total numbers of evaluated tumors are shown. (B) Frequency of 1p loss in WHO grades I, II, and III. The total numbers of evaluated tumors are shown. (C) The Kaplan–Meier curves of recurrence-free survival. Curves are shown for the absence and the presence of 1p loss (n = 40 and 19, respectively; P = .020). Six recurrent events are shown.
To examine the possibility that the 1p loss affects recurrence in the patients, we performed the log-rank tests and constructed the Kaplan–Meier curves. The loss of 1p was found to be significantly associated with recurrence (P = .02; Fig. 7).
In order to examine whether downregulation of 1p genes was associated with 1p loss, we performed a clustering analysis using expression data of 1p recurrence-associated genes (Supplementary Fig. S2). This cluster separated meningiomas with 1p loss from those without it, suggesting that in most cases, downregulation of 1p genes is probably due to 1p loss. In spite of this, some nonrecurrent tumors presented downregulation and loss of 1p genes. These tumors, however, were atypical and anaplastic meningioma, and remarkably, the only WHO grade I tumor that was clustered in the 1p loss group was a recurrent meningioma.
Discussion
An unsupervised clustering of the samples using different filtering criteria revealed that meningiomas can be classified into 2 main molecular groups. One of these groups included all the aggressive malignant meningiomas, most of the recurrent samples, and the 3 original tumors that had relapsed. The other molecular group contained benign and atypical meningiomas. These results are in agreement with those of Carvalho et al., who described 2 main molecular groups of meningiomas, low- and high-proliferative, despite the existence of 3 histopathological grades.17
On the other hand, as our cluster analysis demonstrated, atypical meningiomas did not have a unique molecular signature of their own. Instead, the molecular profiles of atypical meningiomas matched those of either benign or malignant meningiomas, in agreement with the results obtained by other authors.9,18,19 This result is in accordance with the wide variability in clinical behavior that atypical meningiomas exhibit, some tumors having growth patterns similar to those of benign meningiomas, and others having poor clinical outcomes paralleling those of malignant meningiomas.20
Surprisingly, 9 benign tumors were consistently included in the more aggressive group despite the algorithm used. A similar result was found by Watson et al. reporting grade I, II, and III meningiomas clustered together.18 In addition, 5 out of these 9 benign tumors had high proliferation indices, 1p loss, or tumor regrowth. These results may suggest that expression profiles reflect the biological heterogeneity of meningiomas, which is not clearly defined by histopathological criteria, as other authors have pointed out.19
Expression Profiles of Recurrent Meningiomas
Although further studies using greater numbers of tumors are necessary, the gene expression profiling analysis of the original tumor that recurs later and the recurrence revealed no differences of gene expression, suggesting that the biological potential of the tumor to recur is already present in the tumoral sample at diagnosis and is not acquired on recurrence.
Comparison of the gene expression profiles of original and recurrent meningiomas revealed a pattern of 425 known genes and ESTs that distinguished both groups of tumors, most of which were downregulated. Determination of the chromosomal location of the DEGs revealed that the most frequently altered genes were located in 1p, 1q, 6p, 6q, and 14q, and that all these locations featured significantly more frequently among the DEGs than did the genes included in the microarray. Furthermore, an enrichment of underexpressed genes on chromosomes 1p, 6q, and 14q, and of overexpressed genes on chromosomes 1q and 6p in recurrent meningiomas was also found among these DEGs. Our results are consistent with those of other studies that relate these chromosomal regions to the malignant progression and recurrence of meningioma.4,21–25
After monosomy 22 and partial 22q deletion, loss of 1p is the next most frequent chromosomal abnormality observed in meningiomas.26–28 In addition, coexistence of monosomy 14 and deletion of 1p36 in the ancestral tumor cell clone were related to early recurrence in benign grade I meningiomas.4 In agreement with these studies, our analyses confirmed the deletion of 1p in most of the recurrent meningiomas and in all the original tumors that had relapsed. However, some nonrecurrent tumors presented 1p loss, as well as downregulation of 1p genes. These tumors were mainly atypical and anaplastic meningiomas which have been previously described to have 1p loss in around 40–75% and 70–100% of the tumors, respectively, as a mechanism of malignant progression.3
To date, however, the actual targets of these chromosomal losses have remained largely elusive and thus, no tumor suppressor genes have been confirmed as being involved in meningioma pathogenesis, progression, or recurrence.29 The study presented here enabled the identification of candidate genes localized in these regions that could be involved in meningioma recurrence. Candidate 1p, 6q, and 14q genes have been evaluated in this study. LMO4 (1p22.3) expression in original meningiomas was strong compared with normal meningothelial tissue, whereas in recurrences it was similar. LMO4 is highly expressed at multiple sites of mesenchymal–epithelial interactions, and while this gene is known to be overexpressed in breast and pancreatic tumors,30,31 its alteration in meningiomas has not previously been reported. A recent report suggested that LMO4 modulates TGFβ signaling through its interaction with receptor-activated Smads, supporting the finding of the downregulation of the TGFβ pathway in recurrent meningiomas.32
Remarkably, a comprehensive DNA copy number study of chromosome 1 identifies 3 candidate 1p lost regions in meningiomas, 2 of them containing 17 DEGs identified in our study by comparing original and recurrent meningiomas27 (Supplementary Table S3). The involvement of these genes in meningioma recurrence requires further analyses.
Loss of chromosome 6q has been associated with the malignant progression of meningioma.33,34 This region contains the candidate genes CTGF, TMEM30A, and SESN1, which are underexpressed in recurrent meningiomas compared with nontumoral tissue. These genes are transcription factors related to apoptosis, cell growth, DNA repair, damage prevention, and cell cycle. Specifically, the connective tissue growth factor (CTGF) is a multifunctional protein secreted by vascular endothelial cells, the functions of which depend on interactions with other molecules in the microcellular environment. The expression of the CTGF gene is tightly regulated and involves several signaling pathways, including TGFβ–Smad and phosphatidylinositol-3 kinase (PI3K)–AKT signaling pathways.35
The 14q region contains the candidate gene TMEM30B, the second-most DEG between original and recurrent tumors, being downregulated in recurrences compared with nontumoral meningothelial tissue. TMEM30B codes for a transmembrane factor that participates in cell cycle and that has previously been found to be expressed in meningiomas.36 However, its function has not been well established and further studies are needed to elucidate its exact role in meningiomas.
Finally, we observed overexpression of the histone cluster 1 genes, located at 6p. Histones are the major protein components that compact genomic DNA in the nucleus as a highly organized chromatin structure. Histones H2A, H2B, H3, and H4 constitute the nucleosome core, whereas the linker histone H1 enables an additional level of compaction to be achieved, making genes inaccessible to transcription factors and preventing their expression. Recent studies suggest that HIST1H1C (H1.2) can interact with other regulatory factors to ensure its action as a negative modulator of specific genes. Specifically, the physical interaction of the H1.2 complex with p53, most likely through H1.2, represses p53-dependent, p300-mediated chromatin transcription by blocking chromatin acetylation.37 In addition, it has been reported in mammalian cells that linker histones can participate in the epigenetic regulation of gene expression by contributing to the maintenance or establishment of specific DNA methylation patterns.38 The immunohistochemical analyses carried out in our meningioma series confirmed overexpression of HIST1H1C in most recurrences and original tumors that had relapsed, highlighting the relevance of the mechanisms that regulate gene transcription through the linker histone H1 for meningioma recurrence.
We used GSEA to search for cellular pathways that were differentially expressed in original and recurrent meningiomas. The fact that most of these pathways were downregulated in recurrent tumors may be because most of the DEGs were underexpressed in recurrences. Among the enriched molecular pathways related to meningioma recurrence, the association of Notch, TGFβ, Wnt, PDGF, and the insulin receptor signaling pathways with meningioma progression had already been recognized, thus supporting our data.9,14,15,39–41 Interestingly, the most DEG, SFRP1 (8p), is closely associated with the Wnt signaling pathway. SFPR1 belongs to the family of the secreted frizzled-related proteins, which are able to downregulate Wnt signaling by forming an inhibitory complex with the frizzled receptors. The role of SFRP1 as a tumor suppressor has been proposed in many cancers.42,43 In gliomas, lower expression of SFPR1 and promoter hypermethylation has recently been reported.44 However, further studies are needed to elucidate the role of SFPR1 in meningioma recurrence.
In conclusion, in this study, we define a differential gene expression pattern that distinguishes between original and recurrent meningiomas. Furthermore, we propose novel candidate genes that could be involved in meningioma recurrence: SFRP1, TMEM30B, CTGF, SERPINF1, HIST1H1C, and LMO4. Most of these candidate genes are located at chromosomal regions whose loss has previously been associated with a higher risk of recurrence or malignant progression of meningiomas (1p, 6q, and 14q). In general, these genes are underexpressed relative to nontumoral meningothelial tissue, denoting an overall underexpression of genes in meningioma recurrence. Conversely, overexpression of genes of the histone cluster 1 (6p) in recurrent meningiomas is reported here for the first time.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online.
Funding
This work was partially supported by grants SESCAM 06046 and FISCAM_PI-2006/29 from the Instituto de Ciencias de la Salud and Fundación para la Investigación en Castilla-La Mancha (FISCAM) of the Consejería de Consejería de Salud y Bienestar Social, Junta de Comunidades de Castilla-La Mancha and FIS PI070662, FIS081662, INT09/276, and INT09/277 from the Fondo de Investigaciones Sanitarias (FIS), Spain. E.P.-M. is supported by grant MOV-2008_JI/2 from FISCAM and Y.R. by grant CA07/00119 from FIS.
Acknowledgments
We gratefully acknowledge Elena Gómez and Maite Navarro from the Hospital Virgen de la Salud for their technical assistance. We also thank the Spanish Tumour Bank Network of the Centro Nacional de Investigaciones Oncológicas (Madrid, Spain) for providing tumor samples.
References
- 1.Yee G, Rycroft R, Philips C, et al. Hinsdale, IL: CBTRUS; 2009. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in Eighteen States in 2002–2006. [Google Scholar]
- 2.Rohringer M, Sutherland GR, Louw DF, Sima AA. Incidence and clinicopathological features of meningioma. J Neurosurg. 1989;71:665–672. doi: 10.3171/jns.1989.71.5.0665. doi:10.3171/jns.1989.71.5.0665. [DOI] [PubMed] [Google Scholar]
- 3.Perry A, Louis D, Scheithauer B, Budka H, von Deimling A. World Health Organization Classification of Tumours. Lyon: IARC Press; 2007. [Google Scholar]
- 4.Maillo A, Orfao A, Espinosa A, et al. Early recurrences in histologically benign/grade I meningiomas are associated with large tumors and coexistence of monosomy 14 and del(1p36) in the ancestral tumor cell clone. Neurooncology. 2007;9:438–446. doi: 10.1215/15228517-2007-026. doi:10.1215/15228517-2007-026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirimanoff RO, Dosoretz DE, Linggood RM, Ojemann RG, Martuza RL. Meningioma: analysis of recurrence and progression following neurosurgical resection. J Neurosurg. 1985;62:18–24. doi: 10.3171/jns.1985.62.1.0018. doi:10.3171/jns.1985.62.1.0018. [DOI] [PubMed] [Google Scholar]
- 6.Simpson D. The recurrence of intracranial meningiomas after surgical treatment. J Neurol Neurosurg Psychiatry. 1957;20:22–39. doi: 10.1136/jnnp.20.1.22. doi:10.1136/jnnp.20.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stafford SL, Perry A, Suman VJ, et al. Primarily resected meningiomas: outcome and prognostic factors in 581 Mayo Clinic patients, 1978 through 1988. Mayo Clin Proc. 1998;73:936–942. doi: 10.4065/73.10.936. doi:10.4065/73.10.936. [DOI] [PubMed] [Google Scholar]
- 8.Perry A, Stafford SL, Scheithauer BW, Suman VJ, Lohse CM. Meningioma grading: an analysis of histologic parameters. Am J Surg Pathol. 1997;21:1455–1465. doi: 10.1097/00000478-199712000-00008. doi:10.1097/00000478-199712000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Wrobel G, Roerig P, Kokocinski F, et al. Microarray-based gene expression profiling of benign, atypical and anaplastic meningiomas identifies novel genes associated with meningioma progression. Int J Cancer. 2005;114:249–256. doi: 10.1002/ijc.20733. doi:10.1002/ijc.20733. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian A, Tamayo P, Mootha V, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. doi:10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palacios J, Honrado E, Osorio A, et al. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin Cancer Res. 2003;9:3606–3614. [PubMed] [Google Scholar]
- 12.Smith J, Perry A, Borell T, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 13.Gelpi E, Ambros I, Birner P, et al. Fluorescent in situ hybridization on isolated tumor cell nuclei: a sensitive method for 1p and 19q deletion analysis in paraffin-embedded oligodendroglial tumor specimens. Mod Pathol. 2003;16:708–715. doi: 10.1097/01.MP.0000076981.90281.BF. doi:10.1097/01.MP.0000076981.90281.BF. [DOI] [PubMed] [Google Scholar]
- 14.Cuevas I, Slocum A, Jun P, et al. Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res. 2005;65:5070–5075. doi: 10.1158/0008-5472.CAN-05-0240. doi:10.1158/0008-5472.CAN-05-0240. [DOI] [PubMed] [Google Scholar]
- 15.Johnson M, Okediji E, Woodard A. Transforming growth factor-beta effects on meningioma cell proliferation and signal transduction pathways. J Neurooncol. 2004;66:9–16. doi: 10.1023/b:neon.0000013461.35120.8a. doi:10.1023/B:NEON.0000013461.35120.8a. [DOI] [PubMed] [Google Scholar]
- 16.Riemenschneider M, Perry A, Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006;5:1045–1054. doi: 10.1016/S1474-4422(06)70625-1. doi:10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- 17.Carvalho L, Smirnov I, Baia G, et al. Molecular signatures define two main classes of meningiomas. Mol Cancer. 2007;6:64. doi: 10.1186/1476-4598-6-64. doi:10.1186/1476-4598-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson M, Gutmann D, Peterson K, et al. Molecular characterization of human meningiomas by gene expression profiling using high-density oligonucleotide microarrays. Am J Pathol. 2002;161:665–672. doi: 10.1016/S0002-9440(10)64222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fèvre-Montange M, Champier J, Durand A, et al. Microarray gene expression profiling in meningiomas: differential expression according to grade or histopathological subtype. Int J Oncol. 2009;35:1395–1407. doi: 10.3892/ijo_00000457. doi:10.3892/ijo_00000457. [DOI] [PubMed] [Google Scholar]
- 20.Modha A, Gutin P. Diagnosis and treatment of atypical and anaplastic meningiomas: a review. Neurosurgery. 2005;57:538–550. doi: 10.1227/01.neu.0000170980.47582.a5. doi:10.1227/01.NEU.0000170980.47582.A5. [DOI] [PubMed] [Google Scholar]
- 21.Tabernero M, Espinosa A, Maíllo A, et al. Characterization of chromosome 14 abnormalities by interphase in situ hybridization and comparative genomic hybridization in 124 meningiomas: correlation with clinical, histopathologic, and prognostic features. Am J Clin Pathol. 2005;123:744–751. doi:10.1309/D7U997XD2PHBCQCN. [PubMed] [Google Scholar]
- 22.Espinosa A, Tabernero M, Maíllo A, et al. The cytogenetic relationship between primary and recurrent meningiomas points to the need for new treatment strategies in cases at high risk of relapse. Clin Cancer Res. 2006;12:772–780. doi: 10.1158/1078-0432.CCR-05-1480. doi:10.1158/1078-0432.CCR-05-1480. [DOI] [PubMed] [Google Scholar]
- 23.Tabernero M, Maillo A, Gil-Bellosta C, et al. Gene expression profiles of meningiomas are associated with tumor cytogenetics and patient outcome. Brain Pathol. 2009;19:409–420. doi: 10.1111/j.1750-3639.2008.00191.x. doi:10.1111/j.1750-3639.2008.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishino S, Hashimoto N, Fushiki S, et al. Loss of material from chromosome arm 1p during malignant progression of meningioma revealed by fluorescent in situ hybridization. Cancer. 1998;83:360–366. doi:10.1002/(SICI)1097-0142(19980715)83:2<360::AID-CNCR21>3.0.CO;2-Q. [PubMed] [Google Scholar]
- 25.Bello M, de Campos J, Vaquero J, Kusak M, Sarasa J, Rey J. High-resolution analysis of chromosome arm 1p alterations in meningioma. Cancer Genet Cytogenet. 2000;120:30–36. doi: 10.1016/s0165-4608(99)00249-6. doi:10.1016/S0165-4608(99)00249-6. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Nunes F, Stemmer-Rachamimov A, et al. Genomic profiling distinguishes familial multiple and sporadic multiple meningiomas. BMC Med Genomics. 2009;2:42. doi: 10.1186/1755-8794-2-42. doi:10.1186/1755-8794-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckley P, Jarbo C, Menzel U, et al. Comprehensive DNA copy number profiling of meningioma using a chromosome 1 tiling path microarray identifies novel candidate tumor suppressor loci. Cancer Res. 2005;65:2653–2661. doi: 10.1158/0008-5472.CAN-04-3651. doi:10.1158/0008-5472.CAN-04-3651. [DOI] [PubMed] [Google Scholar]
- 28.Lusis E, Gutmann D. Meningioma: an update. Curr Opin Neurol. 2004;17:687–692. doi: 10.1097/00019052-200412000-00008. doi:10.1097/00019052-200412000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Piaskowski S, Rieske P, Szybka M, et al. GADD45A and EPB41 as tumor suppressor genes in meningioma pathogenesis. Cancer Genet Cytogenet. 2005;162:63–67. doi: 10.1016/j.cancergencyto.2005.02.009. doi:10.1016/j.cancergencyto.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Sum E, Segara D, Duscio B, et al. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci USA. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. doi:10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Ohuchida K, Nakata K, et al. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer. 2008;7:93. doi: 10.1186/1476-4598-7-93. doi:10.1186/1476-4598-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Z, Lam K, Wang N, Xu X, Cortes M, Andersen B. LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene. 2006;25:2920–2930. doi: 10.1038/sj.onc.1209318. doi:10.1038/sj.onc.1209318. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki S, Nishizaki T, Ito H, Sasaki K. Comparative genomic hybridization analysis of genetic alterations associated with malignant progression of meningioma. J Neurooncol. 1999;41:167–174. doi: 10.1023/a:1006086723607. doi:10.1023/A:1006086723607. [DOI] [PubMed] [Google Scholar]
- 34.Weber R, Boström J, Wolter M, et al. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94:14719–14724. doi: 10.1073/pnas.94.26.14719. doi:10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cicha I, Goppelt-Struebe M. Connective tissue growth factor: context-dependent functions and mechanisms of regulation. Biofactors. 2008;35:200–208. doi: 10.1002/biof.30. doi:10.1002/biof.30. [DOI] [PubMed] [Google Scholar]
- 36.Katoh Y, Katoh M. Identification and characterization of CDC50A, CDC50B and CDC50C genes in silico. Oncol Rep. 2004;12:939–943. [PubMed] [Google Scholar]
- 37.Kim K, Choi J, Heo K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. doi:10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan Y, Nikitina T, Zhao J, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. doi:10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 39.Baia G, Stifani S, Kimura E, McDermott M, Pieper R, Lal A. Notch activation is associated with tetraploidy and enhanced chromosomal instability in meningiomas. Neoplasia. 2008;10:604–612. doi: 10.1593/neo.08356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson M, Woodard A, Kim P, Frexes-Steed M. Evidence for mitogen-associated protein kinase activation and transduction of mitogenic signals by platelet-derived growth factor in human meningioma cells. J Neurosurg. 2001;94:293–300. doi: 10.3171/jns.2001.94.2.0293. doi:10.3171/jns.2001.94.2.0293. [DOI] [PubMed] [Google Scholar]
- 41.Keller A, Ludwig N, Backes C, et al. Genome wide expression profiling identifies specific deregulated pathways in meningioma. Int J Cancer. 2009;124:346–351. doi: 10.1002/ijc.23942. doi:10.1002/ijc.23942. [DOI] [PubMed] [Google Scholar]
- 42.Chung M, Lai H, Sytwu H, et al. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol. 2009;112:646–653. doi: 10.1016/j.ygyno.2008.10.026. doi:10.1016/j.ygyno.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Caldwell G, Jones C, Gensberg K, et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–888. doi: 10.1158/0008-5472.can-03-1346. doi:10.1158/0008-5472.CAN-03-1346. [DOI] [PubMed] [Google Scholar]
- 44.Götze S, Wolter M, Reifenberger G, Müller O, Sievers S. Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int J Cancer. 2010;126:2584–2593. doi: 10.1002/ijc.24981. [DOI] [PubMed] [Google Scholar]