Abstract
Neuralized (Neurl) is a highly conserved E3 ubiquitin ligase, which in Drosophila acts upon Notch ligands to regulate Notch pathway signaling. Human Neuralized1 (NEURL1) was investigated as a potential tumor suppressor in medulloblastoma (MB). The gene is located at 10q25.1, a region demonstrating frequent loss of heterozygosity in tumors. In addition, prior publications have shown that the Notch pathway is functional in a proportion of MB tumors and that Neurl1 is only expressed in differentiated cells in the developing cerebellum. In this study, NEURL1 expression was downregulated in MB compared with normal cerebellar tissue, with the lowest levels of expression in hedgehog-activated tumors. Control of gene expression by histone modification was implicated mechanistically; loss of 10q, sequence mutation, and promoter hypermethylation did not play major roles. NEURL1-transfected MB cell lines demonstrated decreased population growth, colony-forming ability, tumor sphere formation, and xenograft growth compared with controls, and a significant increase in apoptosis was seen on cell cycle and cell death analysis. Notch pathway inhibition occurred on the exogenous expression of NEURL1, as shown by decreased expression of the Notch ligand, Jagged1, and the target genes, HES1 and HEY1. From these studies, we conclude that NEURL1 is a candidate tumor suppressor in MB, at least in part through its effects on the Notch pathway.
Keywords: deazaneplanocin, Jagged1, Neuralized1, Notch, tumor suppressor
Medulloblastoma (MB), the most common pediatric malignant brain tumor, is an embryonal tumor of the cerebellum. MB occurs when the pathways controlling normal proliferation, differentiation, and cell death in the developing cerebellum are disrupted.1 The Wnt, Sonic hedgehog, and Notch signal transduction pathways are among those frequently dysregulated.2
The Notch pathway affects numerous cellular processes, and downstream effects of receptor-ligand binding are both developmental stage and tissue-specific.3 In the normal cerebellum, Notch2 is highly expressed in the proliferating cells of the external granule layer during the postnatal period and is downregulated as granule cell precursors (GCPs) exit the cell cycle and differentiate.4,5 In vitro studies have confirmed that Notch2 signaling maintains proliferation and prevents differentiation of GCPs.4 These data suggest that Notch signaling may play an important role in MB, as GCPs were demonstrated to be the cell of origin for certain MB tumors.2
Dysregulation of Notch signaling has been implicated in tumor formation in an array of tumor types, having either oncogenic or tumor-suppressive effects depending on the tumor tissue of origin.3,6 Activating mutations of the Notch receptor are very rare in solid tumors, and the ligand-mediated activation of the pathway appears to be the norm.6,7 In MB cell lines, intracellular Notch2 (ICN2) expression increases proliferation, whereas (ICN1) expression has the opposite effect.8 Treatment of both MB cell lines and ex vivo primary tumors with Notch pathway inhibitors results in significantly decreased viable cell numbers compared with controls8,9 and reduced tumor engraftment and growth in nude mice.9,10 Dysregulation in primary tumors may involve either Notch 1 or Notch2 with conflicting reports on the prevalence for each.8,9 However, it is clear that Notch-activated Hes1-positive tumors are associated with a significantly shorter survival time than Hes1-negative tumors.8
The genetic and epigenetic lesions leading to Notch pathway activation in MB have not been characterized. One candidate regulator is the human Neuralized1 (NEURL1) gene, which maps to 10q25.1,11 a region of chromosome 10 with frequent loss of heterozygosity (LOH) in MB.12 Neuralized (DNeur) is an E3 ubiquitin ligase which, in Drosophila, ubiquitinates and promotes activation and degradation of Notch ligands.13 The DNeur and NEURL1 proteins contain 2 neuralized homology regions (NHRs) and a RING domain. The NHR function in Neurl-ligand interactions and the RING domain are required for ubiquitin ligase activity.14,15 In mammals, Neurl1 is expressed at high levels in the differentiated granule cells of the normal adult cerebellum but not in proliferative GCPs in the postnatal cerebellum.16 Neurl1 could therefore suppress proliferation or maintain differentiation in GCPs, and its loss could contribute to tumor formation.
Here, we report that NEURL1 expression is suppressed in MB tumors compared with adult and fetal normal cerebellum and that histone-modifying reagents induce its upregulation. Forced expression of the gene in cell lines resulted in increased apoptosis and decreased cell and tumor growth. An inhibitory effect of NEURL1 on the Notch pathway was demonstrated. Together, these results support NEURL1 as a candidate tumor suppressor, which undergoes epigenetic silencing in MB.
Materials and Methods
Sample Collection
Tumors were collected at Children's Hospital Boston between 1991 and 2006. With institutional review board approval and after obtaining signed informed consent, tumor samples were snap-frozen in liquid nitrogen in the operating room and then stored at −80°C. All diagnoses were confirmed by histological assessment by a neuropathologist. Total RNA from the normal human adult brain and cerebellum was obtained from BD Biosciences, Ambion, Biochain, Clontech, and Stratagene; normal human 33-week fetal cerebellum RNA was from Biochain. Matched samples of normal cerebellar RNA and genomic DNA were from Biochain.
Cell Culture
MB cell lines UW228, UW402, UW426, UW473, R256, R262, R300, and R308 were supplied by Dr Мichael Bobola, University of Washington, Seattle. D283Med, D341Med, D384Med, D425Med, D458Med, and D556Med cells were obtained from Duke University, North Carolina. The DAOY cell line was from the American Tissue Culture Collection (ATCC). Cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) high glucose supplemented with 10% fetal bovine serum (Invitrogen). UW228, UW402, and UW426 were established from the same primary tumor.17 D425 and D458 were derived from the same patient from primary tumor and post-treatment metastatic tumor, respectively.18 Genome-wide SNP data obtained in this laboratory were used to confirm the identity of the cell lines.19
Real-Time PCR
Tumor RNA was extracted using the Trizol method (Invitrogen), and cell line RNA with RNeasy kits (Qiagen). The high-capacity reverse transcription system (Applied Biosystems) was used in subsequent applications. All real-time PCR assays were performed using Taqman primers on the ABI7200 system (Applied Biosystems); the endogenous control was β2-microglobulin. The NEURL1 primers (Hs00184868_m1) targeted exons 1 and 2 of IMAGE clone 4812302 (GenBank BC026336).
Cluster Analysis of Expression Array Data
Unsupervised hierarchical analysis of microarray data has been described elsewhere.2
Sequencing and Promoter Methylation Analysis
Tumor and cell line DNA were extracted using the DNeasy blood and tissue kit (Qiagen). Primers were designed to amplify the exons and intron/exon junctions of the NEURL1 gene for sequencing (Supplementary Material). PCR products were run on a 2% agarose gel, purified with a QIAquick gel extraction kit (Qiagen), and subjected to automated sequencing. NEURL1-associated CpG islands were investigated using CpGplot software available at http://www.ebi.ac.uk/emboss. Bisulfite sequencing was performed as described previously20; MethPrimer (http://www.urogene.org/methprimer/index1.html)-generated semi-nested PCR primers are detailed in the “Supplementary Material” section. Drug treatments for reversal of epigenetic modifications are also described elsewhere.21
Plasmid Construction and Transfections
The cDNA encoding full-length human NEURL1 was amplified by RT-PCR from normal cerebellar RNA (Stratagene). The Superscript III First-Strand Synthesis System (Invitrogen) was used for reverse transcription and Accuprime Pfx polymerase for PCR (Invitrogen). The PCR product was cloned into the KpnI and XhoI sites of pBudCE4.1/V5-His (Invitrogen), distal to the EF1α promoter. Constructs were obtained encoding untagged and C-terminus V5-tagged NEURL1, and a V5-tagged RING domain deletion mutant (ΔRING) was created by PCR using the full-length construct as a template. All sequences were confirmed by automated sequencing.
Transfections
DAOY and R300 cells were transfected for constitutive expression using Fugene (Roche Applied Science), then selected and maintained in the growth medium supplemented with 200 µg/mL zeocin (Invitrogen). Zeocin-resistant clones were analyzed for NEURL1 expression by Western blot (anti-V5 antibody; Invitrogen). DAOY transient transfections were performed by Nucleofection, using 5 × 106 cells, 2 µg of plasmid DNA, and program T-030, according to the manufacturer's instructions. For UW228 transfections, program T-020 was used.
Western Blot
RIPA buffer (Boston Bioproducts) was used for protein extraction and 20 or 50 µg of protein run on a 10% polyacrylamide gel under reducing conditions. Following transfer, membranes were blocked in 5% nonfat dried milk. ECL Plus detection reagent was used for signal development (GE Healthcare). Antibodies and concentrations were as follows: anti-V5 (Invitrogen) 1:5000, anti-Jagged1 1:200 (Santa Cruz), anti-β-actin (Abcam) 1:4000, HRP-conjugated secondary antibodies (Jackson Immunoresearch Laboratories) 1:10 000.
Cell Growth Assays
The CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) was used for MTS assays. Cells were plated in the 96-well cell culture plates (Costar) at 1000 cells/well (DAOY) and at 500 cells/well (R300) in the 100 µL of growth medium. The medium was replaced every 2–3 days. The assay was performed according to the manufacturer's instructions. siRNA knockdown of NEURL1 was achieved using siPORT NeoFX Transfection Agent and Ambion NEURL1 Silencer siRNA ID:s17488 with a scrambled control (Applied Biosystems). Twenty-four hours post-transfection, cells were counted and redistributed for MTS assay and expression analysis. For colony-forming assays, stable transfectants were seeded in 10-cm plates at 5000 (R300) or 10 000 cells/plate (DAOY), and cultured for 10–14 days. Cells were fixed with 2% paraformaldehyde (Electron Microscopy Sciences), and stained with crystal violet (Sigma). For neurosphere growth assays, the medium was prepared from DMEM/F12, B27 (1×), bFGF (20 ng/mL), and EGF (20 ng/mL), all from Invitrogen. One hundred cells per well were plated into the 100 µL of neurosphere medium in a 96-well plate; 100 µl of medium was added after 1 week, and spheres were counted at 2 weeks. For xenograft studies, 5 × 106 cells were suspended in 100-µL culture medium plus 100 µl Matrigel (BD Biosciences). Cells were injected into the flanks of nude mice (Crl:NU/NU-Foxn1; Charles River Laboratories). After 9 weeks, the animals were euthanized and the tumors excised and weighed. Tumors were processed for routine histological analysis by hematoxylin and eosin (H&E) staining of paraffin-embedded tissue sections. Institutional approval was obtained prior to in vivo experiments.
Cell Death Analysis
For cell cycle analysis, cells were plated at 30% confluence, collected 24 hours later, and fixed in 0.25% paraformaldehyde (Sigma) for 30 minutes at 4°C. After washing with PBS, the cells were resuspended in 50 µg/mL propidium iodide (Sigma) and processed on a FACS Vantage SE using CellQuant software (BD Biosciences). The Cell Death ELISAPLUS (Roche Applied Systems) was used to quantify mono- and oligonucleosomes in stable transfectants, according to the manufacturer's instructions.
Immunocytochemistry
DAOY stable transfectants were transiently transfected with a Jagged1-HA construct (G. Weinmaster). Cells were collected at 24 hours and fixed in 2% paraformaldehyde (Electron Microscopy Sciences) for 1 hour. Processing for immunocytochemistry was as described previously.22 Reagents used were: anti-V5 antibody (Invitrogen) 1:200, anti-Jagged (Santa Cruz) 1:50, and Alexa Fluor–labeled secondary antibodies (Invitrogen).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism. A P-value of <.05 was considered to be statistically significant. The mean and the standard error were used in all graphs, unless otherwise stated.
Image Acquisition Tools and Image Processing Software
Western blot images were captured from X-ray film using a HP3180 scanner (Hewlett Packard). Gels were visualized and photographed with the Gene Genius Bioimaging System (Syngene). The confocal microscope was a Zeiss LSM510 META 2-Photon and images obtained were analyzed in LSM Image Browser (Zeiss). Images were processed in Adobe Photoshop CS (Adobe).
Results
Reduced Expression of NEURL1 in MB
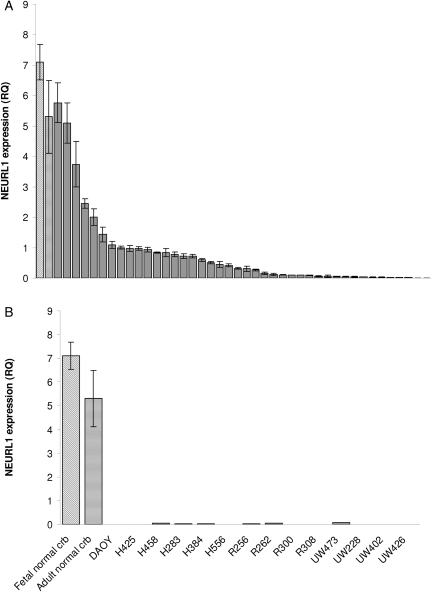
To test NEURL1 as a candidate tumor suppressor in MB, we examined NEURL1 relative expression by real-time PCR among 42 primary tumor samples compared with adult and fetal normal cerebellum. Ninety percent of the primary tumor samples tested had NEURL1 expression less than half that of the normal cerebellum, whereas 57% had levels less than one-tenth that of the cerebellar samples (median: 0.068, mean: 0.173, range: 0–1.259; Fig. 1A). Maximum expression in all the cell lines tested was 1.4% that of the normal samples (Fig. 1B). Together these results demonstrate significant downregulation of NEURL1 in primary tumors and stable cell lines, 21 primary tumors from a published cohort2 were assigned to subgroups by cluster analysis of microarray data and ranked by NEURL1 expression (Table 1). Group III consisted of hedgehog-activated tumors including, but not limited to, those of the desmoplastic subtype. These tumors fell almost exclusively below the median (0.130), and the association between hedgehog-activated tumors and low NEURL1 expression was significant (P = .02; Fisher's exact test). From the array expression data of tumors in which we analyzed NEURL1 expression by real-time PCR (Fig. 1A), we observed that the hedgehog-regulated oncogenes and signaling markers, GLI1 and GLI2 were negatively correlated with NEURL1 expression. GLI1 expression is negatively correlated with NEURL1 expression, with Spearman's rank correlation coefficient ρ = −0.42 and permutation P-value of <.02; GLI2 expression is negatively correlated with NEURL1 expression, with Spearman's rank correlation coefficient ρ = −0.33 and permutation P-value of <.07.
Fig. 1.
NEURL1 is downregulated in MB tumors and cell lines. (A) NEURL1 expression in 42 primary tumors. Real-time PCR was performed with exon 1/2 NEURL1 primers; RQ, relative quantity; angled-stripe bar, fetal NC; horizontal-stripe bar, adult NC; the mean of 4 samples. (B) NEURL1 expression in MB cell lines compared with fetal NC and adult NC; NC, normal cerebellum. Each sample was analyzed in triplicate.
Table 1.
Association between hedgehog-activated tumors and low NEURL1 expression
| ID | NEURL1 expression | Age at Diagnosis (mos) | Subgroup (n = 4) | Subtype | Relapse | Current status | M stage at diagnosis |
|---|---|---|---|---|---|---|---|
| MD_99 | 1.259 | 7 | II | C | Y | D | M0 |
| MD_111 | 1.113 | 156 | II | C | N | A | M0 |
| MD_129 | 0.816 | 180 | II | C | N | A | M0 |
| MD_90 | 0.312 | 108 | II | C | N | A | M0 |
| MD_137 | 0.218 | 174 | III | C | N | A | M0 |
| MD_131 | 0.212 | 84 | II | C | Y | A | M+ |
| MD_96 | 0.211 | 150 | II | C | N | A | M0 |
| MD_107 | 0.183 | 456 | IV | C | Y | D | M3 |
| MD_118 | 0.181 | 114 | II | C | N | LTFU | M1 |
| MD_106 | 0.158 | 30 | I | C | N | A | M0 |
| MD_112 | 0.13 | 96 | IV | C | N | A | M0 |
| MD_97 | 0.098 | 24 | III | D | N | A | M0 |
| MD_113 | 0.09 | 78 | III | C | Y | LTFU | M2 |
| MD_109 | 0.058 | 20 | III | D | N | A | M0 |
| MD_145 | 0.023 | 84 | I | C | N | A | M0 |
| MD_101 | 0.012 | 138 | III | D | N | A | M0 |
| MD_91 | 0.01 | 138 | III | C | Y | D | M3 |
| MD_86 | 0.008 | 336 | III | D | N | A | M0 |
| MD_98 | 0.002 | 72 | I | C | Y | D | M0 |
| MD_102 | 0.001 | 16 | III | D | Y | A | M0 |
| MD_110 | 0 | 156 | IV | C | Y | D | M0 |
| *P = .02 |
Abbreviations: Group III, hedgehog-activated tumors. Subtypes: C, classic; D, desmoplastic. Relapses: Y, yes; N, no. Status: D, dead; A, alive; LTFU, lost to follow-up. M stage, Chang's metastatic stage. MB primary tumors are ranked by NEURL1 expression from high to low (measured by real-time PCR). Expression in the normal cerebellum =1. MB subgroups I–IV were derived by unsupervised hierarchical analysis of primary tumors from reference.2
*There are significantly more tumors from Group III below the median than above (Fisher's exact test, P = .02).
Epigenetic Regulation of NEURL1 Expression
To gain insight into the mechanism of NEURL1 inactivation, we first set out to identify the coding mutations in 7 unrelated cell lines. Sequencing revealed 2 previously recognized polymorphisms but no mutations in the gene. In a previous study, 10q loss was demonstrated in 3 of the 7 cell lines tested in this study (10q loss, DAOY, D283, and UW228-related lines; no loss detected, D341, D384, D425/458, D556).12 However, loss of 1 allele alone does not account for the observed reduction in expression (Fig. 1B), which is comparable between cell lines. No homozygous deletions were detected during sequencing. In addition, in 17 MB tumors with known karyotypic information, low NEURL1 expression did not relate to loss of 10q (P = .78, nonparametric Wilcoxon rank-sum test). When this cohort was sorted based on NEURL1 expression, the 4 tumors with chromosome 10q loss were observed at 65%, 59%, 41%, and 29% percentiles.
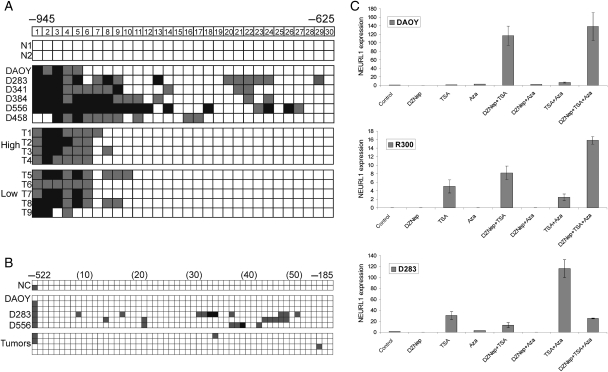
We next investigated the contribution of promoter hypermethylation to NEURL1 transcriptional repression, first defining the position of the transcription start site (TSS) of the NEURL1 gene by RACE PCR. Using CpGPlot software, we identified a CpG island (CGI) at the promoter that extends from −940 to +197 relative to the TSS. As 5-aza-2′-deoxycytidine (AzaCdR) treatment of MB cell lines led to differing degrees of re-expression of NEURL1 in the 5 cell lines tested (data not shown), bisulfite sequencing of cell lines and primary tumors was performed. Tumor-specific methylation was discovered at the distal end of the CGI, but further analysis revealed methylation in all samples, irrespective of the level of NEURL1 expression (Fig. 2A). In the more proximal regions, sporadic methylation was detected in cell lines and virtually none in tumors (Fig. 2B).
Fig. 2.
Analysis of epigenetic mechanisms controlling NEURL1 expression. Promoter hypermethylation analysis of the NEURL1 promoter by bisulfite sequencing. (A) −945 to −625 and (B) −522 to −185 from the transcription start site. The CpG dinucleotides are numbered. N, normal cerebellum; T, primary tumor; high, expression equivalent to that in normal cerebellum; low, expression <10% that in normal cerebellum. Black squares, >75% methylation; gray squares, 25%–75% methylation; white squares, <25% methylation. Representative results for the tumors analyzed are shown in (B); NC, normal cerebellum. (C) Histone modification analysis is depicted for DAOY, R300, and D283 MB cell lines. NEURL1 expression was measured following drug treatments. Control, vehicle treatment; DZNep, 5 µM 3-deazaneplanocin A; TSA, 200 nM trichostatin A; Aza, 5 µM 5-aza-2′-deoxycytidine.
Recent studies have shown that DNA methylation-independent processes, such as histone methylation and acetylation, may achieve chromatin remodeling at gene promoters and thus the regulation of expression.21,23,24 We therefore treated MB cell lines with AzaCdR, trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, and 3-deazaneplanocin A (DZNep), a histone methylation inhibitor,25 alone or in combination, and monitored expression of NEURL1. In the DAOY, R300, D283 (Fig. 2C), UW228 cells, and D556 cell lines (Supplementary Material, Fig. S1A and B), the individual drugs had little effect on gene expression. However, DZNep and TSA together functioned synergistically in the DAOY, R300, and UW228 cells, increasing expression by 10- to 120-fold over untreated cells. Expression was increased to a similar extent in UW228 cells by the combination of AzaCdR and TSA. In contrast, only the AzaCdR-TSA combination was effective in the D283 (Fig. 2C) and D556 cell lines (Supplementary Material, Fig. S1B). The results of these experiments indicate that NEURL1 expression may be silenced by epigenetic events other than promoter DNA hypermethylation.
Reduction in Tumor Phenotype on NEURL1 Re-Expression
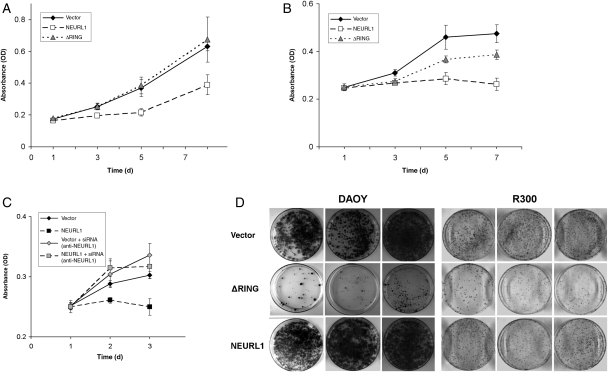
To examine the consequences of NEURL1 expression in MB cell lines, expression constructs encoding NEURL1 and a RING domain deletion mutant were created (see “Materials and Methods” section). A RING domain deletion mutant was used as control to exclude nonspecific effects by the overexpression of NEURL1, as a functional RING domain is required for E3 ubiquitin ligase activity.26 The DAOY and R300 MB cell lines were stably transfected with each of the expression constructs and an empty vector control. To assess the growth capacity of the transfected cells, MTS assays were performed for each of the 2 cell lines, with multiple clones representing each construct. NEURL1 expression was found to inhibit the growth of DAOY and R300 cells (Fig. 3A and B), and transient transfection of the constructs into UW228, D458, and D425 lines also resulted in a modest decrease in cell growth (Supplementary Material, Fig. S3, and data not shown). siRNA experiments to knock down overexpressed NEURL1 in DAOY achieved a 61% reduction in expression at 48 hours (data not shown). The knockdown experiment restored growth and confirmed that the effect of NEURL1 was gene-specific (Fig. 3C). NEURL1-expressing cells also demonstrated a greatly reduced colony number and smaller colony size in colony-forming assays (Fig. 3D).
Fig. 3.
Expression of NEURL1 inhibits cell growth. (A, B, and D) NEURL1-expressing DAOY and R300 stable transfectants grow more slowly than vector-only or ΔRING transfectants. In (A) and (B), viable cells were measured by MTS assay at the time points shown. (A) DAOY cells: NEURL1 (n = 5), ΔRING (n = 5), and vector only (n = 5); (B) R300 cells: NEURL1 (n = 4), ΔRING (n = 4), and vector only (n = 4); n, number of individual stable clones. (C) siRNA-mediated knockdown of NEURL1 restores the cell growth properties of the DAOY line. Expression of NEURL1 was reduced by 32% and 61% at 24 and 48 hours, respectively (data not shown). NEURL1 (black squares) and vector-only (black diamonds) transfectants treated with scrambled siRNA control, and NEURL1 (gray squares) and vector-only (gray diamonds) transfectants treated with anti-NEURL1 siRNA. (D) Colony-forming assay with DAOY and R300 tansfectants: NEURL1 (n = 3), ΔRING (n = 3), and vector only (n = 3); n, number of individual stable clones; 10 000 cells (DAOY) and 5000 cells (R300) were seeded into the 10-cm plates in triplicate and cultured for 2 weeks. Images of 3 individual clones in each group of transfectants are shown for each cell line.
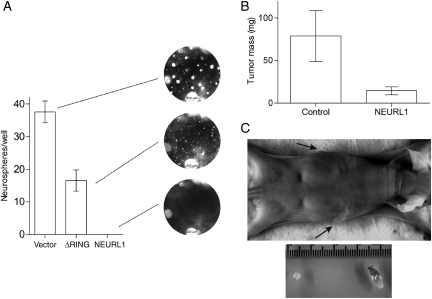
Single-cell suspensions of the stable transfectants were plated in a serum-free neurosphere medium. Under these conditions, DAOY generates tumor spheres which are competent for serial propagation.27 Spheres readily formed in the vector-only transfectants, whereas ΔRING transfectants formed free-floating spheres and/or became adherent and grew rapidly attached to the plate. In contrast, tumor sphere formation in NEURL1-expressing cells was virtually abolished (Fig. 4A). A small fraction of the cells remained viable but did not form spheres >50 µm in diameter. From this and the previous assays, we predicted that xenograft formation from NEURL1-expressing cells would produce smaller tumors than control cells. The NEURL1-expressing tumors were significantly smaller than those in the control group (1-tailed t-test, P = .039; Fig. 4B and C). These results support the idea that NEURL1 reactivation is sufficient to block MB cell growth. Tumors were assessed by H&E staining for possible differences in tumor histology, content of stromal cells, and residual matrix. Examined xenografts from both control and NEURL1 groups showed a high degree of cellularity with a thin stromal tissue surrounding highly cellular tumor, and a similar degree of collagen deposition between cellular regions was observed in ∼20% of xenografts from control and NEURL1 groups.
Fig. 4.
NEURL1 expression reduces neurosphere and tumor growth. (A) Tumor sphere assay (DAOY cells). One hundred cells/well of representative transfectants NEURL1 (n = 3), ΔRING (n = 3), and vector only (n = 3) were plated in the 96-well plates in a neurosphere medium and grown for 2 weeks. Spheres >50 µm were counted in 7 replicate wells. Photographs of representative wells are shown. (B) Xenograft assay (DAOY cells). Controls consist of vector-only (n = 2) and ΔRING (n = 2) transfectants, NEURL1 (n = 4); n, number of individual stable clones. Seven and 6 mice were injected with controls and NEURl1 transfectants, respectively; P = .039, 1-tailed t-test. (C) The photographs show the representative views of NEURL1-expressing (top arrow, left tumor) and control (bottom arrow, right tumor) tumors.
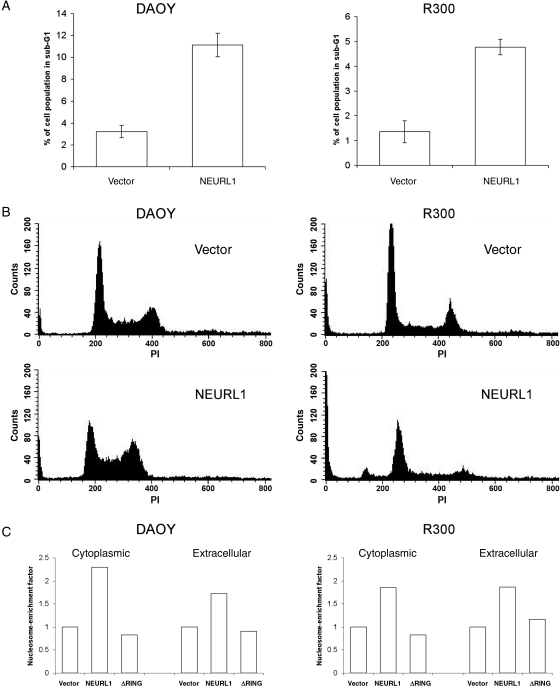
NEURL1 Expression Increases Apoptosis
Cell cycle analysis of DAOY and R300 representative stably expressing clones (Fig. 5A and B) revealed a consistent and significant increase in the sub-G1 fraction in NEURL1 transfectants compared with controls (P = .03 [DAOY] and P = .033 [R300]; unpaired t-test), and a nucleosomal ELISA confirmed that NEURL1-expressing cells undergo increased apoptosis (Fig. 5C). Fan et al.10 described a 25% increase in the percentage of cells in G1/G0, and differentiation of DAOY cells on treatment with a gamma secretase inhibitor. In this study, we did not see a consistent increase in the G1/G0 fraction, and real-time PCR for the differentiation markers used, TUBB3 and GABRA6, revealed no difference between the transfectants (data not shown).
Fig. 5.
NEURL1 expression increases apoptosis. (A) Apoptosis was measured by propidium iodide staining and flow cytometry quantifying the sub-G1 cell population. NEURL1 expression leads to a significant increase in apoptotic cells compared with controls (DAOY, P = .03; R300, P = .033; unpaired t-test). Cell cycle analysis was performed for NEURL1 (n = 3) and vector only (n = 3) in triplicate for each transfectant. (B) Representative histograms are shown beneath the bar chart. (C) Results from cell death ELISA to measure cytoplasmic and extracellular mono- and oligonucleosomes. The nucleosome-enrichment factor compares each result with the vector-only control. Cytoplasmic nucleosomes result from apoptosis and extracellular nucleosomes from primary or secondary necrosis. NEURL1 (n = 2), ΔRING (n = 2), and vector only (n = 2); n, number of individual stable clones.
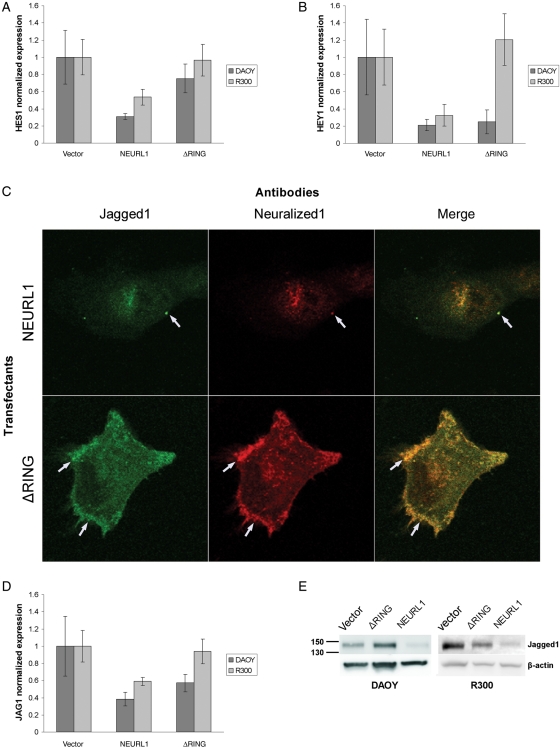
NEURL1 Expression Decreases Notch Pathway Activation
Given the association between DNeur and the Notch pathway, Notch target gene expression was analyzed in all clones of the stable transfectants of DAOY and R300 cell lines. Cells expressing full-length NEURL1 produced less HES1 and HEY1 than vector-only controls (Fig. 6A and B), the opposite effect to that described by Fan et al.8 after DAOY transfection with Notch2. Transient transfection of UW228 with NEURL1 also decreased HES1 levels (Supplementary Material, Fig. S3B). Expression of the ΔRING transfectants resulted in a reduction in HEY1 levels in DAOY, but not in R300 (Fig. 6B). Effects of the mutant protein may be a consequence of Jagged1 sequestration at the cell surface (see below).
Fig. 6.
NEURL1 inhibits the Notch pathway and colocalizes with Jagged1. Stable transfectants of DAOY and R300 show reduced Notch pathway activation with decreased expression of target genes (A) HES1 and (B) HEY1. In (A) and (B), DAOY: NEURL1 (n = 5), ΔRING (n = 5), and vector only (n = 5); R300: NEURL1 (n = 4), ΔRING (n = 4), and vector only (n = 4); n, number of individual stable clones. (C) Immunofluorescence microscopy of DAOY NEURL1 and ΔRING stable expressing clones transiently transfected with JAG1 shows reduced staining of Jagged1 in the NEURL1 transfectant. Arrows show colocalization of Neuralized1 and Jagged1 in punctate clusters at the cell surface. (D) Jagged1 mRNA levels are also decreased through the expression of NEURL1. DAOY cells: NEURL1 (n = 5), ΔRING (n = 5), and vector only (n = 5); R300 cells: NEURL1 (n = 4), ΔRING (n = 4), and vector only (n = 4); n, number of individual stable clones. (E) The Western blots show decreased expression of endogenous Jagged1 in NEURL1 stable transfectants compared with controls.
Koutelou et al. recently demonstrated Neurl1/Jagged1 interaction by immunoprecipitation and RING-dependent ubiquitination of the Notch ligand. Their results indicated Neurl1-dependent lysosomal degradation of Jagged1 with inhibition of Notch signaling.28 In Drosophila, DNeur colocalizes with Delta and Serrate at the cell membrane prior to internalization of both DNeur and the Notch ligand.13 To add to these results, we performed immunofluorescence experiments on representative DAOY NEURL1 stable transfectants transiently transfected with JAG1 and found that Jagged1 staining is significantly reduced or absent in NEURL1-expessing cells and not in ΔRING transfectants (Fig. 6C), confirming our observation of endogenous Jagged1 protein and mRNA downregulation in NEURL1 transfectants (Fig. 6D and E). In NEURL1-expressing cells, the 2 proteins appear to colocalize mostly in fine intracellular punctate staining and few distinct punctate clusters at the cell membrane. In ΔRING mutant transfectants, colocalized Jagged1/ΔRING punctate staining is more intense at the cell surface and the leading edges of the cell membrane (Fig. 6C).
Discussion
Neurl1 is a Notch ligand-associated E3 ubiquitin ligase. The effect of Neurl1 on MB tumor growth was investigated because the gene is in a common region of loss on chromosome 10q,12 Notch activation is important in tumor growth in a proportion of MBs,8–10 and Neurl1 is expressed in differentiated granule cells of the cerebellum, but not proliferating GCPs.16
The NEURL1 transcript was downregulated in MB primary tumors, and minimally expressed in all the MB cell lines tested. The mechanism behind NEURL1 repression was investigated in MB cell lines, and no evidence for coding mutations or homozygous deletion was found. LOH at the NEURL1 locus is unlikely to be the sole factor in the regulation of expression as all cell lines and 57% of primary tumors expressed less than 10% of the normal levels of NEURL1. Consequently, epigenetic mechanisms behind transcriptional repression were investigated. Tumor-specific methylation was detected at the distal end of the promoter (900 bp upstream of the TSS) by bisulfite sequencing, but there was no variation in methylation between high- and low-expressing tumors. In the proximal promoter regions, sporadic methylation was detected in cell lines such as D283 and D556, with essentially none found in tumors.
In polycomb group (PcG) target genes, epigenetic control of transcription can occur by histone modification alone.29,30 In addition, the combinatorial effects of different chromatin-modifying reagents are often required for the reversal of epigenetic silencing.31 Cell lines were therefore treated with AzaCdR, a DNA methylase inhibitor; TSA, a HDAC inhibitor; and DZNep, a newly identified histone methylation inhibitor,25 individually or in combination, to assess the effect on NEURL1 expression.
In DAOY, R300, and UW228 cells, DZNep and TSA were found to act synergistically to increase NEURL1 expression up to 120-fold over control levels. In a previous study, the DZNep-TSA combination was shown to convert a repressive histone code to an active one, but could not restore expression of genes repressed by promoter DNA hypermethylation.21 AzaCdR and TSA together dramatically increased expression in the UW228, D283, and D556 cell lines. AzaCdR action may resolve the sparse DNA methylation identified in D283 and D556 (Fig. 2B) resulting, in combination with TSA, in transcription factor access to the methylated regions of promoter DNA. Alternatively, at specific loci, AzaCdR can promote activating alterations of the histone code, without concurrent DNA demethylation.32,33 These results suggest a possible role for histone modifications in NEURL1 silencing. As corroborative evidence, in 3 independent studies, NEURL1 was marked as a PcG target gene with the repressive histone modification H3K27me3 (trimethylation of lysine 27 at histone H3).34–36 Coincidentally, the importance of altered control of histone modifications in MB pathogenesis has been reported.37
A recent genome-wide screen for genes silenced by promoter hypermethylation in colorectal cancer identified NEURL1.38 Extensive tumor-specific methylation of the region 300–450 bp upstream of the NEURL1 TSS was subsequently detected and shown to correlate with expression in cell lines. A separate study of DZNep/TSA treatment of colon cancer cell lines showed that a number of genes were significantly upregulated.21 Our analysis of the array results (GEO public database GSE10972) showed that NEURL1 expression was not affected (data not shown), as would be expected for a highly methylated promoter. A spectrum of expression control exists for PcG target genes, from tightly silenced, through silenced but poised for expression, to actively expressed, as determined by the histone code at the promoter.29,30 Tightly silenced genes have a repressive histone code together with DNA hypermethylation, but downregulated genes may be regulated by histone modifications alone. Thus, in different tumor types, NEURL1 expression appears to be regulated by essentially the same mechanism.
To determine the biological properties of NEURL1, and the implications of decreased expression in tumors, the gene was re-expressed in MB cell lines. Reintroduction of NEURL1 had a significant effect on tumor growth properties as shown by decreased cell growth in an MTS viability assay, decreased colony formation in vitro, and decreased xenograft size in vivo. Tumor sphere formation, as a measure of cancer stem/progenitor cell tumor-forming potential, was virtually abolished in NEURL1-expressing cells. Cell cycle and cell death analysis showed increased apoptosis over controls, whereas differentiation was excluded as an NEURL1-induced process. The induction of apoptosis and lack of differentiation have been noted (data not shown) in a previous study.39
NEURL1 therefore has tumor suppressor properties in MB cells and appears to be a PcG target gene regulated by histone modifications in MB. Neurl1-null mice, however, do not develop spontaneous tumors,39,40 probably because cooperative mutations are required. Loss of PC3, a gene which promotes differentiation in GCPs, only causes transformation in combination with mutant genes (eg, Ras, Ptch1).41–43 Mind bomb 2, another Notch ligand–associated E3 ligase, also has tumor suppressor activity44 but no phenotype during mouse development.45
The link between the tumor suppressor properties of NEURL1 in MB and the Notch pathway was subsequently investigated. We found that NEURL1 transfectants had decreased mRNA levels of the HES1 and HEY1 Notch target genes, the opposite to the effect of Notch2 on DAOY cells, where increased transcription of HES1 and HEY1 occurred.8 NEURL1 also has the reverse effect to Notch2 on tumor growth, as described above. Cancer stem cells (CSCs), the small fraction of tumor cells proposed to initiate and maintain tumor growth, are depleted from MB cell line populations by Notch blockade-induced apoptosis.10,46 Tumor sphere formation studies indicate that NEURL1 expression may also decrease the CSC population in MB.
NEURL1 appears to control the Notch pathway at the protein level. A recent functional study showed Neurl1 co-immunoprecipitation with and specific ubiquitination of the Notch ligand, Jagged1, leading to decreased levels of Jagged1 protein.28 HES1 expression was diminished as a result, the opposite effect to that expected from Drosophila studies, where DNeur–ligand interactions caused pathway activation and ligand degradation.13 In our study, confirmatory evidence was provided of both a direct effect of NEURL1 on the Notch pathway via Jagged1 and consequent pathway inhibition. Interestingly, decreased Jagged1 mRNA expression levels were observed in NEURL1 and ΔRING transfectants, most likely due to the disruption of the Jagged1-positive autoregulatory feedback loop.47
Although NEURL1 has the reverse effects to Notch2 on both tumor growth and target gene expression, it did not have the effect of Notch pathway blockade in inducing neuronal differentiation in DAOY cells.10 This lack of differentiation was previously noted in different cell lines39 and corresponds to the phenotype of Jagged1 loss in the developing cerebellum,48 where increased apoptosis occurs but GCP differentiation is not affected. The difference could be that total Notch blockade affects the aspects of the Notch pathway other than those initiated by Jagged1 signaling. In addition, NEURL1 most probably plays a role in other signaling pathways that also contribute to the final outcome of gene expression. Alternatively, NEURL1 may play a role in the differentiation process alongside other differentiation-associated proteins.
The fact that Notch2 and NEURL1 have opposing effects in MB cell lines is reflected in their expression patterns in primary tumors (Supplementary Material, Fig. S4; GEO expression array data GSE10237).49 In general, where Notch2 is high, NEURL1 is low, and vice versa, suggesting coordinated regulation at the transcriptional level of the 2 genes. From this data, it can also be seen that Notch2 expression is high and NEURL1 expression is low in hedgehog-activated tumors (Supplementary Material, Fig. S5). This confirms the findings from our study where low NEURL1 expression was significantly associated with hedgehog-activated tumors (Table 1), and again recapitulates the situation in GCPs in the developing cerebellum, the apparent cell of origin of hedgehog-activated tumors.2 We would like to note that the possibility that some MBs may arise from a rare cell type that does not express NEURL1 cannot be excluded.
In summary, NEURL1 is expressed in the normal cerebellum but only at low levels in tumors. The observed NEURL1 expression deficits in tumors appear to be functionally significant, as re-expression of the gene in cell lines downregulates the Notch pathway and inhibits growth and tumor-forming capacity. NEURL1 may function as a tumor suppressor gene in MB, at least in part, through the regulation of the Notch pathway. The failure of GCPs to acquire NEURL1 at the proper developmental stage may be a relevant step in MB tumorigenesis.
Funding
This work was supported by NIH grant CA; the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick; and an American Brain Tumor Association Fellowship, in memory of Stephanie Lee Kramer. This work was also supported by NIH grants R01CA109467 and P30 HD018655.
Supplementary Material
Supplementary Material is available at Neuro-Oncology online.
Acknowledgments
With thanks to GA Weinmaster for the Jagged1-HA construct, Juliana Brown for assistance with xenograft experiments, and Laurie Jackson-Grusby for manuscript revisions.
Conflict of interest statement. None declared.
References
- 1.Kim JY, Nelson AL, Algon SA, et al. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev Biol. 2003;263:50–66. doi: 10.1016/s0012-1606(03)00434-2. doi:10.1016/S0012-1606(03)00434-2. [DOI] [PubMed] [Google Scholar]
- 2.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. doi:10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 3.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. doi:10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 4.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. doi:10.1016/S0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 5.Irvin DK, Zurcher SD, Nguyen T, Weinmaster G, Kornblum HI. Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J Comp Neurol. 2001;436:167–181. doi:10.1002/cne.1059. [PubMed] [Google Scholar]
- 6.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. doi:10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Lee SH, Jeong EG, Yoo NJ, Lee SH. Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias. Apmis. 2007;115:1357–1363. doi: 10.1111/j.1600-0463.2007.00751.x. doi:10.1111/j.1600-0463.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 8.Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. doi:10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 9.Hallahan AR, Pritchard JI, Hansen S, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. doi:10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 10.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. doi:10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura H, Yoshida M, Tsuiki H, et al. Identification of a human homolog of the Drosophila neuralized gene within the 10q25.1 malignant astrocytoma deletion region. Oncogene. 1998;16:1009–1019. doi: 10.1038/sj.onc.1201618. doi:10.1038/sj.onc.1201618. [DOI] [PubMed] [Google Scholar]
- 12.Scott DK, Straughton D, Cole M, Bailey S, Ellison DW, Clifford SC. Identification and analysis of tumor suppressor loci at chromosome 10q23.3-10q25.3 in medulloblastoma. Cell Cycle. 2006;5:2381–2389. doi: 10.4161/cc.5.20.3360. [DOI] [PubMed] [Google Scholar]
- 13.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. doi:10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 14.Commisso C, Boulianne GL. The NHR1 domain of neuralized binds delta and mediates delta trafficking and Notch signaling. Mol Biol Cell. 2007;18:1–13. doi: 10.1091/mbc.E06-08-0753. doi:10.1091/mbc.E06-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh E, Dermer M, Commisso C, Zhou L, McGlade CJ, Boulianne GL. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr Biol. 2001;11:1675–1679. doi: 10.1016/s0960-9822(01)00527-9. doi:10.1016/S0960-9822(01)00527-9. [DOI] [PubMed] [Google Scholar]
- 16.Timmusk T, Palm K, Belluardo N, Mudo G, Neuman T. Dendritic localization of mammalian neuralized mRNA encoding a protein with transcription repression activities. Mol Cell Neurosci. 2002;20:649–668. doi: 10.1006/mcne.2002.1148. doi:10.1006/mcne.2002.1148. [DOI] [PubMed] [Google Scholar]
- 17.Keles GE, Berger MS, Srinivasan J, Kolstoe DD, Bobola MS, Silber JR. Establishment and characterization of four human medulloblastoma-derived cell lines. Oncol Res. 1995;7:493–503. [PubMed] [Google Scholar]
- 18.He XM, Wikstrand CJ, Friedman HS, et al. Differentiation characteristics of newly established medulloblastoma cell-lines (D384 Med, D425 Med, and D458 Med) and their transplantable xenografts. Lab Invest. 1991;64:833–843. [PubMed] [Google Scholar]
- 19.Langdon JA, Lamont JM, Scott DK, et al. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes Chromosomes Cancer. 2006;45:47–60. doi: 10.1002/gcc.20262. doi:10.1002/gcc.20262. [DOI] [PubMed] [Google Scholar]
- 20.Lindsey JC, Lusher ME, Anderton JA, et al. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. doi:10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Tan J, Li J, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi: 10.1016/j.ccr.2008.04.019. doi:10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weeraratne SD, Valentine M, Cusick M, Delay R, Van Houten JL. Plasma membrane calcium pumps in mouse olfactory sensory neurons. Chem Senses. 2006;31:725–730. doi: 10.1093/chemse/bjl014. doi:10.1093/chemse/bjl014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caslini C, Capo-chichi CD, Roland IH, Nicolas E, Yeung AT, Xu XX. Histone modifications silence the GATA transcription factor genes in ovarian cancer. Oncogene. 2006;25:5446–5461. doi: 10.1038/sj.onc.1209533. doi:10.1038/sj.onc.1209533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. doi:10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 25.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. doi:10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson PK, Eldridge AG, Freed E, et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. doi:10.1016/S0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. doi:10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutelou E, Sato S, Tomomori-Sato C, et al. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. doi:10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- 29.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. doi:10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 30.Pietersen AM, van Lohuizen M. Stem cell regulation by Polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. doi:10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. doi:10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. doi:10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 34.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. doi:10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. doi:10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. doi:10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 37.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollrath B, Pudney J, Asa S, Leder P, Fitzgerald K. Isolation of a murine homologue of the Drosophila neuralized gene, a gene required for axonemal integrity in spermatozoa and terminal maturation of the mammary gland. Mol Cell Biol. 2001;21:7481–7494. doi: 10.1128/MCB.21.21.7481-7494.2001. doi:10.1128/MCB.21.21.7481-7494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan Y, Tecott L, Jiang MM, Jan LY, Jan YN. Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized. Proc Natl Acad Sci USA. 2001;98:9907–9912. doi: 10.1073/pnas.171321098. doi:10.1073/pnas.171321098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. doi:10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farioli-Vecchioli S, Tanori M, Micheli L, et al. Inhibition of medulloblastoma tumorigenesis by the antiproliferative and pro-differentiative gene PC3. FASEB J. 2007;21:2215–2225. doi: 10.1096/fj.06-7548com. doi:10.1096/fj.06-7548com. [DOI] [PubMed] [Google Scholar]
- 43.Park S, Lee YJ, Lee HJ, et al. B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/smad signaling. Mol Cell Biol. 2004;24:10256–10262. doi: 10.1128/MCB.24.23.10256-10262.2004. doi:10.1128/MCB.24.23.10256-10262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi T, Adachi Y, Sonobe H, Furihata M, Ohtsuki Y. A ubiquitin ligase, skeletrophin, is a negative regulator of melanoma invasion. Oncogene. 2006;25:7059–7069. doi: 10.1038/sj.onc.1209688. doi:10.1038/sj.onc.1209688. [DOI] [PubMed] [Google Scholar]
- 45.Koo BK, Lim HS, Song R, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. doi:10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- 46.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 47.Ascano JM, Beverly LJ, Capobianco AJ. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J Biol Chem. 2003;278:8771–8779. doi: 10.1074/jbc.M211427200. doi:10.1074/jbc.M211427200. [DOI] [PubMed] [Google Scholar]
- 48.Weller M, Krautler N, Mantei N, Suter U, Taylor V. Jagged1 ablation results in cerebellar granule cell migration defects and depletion of Bergmann glia. Dev Neurosci. 2006;28:70–80. doi: 10.1159/000090754. doi:10.1159/000090754. [DOI] [PubMed] [Google Scholar]
- 49.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. doi:10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]