Abstract
Traditional in vitro culturing of tumor cells has been shown to induce changes so that cultures no longer represent the tumor of origin. Serum-free culturing conditions are used in a variety of cancers to propagate stem-like cells in vitro. Limited reports, however, exist on the effects of such propagation. We have compared cells from brain tumor biopsies cultivated under serum-free conditions at passages 2 and 10 to describe the effects of in vitro culturing. We were able to establish cell lines from 7 of 10 biopsies from patients with glioblastoma. The cell lines adapted to conditions and had 2.2 times increased population doubling rate at later passages. Karyotyping and comparative genomic hybridization analysis revealed that all examined cell lines had cytogenetic aberrations commonly found in glioblastomas, and there were only minor differences between tumor and early and late passages in the same culture. Whole-transcriptome analysis shows that tumors had interindividual differences. Changes in the overall expression patterns through passaging were modest, with a significant change in only 14 genes; the variation among cultures was, however, reduced through passages. The ability to differentiate differed among tumors but was maintained throughout passaging. The cells initiated tumors upon transplantation to immunodeficient mice with differing phenotypes, but a given cell culture maintained tumor phenotype after serial cultivation. The cultures established maintained individual characteristics specific to culture identity. Thus, each cell culture reflects an image of the tumor—or a personalized model—from which it was derived and remains representative after moderate expansion.
Keywords: adaptation, brain tumor stem cell, culture media, glioblastoma multiforme, serum-free, tumor sphere
Glioblastoma multiforme is the most common primary brain tumor. It has a dismal prognosis. Despite a multimodal strategy with surgery and chemo- and radiotherapy, the median survival is only 15 months.1 Although a significant effort has been put into understanding the initiation and development of these cancers, little success has been achieved in converting basic science into better treatment options for patients. One of the major obstacles for this progress is the lack of good preclinical models representing more accurately the tumor and its ability to act within a modified microenvironment.
Since Reynolds and Weiss2 established that neural stem cells could be cultivated in vitro in epidermal growth factor (EGF)- and basic fibroblast growth factor (bFGF)-enriched serum-free medium, we and other groups have shown that brain tumor biopsies can be cultivated under similar conditions and that these tumor cells have stem cell characteristics.3–8 Two recent studies showed that the ability to form tumor spheres under serum-free conditions from tumor biopsies is an independent prognostic predictor of clinical outcome.9,10 Similar serum-free culturing conditions have been used to study the stem-like cell populations in breast, pancreatic, sarcomatic, and other tumors.11–13
So far, studies evaluating tumor stem cells have been done after the first formation of tumor spheres4–6 or after serial passages (typically >1514–16). To be able to do more advanced studies (ie, fluorescence-activated cell sorting of small populations, proteomics, and evaluation of epigenetic changes), in vitro propagation would be beneficial for increasing the amount of tumor material. Adaptation to in vitro culturing conditions is, however, a well-described phenomenon. While making the cells easier to propagate, adaptation could affect the phenotype, tumorigenicity, and expression pattern of tumor stem cells. The major concern is that the changes imposed on the cells are such that they may no longer represent the population of tumor cells originally present in situ. For cells cultivated under serum-containing conditions, recent studies have shown that during in vitro expansion patient-derived cells change karyotype, phenotype, and most important, the ability to form invasive, glioma-like tumors upon xenotransplantation.17,18
Studies examining the effect of in vitro propagation in serum-free conditions upon growth rate, karyotype, transcriptome, and ability to differentiate and to induce tumors with cellular heterogeneity have been lacking.9 Herein, we report a comparative analysis of brain tumor stem cells propagated under serum-free conditions at early (passage 2) and late (passage 10) passages.
Materials and Methods
Biopsies and Cell Culturing
Tumor biopsy specimens were obtained from informed and consenting patients, and the tissue harvesting was approved by the Norwegian National Committee for Medical Research Ethics (07321b). Biopsy specimens were put in ice-cold Leibowitz-15 medium (L-15, Invitrogen). Typically, a biopsy specimen was washed in L-15 and mechanically dissociated using 2 scalpels. Dissociation into single cells was achieved by incubation in trypsin-EDTA (Invitrogen) and mechanical dissociation. Thereafter, trypsin was blocked using 2 mg/mL human albumin (Octapharma Pharmazeutika Produktionges) and washed in L-15 twice, before the cells were transferred to a proliferation medium containing bFGF 10 ng/mL, EGF 20 ng/mL (both R&D Systems), leukemia inhibitory factor 10 ng/mL (Millipore, Billerica, MA), B27-supplement 1:50 (Invitrogen), penicillin and streptomycin 100 U/mL of both (Lonza), heparin 1 ng/mL (Leo Pharma), and HEPES 8 mM (Lonza) in Dulbecco's modified essential medium with nutrient mix F-12 and Glutamax (DMEM/F12) (Invitrogen). Cells were then incubated at 105 cells/mL in a 75-cm2 nontreated cell culturing flask (Nunc). After 7–21 days, cells formed tumor spheres, and these were dissociated into single cells using trypsin-EDTA 0.05% for 5 minutes. Cells were then replated at 5 × 104 cells/mL. This dissociation and replating was defined as a passage. When spheres reached a size where the core of the spheres turned dark (70–100 µm), cultures were trypsinized to single cells. Spheres formed after the second passage (P2, termed “early”) were used for a direct comparison with those formed after 10 passages (P10, termed “late”) to elucidate any changes occurring during in vitro culturing. Cells were quantified at each passage using Trypan blue (Sigma-Aldrich) and a hematocytometer (Paul Marienfeld). For differentiation, 4-well plates (BD Biosciences) and glass-bottomed dishes (Corning) were coated with poly-l-ornithine 15 µg/mL (Sigma-Aldrich) for 48 hours. Cells were dissociated into single-cell suspensions and plated with DMEM/F12 containing 4% fetal calf serum (PAA Laboratories), Hepes, penicillin/streptomycin, and B27-supplement with retinoic acid at 1:2000 dilution (Invitrogen). Cells were fixed in 4% paraformaldehyde after 7 days.
Karyotyping
Following the addition of fetal calf serum to the cultured cells, colchicine (Sigma-Aldrich) was added for the last 4 hours of culturing and the cells were harvested according to previously published protocols.19 The chromosomes in the dividing cells were then G-banded, and a karyotype was established in accordance with “An International System for Human Cytogenetic Nomenclature” (ISCN) recommendations.20
High-Resolution Comparative Genomic Hybridization
To evaluate the genomic imbalances in more detail, DNA from tumor, as well as from early and late passages from the same tumors, was isolated using the Maxwell 16 system with the Maxwell 16 tissue DNA purification kit according to the manufacturer's instructions (Promega Corporation). Material from tumors G1 and G2 and the early and late passages of these and G3 were not available for DNA extraction and comparative genomic hybridization (CGH) analysis. CGH was performed on DNA from these samples, and the results were analyzed as described previously.21 Final evaluations of the CGH results used dynamic standard reference intervals as described by Ribeiro et al.22 Data obtained from 10–15 cells were combined to generate average ratio profiles for each chromosome, and aberrations were scored whenever the case profile and the reference profile did not overlap with a significance level of 99%. When analyzing the high-resolution CGH (HR-CGH) data, acrocentric chromosome arms (13p, 14p, 15p, 21p, and 22p) and the Y chromosome were excluded from the analysis, as these contain large repetitive sequences that are known to produce unreliable results. Positive and negative controls were included in each experiment. HR-CGH copy number changes were described according to the recommendations of the ISCN.20
RNA Extraction and Transcriptome Analysis
Cultures forming spheres after P2 and P10 were spun down, and the total RNA was extracted using Qiazol and the RNeasy Kit (Qiagen). RNA was quantified and qualitatively evaluated using Nanodrop (Thermo Scientific) and Bioanalyzer (Agilent Technologies). Five hundred nanograms of RNA of each sample were reverse transcribed and amplified using the NanoAmp RT-IVT Labeling Kit (Applied Biosystems). The DIG-labeled cRNA (10 µg) was fragmented and hybridized to Applied Biosystems Human Genome Survey Microarray V2.0 (Applied Biosystems) according to the manufacturer's protocol (Applied Biosystems Chemiluminesence Detection Kit). The slides were scanned using ABI 1700 Chemiluminescent Microarray Analyzer. Analysis and statistics were done in Microsoft Office Excel 3.0 and J-Express 2.8b (Molmine). After scanning, flagged and control spots were removed leaving 32 869 spots. The signals were log2 transformed and controlled for equal mean and distribution levels before further analysis.
Immunocytochemistry
Differentiated fixed cells were first washed in phosphate-buffered saline (PBS) and blocked in 1% bovine serum albumin (BSA) solution for 1 hour at room temperature, before being incubated in primary antibody overnight at 4°C. The antibodies used were reactive against β3-tubulin (mouse, 1:1000, Sigma-Aldrich), CD133 (mouse, 1:10, Miltenyi Biotec), CNPase (mouse, 1:200, Millipore), CXCR4 (goat, 1:50, Santa Cruz Biotechnology), glial fibrillary acidic protein (GFAP; rabbit, 1:1000, Dako), human-specific Nestin (mouse, 1:500, R&D systems), Tenascin C (goat, 1:50, Santa Cruz), maternal embryonic leucine zipper kinase (MELK; goat, 1:50, Santa Cruz), Sox2 (mouse, 1:200, R&D systems), and Vimentin (rabbit, 1:50, Thermo Fischer Scientific). After washing, cultures were incubated with secondary antibodies for 1 hour at 4°C (anti-rabbit Alexa 488, 1:500, Molecular Probes, Invitrogen; anti-mouse-Cy3 or anti-goat-Cy3, 1:500, Jackson Immunoresearch). Nuclear staining was done with Hoechst 33358 (1:5000, Sigma-Aldrich). Analysis and image acquisition were done on an Olympus BV 61 FluoView confocal microscope (Olympus).
Transplantation and Tissue Processing
All animal procedures were approved by the National Animal Research Authority. C.B.-17 severe combined immunodeficient (SCID) mice (7–9 weeks old, Taconic, Ejby, Denmark) were anesthetized with a subcutaneous injection of Hypnorm (10 mg/mL fluanisone and 0.315 mg/mL fentanyl citrate; Veta Pharma) and Dormicum (5 mg/mL midazolam; Roche) and placed in a stereotactic frame (David Kopf Instruments). Immediately prior to transplantation, suspensions of single cells were prepared in the L-15 medium. A 2-µL suspension containing 100 000 cells/µL was injected into the right striatum just below the corpus callosum using a cannula (Plastic One) attached to a Hamilton syringe (Hamilton Bonaduz). The needle was left in situ for 2 minutes after injection before being slowly removed. The mice were killed by transcardial perfusion with 4% buffered formaldehyde after 12 weeks or sooner if weight loss or neurological symptoms developed. The fixed brains were cut into 10-µm sections on a freezing microtome (Leica), thawed onto Super Frost/Plus glass slides (Menzel-Gläser), and stored at −20°C. Brain sections were stained with hematoxylin and eosin (H&E). Immunohistochemistry on sections was performed as described previously.7 Briefly, sections were air dried at room temperature for 1 hour, rehydrated in PBS and blocked with a solution of PBS containing 1% BSA, 0.3% Triton X-100, and 0.1% sodium azide (all Sigma-Aldrich) for 1 hour at room temperature. The H&E-stained sections were analyzed using a Zeiss Axioskop 2 microscope (Zeiss), and the immunolabeled sections were studied under an Olympus BV 61 FluoView confocal microscope (Olympus).
Results
Cell Lines Were Efficiently Established from Human High-Grade Astrocytomas; They Differed in Growth Rate and Adapted to In Vitro Expansion
Ten high-grade glioma samples were cultivated, and we were able to passage 7 of these for at least 10 generations of tumor spheres. These 7 cell lines were named G1–G7 and all were cultivated beyond passage 20 without changes in the phenotype of the spheres and without signs of diminished ability to re-establish spheres after dissociation. The median number of cells available at P2 was 1.6 × 106 cells (range: 0.8–70 × 106 cells).
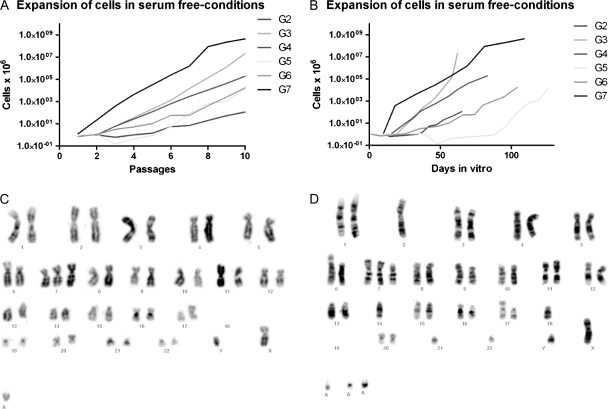
At early passages, the cultures contained single cells not included in spheres, but after further passages most single cells disappeared and almost all cells in culture were contained within spheres. The morphology of spheres was very similar between cell lines, but they varied highly in their ability to expand in number. Whereas some tumors could grow exponentially from the time of isolation (G7), the others acquired similar growth kinetics only after being kept in vitro for quite some time (G5). The growth rate at passages 2–4 and passages 8–10 was significantly different (mean ± SEM; 3.7 ± 1.7 and 8.3 ± 2.2 cell doublings per passage, respectively; paired samples t-test, P = .018) (Fig. 1). This suggested that even a lower number of passages will allow for the adaptation of cells to in vitro conditions.
Fig. 1.
Expansion rates of brain tumor stem cells in culture. Cultures expanded at exponential rates between passages (A), but the time between passages was different among the tumors (B). Karyograms from early (C) and late (D) passages of tumor G4.
Cell Culturing Demonstrated Karyotypic Variation Between Tumors, but the Chromosomal Aberrations Were Maintained Through Culturing
A karyotype was successfully established from cultures G3, G4, G6, and G7 (Fig. 1, Supplementary Material, Table S2). All cell lines had an aberrant chromosome constitution, but no chromosomal aberrations were common to them. In the 3 cultures (G4, G6, and G7) where both early and late passages were examined, the chromosomal aberrations were very similar in early and late passages. Several identifiable aberrations were found, but the number of abnormalities involving at least partly unidentifiable chromosomal material was higher. Some tumor cell heterogeneity was found, that is, all aberrations were not found in all cells examined. From 1 culture (G3), mitotic chromosomes could be obtained only from the late passage and a very aberrant chromosome constitution with mostly unrecognizable acrocentric chromosomes was found.
The gain/loss profiles for all tumors and corresponding early and late passages were very similar as analyzed by HR-CGH (Supplementary Material, Fig. 1 and Table S3). Although the scored gained and lost regions were not entirely identical within different samples from the same tumor (tumor, early and late passages), the profiles looked very much alike. Because of the large number of not fully characterizable chromosomes by karyotyping, the comparison of HR-CGH results for each sample with the corresponding karyotype was hard. Nonetheless, the gain of (parts of) chromosome 7 in all samples and the losses of (parts of) chromosome 10, parts of chromosome arm 4q, and parts of chromosome arm 9p correspond well with the karyotypic findings. Furthermore, the extremely aberrant late passage of tumor 3 was found to have a correspondingly large number of gained and lost regions as analyzed by HR-CGH.
Whole-Transcriptome Analysis Indicated Interindividual Differences: Changes Through Passaging Were Not Significantly Different, but the Variation Was Reduced
Purified total RNA from G2–G7 cell cultures was analyzed using an Applied Biosystems microarray system for transcriptome analysis. From these results, we examined heterogeneity between cultures derived from different patients and whether variation between cultures was maintained upon in vitro culturing. Further, we analyzed significant changes in all cultures at P2 and P10.
The overall expression levels did not vary significantly among individual cultures at P2 or P10 (ANOVA, F = 0.66 and 0.38, critical F = 2.098, P = .68 and .89); thus, no individual cell cultures were found to differ statistically from the others regarding the overall hybridization signal at the given passage. When comparing the overall expression levels at early and late passages in individual cultures, however, culture G3 was found to have a significant reduction in the overall expression level upon cultivation (Student's t-test, 2-tailed, paired, P = 2.3 × 10−5). The overall expression-level changes in the other cell lines were not significantly different (P > .05).
To analyze whether interindividual variation is more profound in the early than the late passages, the mean variance of individual probes was analyzed at the 2 time points. This showed that the mean variance was higher at P2 than at P10 and that this difference was significant (F-test and ANOVA, P < .0001).
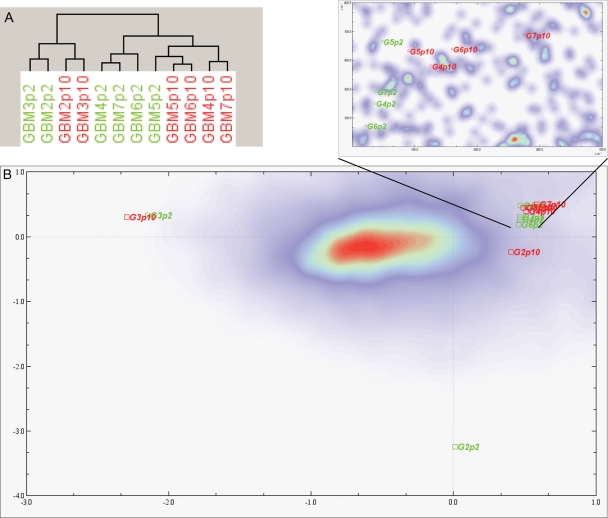
Unsupervised hierarchical clustering showed that G2 and G3 grouped together, but no clustering was apparent for early and late passages (Fig. 2). Also, with all 6 cultures compared by paired analysis, there was a high correlation of early and late expression levels (r2 = .96). This was confirmed using correspondence analysis, where all samples clustered together except for G2 and G3 that were found segregated from the others. For G3, both P2 and P10 grouped together, at a distance from the main cluster. With regard to the G2 culture, only the P2 sample showed a high distance on the correspondence analysis plot, whereas sample P10 was closer to the main cluster (Fig. 2B).
Fig. 2.
Unsupervised hierarchical clustering of transcripts of 6 cell cultures at early (P2) and late (P10) passages. Samples G2 and G3 cluster away from the other samples (A). Correspondence analysis showing the clustering of samples G2 and G3 away from the other samples. For the G3 culture, P2 and P10 samples group together. For the G2 culture, P10 clusters more closely to the remaining 4 cultures than the sample at P2 (B). An enhanced area from (B) shows that in this analysis P2 and P10 cluster separately.
We then identified the differentially expressed genes at P2 and P10 first by pairwise comparison of all 6 samples. Using a cutoff for q-value of <20, we revealed 22 differentially expressed genes within the samples by 2 class-paired significance analysis for microarray (SAM) analysis. By the rank product test, 36 genes were found significant differentially expressed (q-value < 0.05). The 14 genes overlapping in both tests are presented in Table 1. To increase the sensitivity, outlier samples G2 and G3 were removed from the analysis. This unmasked 92 and 93 genes by SAM and rank product, accordingly, using the same α-values. All the 14 genes discovered comparing all 6 samples were found by both analyses, and an additional 18 genes were found for these 4 samples using both SAM and rank product (Supplementary Material, Table S3).
Table 1.
Differentially regulated genes at early (P2) and late (P10) passages in 6 different cell cultures
| Short name | Fold change | Gene id | |
|---|---|---|---|
| Downregulated | |||
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 26.94 | hCG16977.3 |
| Microsomal glutathione S-transferase 1 | MGST1 | 17.38 | hCG25167.4 |
| Integrin β4 | ITGB4 | 10.62 | hCG27538.2 |
| Lymphocyte antigen 96 | LY96 | 9.65 | hCG21314.3 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 8.00 | hCG29298.3 |
| G protein-coupled receptor 109A | GPR109A | 7.75 | hCG2016371.1 |
| Major histocompatibility complex, class II, DR β1 | HLA-DRB1 | 7.64 | hCG1791131.3 |
| Neurofascin homolog (chicken) | NFASC | 6.84 | hCG2025385 |
| Chemokine (C-X-C motif) ligand 14 | CXCL14 | 6.83 | hCG39697.3 |
| 2′-5′-Oligoadenylate synthetase 2, 69/71 kDa | OAS2 | 6.81 | hCG38536.2 |
| Guanylate binding protein 1, interferon-inducible, 67 kDa | GBP1 | 6.16 | hCG1774285.4 |
| Null | DKFZP566N034 | 6.10 | hCG2006852 |
| Null | RAFTLIN | 5.83 | hCG27653.3 |
| Upregulated | |||
| PTPRF interacting protein, binding protein 2 (liprin β2) | PPFIBP2 | 3.81 | hCG22748.3 |
Genes significantly different both by SAM (q-value < 20) and rank product (q-value < 0.05) analyses. Fold-change differences at late vs early passages.
The most significant change was the reduction in the expression levels at P10 compared with P2. Only the relatively uncharacterized protein tyrosine phosphatase, receptor-type, F interacting protein, binding protein 2 (liprin beta 2) was significantly upregulated.23 Differentially expressed genes were associated with function in immune response (11 genes), metabolism (8 genes), cell adhesion/motility (7 genes), cell proliferation/growth (5 genes), and apoptosis (3 genes). Specifically, the neural lineage-related genes GFAP and integrin β4 were downregulated.
Ability to Differentiate Differed Among Tumors, but Was Maintained Throughout Passaging
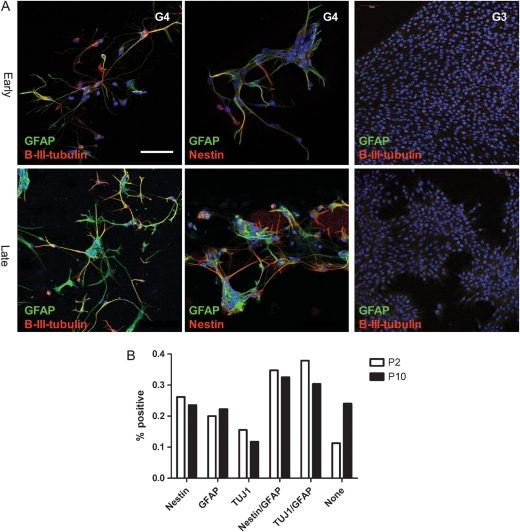
To examine the multilineage potential of the cells at P2 and P10, cells were transferred to differentiating conditions by removal of mitogens and addition of serum. Upon differentiation, the cells adhered and showed a reduction in proliferation. Similar to earlier reports,3,5,7 cells had aberrant morphology and staining patterns. Nuclear staining showed a wide variety in nuclear size and morphology, with many cells containing multiple nuclei. Cells had chaotic arborizations and tortuous processes, but stained concomitantly for the stem cell marker (nestin) and the markers of neuronal (β3-tubulin) and glial (GFAP) differentiation, a phenotype not present in differentiated normal stem cells (Fig. 3).2,7,24 As described previously,7 differentiated cells were negative for oligodendrocyte markers (CNPase, not shown). For culture G3, no certain differentiation was present at either early or late passage. In the other 5 samples, cells were able to differentiate into neuronal, glial, and combined phenotypes at early as well as late passages. There were no significant changes in the population of immunopositive cells at P2 and P10 overall. However, the morphologies of differentiated cells differed between early and late passage. Early passages had more cells adhering with a more arborized morphology. Later passage cells tended to grow more in clusters, producing less widespread branching of cells.
Fig. 3.
Differentiation of G4 at early (P2) and late (P10) passages of G3 at P2 (A). Cells were stained with antisera against GFAP (green), β3-tubulin (red: left and right column), and nestin (red: middle column). Scale bar, 50 µm. Quantification of GFAP, β3-tubulin, double-positive cells, and nonstaining cells after differentiation at early (P2, white) and late (P10, black) passages (B).
Tumors Formed Upon Xenotransplantation Differed Among Patients, but the Tumor Phenotype Was Maintained Throughout Passaging
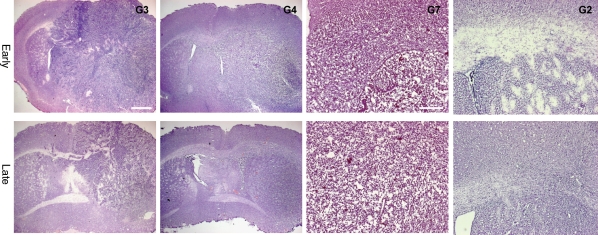
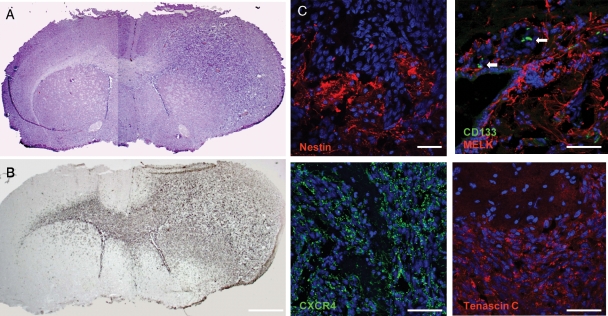
To test for the ability to reinitiate tumor growth in vivo, cells were orthotopically transplanted to the putamen of SCID mice. In all cases, transplanted cells formed tumors. Tumors all grew with an infiltrative pattern, with the main tumor bulk around the site of injection. Tumor cells infiltrated within white matter tracts, but some tumor cells were also found in grey matter. The tumors contained neovascularization, but the areas of necrosis were scarce and did not show pseudopalisading necrosis (Fig. 4). Tumor cells had atypical and pleomorphic nuclei, and such abnormal nuclei were found in subventricular zones both ipsi- and contralateral to the site of injections. The tumors contained cells immunopositive for the markers of reactive astrocytes (GFAP, vimentin, MELK) and stem cells (nestin, Sox2, tenascin C; Fig. 5). Weak CD133-positive staining was found along the ependymal layer of the ventricle walls, whereas strongly positive cells were found in cluster-like formation in the vicinity of the subventricular zone.
Fig. 4.
Comparison of tumors developed in SCID mice after transplantation of early (P2; top) or late (P10; bottom) passaged cells in cultures G3, G4, G7, and G2. Scale bars, 1 mm (G3 and G4) and 0.5 mm (G7 and G2).
Fig. 5.
Tumors developed in SCID mice after transplantation of 200 000 cells (A, H&E), harboring highly invasive cells crossing the corpus callosum and over into the contralateral hemisphere (B, vimentin, DAB-staining). Tumors were immunopositive for human-specific nestin (red: top left) (C), maternal embryonic zipper leucine kinase (MELK) (red), CD133 (green, white arrows) (top right), CXCR4 (green: bottom left), and tenascin C (red: bottom right). Nuclei (Hoechst, blue). Scale bars, 1 mm (A and B) and 130 µm (C).
To confirm the tumor-initiating properties of the cultured cells, serial transplantation for 3 generations was performed. Between each generation of in vivo tumor initiation, the sphere-forming ability was confirmed by 2 passages in vitro before new orthotopic transplantation was performed. These transplantations produced tumors similar to the primary xenotransplant, with a similar invasive phenotype, nuclear atypia, and immunopositivity for reactive astrocyte and stem cell markers.
The tumors' phenotypes differed between the individual tumor cultures; that is, G2 had a diffusely invasive phenotype, whereas G3 contained spindle-like cells, the presence of extracellular matrix, and compact tumor bulk. The comparison of transplanted cells from early and late passages indicated that the established tumors had maintained their original phenotype (Fig. 4). For 5 of the cultures, the SCID mice were sacrificed at 12 weeks both for cells transplanted from late and early transplants; only for G3 did the mice develop symptoms before this time point. In these animals, the survival was different in early (6 and 10 weeks) and late passages (all died after 3 weeks).
Discussion
To our knowledge, this report is the first studying the early effects of in vitro cultivation of tumor stem cells. We show that in vitro expansion in serum-free conditions is possible in a high fraction of glioblastoma tumor samples and that this in vitro propagation can produce a large number of heterogeneous cells. Although the overall chromosomal organization is stable, the expression heterogeneity of samples is slightly reduced. Even so, the differentiation potential and the ability to form heterogeneous tumors upon orthotopic xenografting are not changed.
The rate of successful propagation of biopsies as free-floating spheres has been variable (12 of 24 samples,3 19 of 40 samples,15 11 of 15 samples [4 of these 15 were as adherent cultures],14 11 of 15 samples,9 14 of 44 samples,10 and 0 of 16 samples beyond passage 3).25 The results presented here, with multiple passages possible in 7 out of 10 samples, are comparable with these reports.
After dissociation of a tumor biopsy, the sample contains a large number of cells, but only a very limited number of these cells take part in further sphere formation. The majority of the cells present in the original tumor biopsy do not survive passaging and serum-free cultivation. Whereas the number of cells is reduced through the first passages, the fraction of cells taking part in sphere formation is increased compared with single and adherent cells. After ∼2 passages, cells within spheres dominate the culture, and this allows for the use of all cells in the culture for studying the sphere-forming cells. The critical transition for adaptation to in vitro conditions when culturing adult human neural stem cells and brain tumor samples is, based on our experience and that of others, probably around passages 3–5.7,24–26 Therefore, a culture well beyond this passage number, but still at a lower number of passages when compared with the established cell lines, was chosen (passage 10). This presumption fit with the observed changes in the growth rate at early compared with late passages. To our knowledge, this is the first report comparing cell adaptation to in vitro conditions at such early passages on more than expression levels.17 Optimally, all examinations should be undertaken of all samples both at early and late passages, but due to the lower number of cells at P2, this has not been possible.
The genomic aberrations differed among cultures, but we found that the distinct aberrations of the individual cultures were quite stable when cultured. Cultures were found to be representative of the tumor of origin and were stable from early to late passages evaluated by karyotyping and HR-CGH. This implies that additional genomic changes are not selected for through propagation. Our data thereby contrast with findings in established serum-propagated glioma cell lines where there is, through culturing, a selection of cells with chromosomal aberrations not common in glioblastomas, but the accumulation of aberrations typically found in other serum-propagated cell lines from other types of tumors.18 Such conservation of cytogenetic changes present in the parent tumor under propagation in serum-free conditions has been described previously by others.17,27
When passaged, the cultures adapt to the in vitro conditions and changes in the overall transcriptome are seen. These changes are minor compared with the differences found between adult human neural stem cells and tumor stem cells (>3400 genes differentially regulated, Sandberg et al., in preparation), but several important changes occur. In most samples, there was no reduction in total hybridization signal, but in G3, there was a marked reduction in signal, showing that in vitro passaging also can affect this. Upon passaging, the variation in the transcriptome among cultures is reduced, causing less diversity among cell cultures. The reduction in intrasample phenotypic heterogeneity (variance) is followed by a similar reduction in intersample heterogeneity demonstrated by the closer resemblance of G2 to the cluster of G4–G7 at P10 than G2 at P2 (correspondence analysis). The consequence of such a reduced variability could be that cultures are more easily targeted in preclinical trials, thus underestimating tumor plasticity and its ability to resist chemo- and radiotherapy when novel therapeutics are translated to the clinical setting. The adaptation to in vitro sphere-forming conditions by a reduced expression of genes related to adhesion, invasion, and immune defense is similar to the differences between adherent and sphere-forming tumor stem cell cultures shown earlier.14,17 Of particular interest is the reduction in genes related to neural stem cells and lineage differentiation, such as β4-integrin, neurofascin, GFAP, and secreted phosphoprotein 1 (osteopontin), and we speculate that the reduction in these genes may be part of a transition toward a more mesenchymal phenotype.14,28–30
The effect of in vitro propagation on the ability of neural stem cells to differentiate has been described, but with conflicting results. Whereas 1 group31 found a stable distribution of phenotype throughout passaging, a reduction in the frequency of neuronally differentiated cells has been shown by others.32 The data presented here show that in vitro culturing has a limited effect on cellular fractions as there is no orderly change in the differences. The ability to differentiate is present even after repeated passages. Also, the 1 sample with a very limited capability for differentiation, G3, had similar ability both at P2 and P10. This fits with the more mesenchymal phenotype of the xenotransplant. Thus, even though it failed to express neural lineage-specific differentiation, it was able to produce a heterogeneous tumor cell population in vitro. The ability of serum-free propagation of biopsy specimens to represent the tumor of origin is further supported by recent data showing the maintenance of a specific gliosarcoma phenotype under these conditions.33
With a possible effect on the differentiation properties of the tumors, it is interesting that the phenotype is preserved for any given culture between cells transplanted at P2 and P10. Specifically, all cultures maintained the propensity to produce infiltrating tumors. These tumors3,17 much more convincingly recapitulate this important phenotypic characteristic of glial tumors than traditionally established cell lines do, even after the transition of these lines into a serum-free medium.34,35 However, although highly invasive and containing pathological vessels, these tumors did not contain the typical necrosis or pseudopalisading often found in glioblastomas.
At both early and late passages, there was a high variability among cultures derived from different patients. In these 7 cultures, G3 diverges from the others having a more complex karyotype, revealing singular clustering in transcriptome analysis, having a reduced ability to differentiate into neural lineages, and producing a tumor with a more immature phenotype. This culture may represent a hypermutated subtype such as found after temozolomide treatment, although this patient had not undergone such a treatment.36,37
Overall, the 7 cultures established maintained their specific characteristics separating the different cultures from each other, that is, the differences in growth rate, karyotype, transcriptome, differentiation abilities, and tumor phenotype were overall more similar at P2 and P10 than among different tumors. Thus, each of the cultures reflects an image—or a personalized model—of the tumor from which it was derived. The serum-free in vitro cultivation makes patient-specific cultures possible, but even at these low passage numbers all cultures show moderate changes. Therefore, findings in high-passage primary cultures should ideally be confirmed in low-passage cultures. Also, because there seems to be heterogeneity among brain tumor stem cell cultures, multiple samples should be tested to ensure extrinsic biological validity for any therapeutic approach.
Supplementary Material
Supplementary Material is available at Neuro-Oncology online.
Funding
Departments of Neurosurgery at Oslo University Hospital, the South-Eastern Norway Regional Health Authority, and the Research Council of Norway through the Cancer Stem Cell Innovation Center (CAST-SFI).
Acknowledgments
We are grateful for the support from Professor Ola Myklebost, Department of Tumor Biology, Oslo University Hospital, as well as excellent technical assistance from Thea Charlotte Smedsrud, Norwegian Microarray Consortium, Department of Molecular Bioscience, University of Oslo, and Elin Kampenhaug, Institute for Surgical Research, Oslo University Hospital. We would like to thank Sissel Reinlie, Head of Department of Neurosurgery, and Professor Ansgar Aasen, Director of Institute for Surgical Research, Oslo University Hospital, for excellent working conditions.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. doi:10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. doi:10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 3.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. doi:10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 4.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. doi:10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. doi:10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 6.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 7.Varghese M, Olstorn H, Sandberg C, et al. A comparison between stem cells from the adult human brain and from brain tumors. Neurosurgery. 2008;63:1022–1033. doi: 10.1227/01.NEU.0000335792.85142.B0. doi:10.1227/01.NEU.0000335792.85142.B0. [DOI] [PubMed] [Google Scholar]
- 8.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. doi:10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 9.Laks DR, Masterman-Smith M, Visnyei K, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. doi:10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pallini R, Ricci-Vitiani L, Banna GL, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. doi:10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hajj M, Wicha MS, ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. doi:10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs CP, Kukekov VG, Reith JD, et al. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7:967–976. doi: 10.1593/neo.05394. doi:10.1593/neo.05394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo S, Liu T, Wang C, et al. Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas. 2007;34:429–435. doi: 10.1097/MPA.0b013e318033f9f4. [DOI] [PubMed] [Google Scholar]
- 14.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. doi:10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 15.Gunther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. doi:10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 16.Salmaggi A, Boiardi A, Gelati M, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54:850–860. doi: 10.1002/glia.20414. doi:10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. doi:10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. doi:10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 19.Mandahl N. Methods in solid rumors cytogenetics. 1992. pp. 155–187. In: Rooney DE, Czepulkowski BH eds. Human Cytogenetics: A Practical Approach. Vol. 2, Malignancy and Acquired Chromosome Abnormalities. Eynsham, Oxon: IRL Press. [Google Scholar]
- 20.Schaffer LG, Tommerup N. ISCN: an International System for Human Cytogenetic Nomenclature. 2005 Basel, Switzerland: S. Karger. [Google Scholar]
- 21.Teixeira MR, Ribeiro FR, Eknaes M, et al. Genomic analysis of prostate carcinoma specimens obtained via ultrasound-guided needle biopsy may be of use in preoperative decision-making. Cancer. 2004;101:1786–1793. doi: 10.1002/cncr.20527. doi:10.1002/cncr.20527. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro FR, Jeronimo C, Henrique R, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–3970. doi: 10.1158/1078-0432.CCR-05-1977. doi:10.1158/1078-0432.CCR-05-1977. [DOI] [PubMed] [Google Scholar]
- 23.Serra-Pages C, Kedersha NL, Fazikas L, Medley Q, Debant A, Streuli M. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 1995;14:2827–2838. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moe MC, Varghese M, Danilov AI, et al. Multipotent progenitor cells from the adult human brain: neurophysiological differentiation to mature neurons. Brain. 2005;128:2189–2199. doi: 10.1093/brain/awh574. [DOI] [PubMed] [Google Scholar]
- 25.Fael Al-Mayhani TM, Ball SL, Zhao JW, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Varghese M, Olstorn H, Berg-Johnsen J, Moe MC, Murrell W, Langmoen IA. Isolation of human multipotent neural progenitors from adult filum terminale. Stem Cells Dev. 2009;18:603–613. doi: 10.1089/scd.2008.0144. doi:10.1089/scd.2008.0144. [DOI] [PubMed] [Google Scholar]
- 27.Ernst A, Hofmann S, Ahmadi R, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res. 2009;15:6541–6550. doi: 10.1158/1078-0432.CCR-09-0695. doi:10.1158/1078-0432.CCR-09-0695. [DOI] [PubMed] [Google Scholar]
- 28.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. doi:10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Rieske P, Golanska E, Zakrzewska M, et al. Arrested neural and advanced mesenchymal differentiation of glioblastoma cells-comparative study with neural progenitors. BMC Cancer. 2009;9:54. doi: 10.1186/1471-2407-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tso CL, Shintaku P, Chen J, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–619. doi: 10.1158/1541-7786.MCR-06-0005. doi:10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 31.Foroni C, Galli R, Cipelletti B, et al. Resilience to transformation and inherent genetic and functional stability of adult neural stem cells ex vivo. Cancer Res. 2007;67:3725–3733. doi: 10.1158/0008-5472.CAN-06-4577. doi:10.1158/0008-5472.CAN-06-4577. [DOI] [PubMed] [Google Scholar]
- 32.Machon O, Backman M, Krauss S, Kozmik Z. The cellular fate of cortical progenitors is not maintained in neurosphere cultures. Mol Cell Neurosci. 2005;30:388–397. doi: 10.1016/j.mcn.2005.08.003. doi:10.1016/j.mcn.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Decarvalho AC, Nelson K, Lemke N, et al. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28:181–190. doi: 10.1002/stem.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappa G, Mercapide J, Anzanello F, et al. Growth of cancer cell lines under stem cell-like conditions has the potential to unveil therapeutic targets. Exp Cell Res. 2008;314:2110–2122. doi: 10.1016/j.yexcr.2008.03.008. doi:10.1016/j.yexcr.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu SC, Ping YF, Yi L, et al. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124–134. doi: 10.1016/j.canlet.2008.02.010. doi:10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. doi:10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]