Abstract
Background
Determination of HIV-1 tropism is a pre-requisite to the use of CCR5 antagonists. This study evaluated the potential of population genotypic tropism tests (GTTs) in clinical practice, and the correlation with phenotypic tropism tests (PTTs) in patients accessing routine HIV care.
Methods
Forty-nine consecutive plasma samples for which an original TrofileTM assay was performed were obtained from triple-class-experienced patients in need of a therapy change. Viral tropism was defined as the consensus of three or more tropism calls obtained from the combination of two independent population PTT assays (Trofile Biosciences, San Francisco, CA, USA, and Virco, Beerse, Belgium), population GTTs and GTTs based on ultra-deep sequencing. If no consensus was reached, a clonal PTT was performed in order to finalize the tropism call. This two-step approach allowed the definition of a reference tropism call.
Results
According to the reference tropism result, 35/49 samples were CCR5 tropic (R5) (patients eligible for maraviroc treatment) and 14/49 were assigned as non-R5 tropic. The non-R5 samples [patients not eligible for maraviroc treatment according to the FDA/European Medicines Agency (EMEA) label] group included both the CXCR4 (X4) samples and the dual and mixed CCR5/CXCR4 (R5/X4) samples. Compared with TrofileTM population PTTs, population GTTs showed a higher sensitivity (97%) and a higher negative predictive value (91%), but almost equal specificity and an equal positive predictive value.
Conclusions
In line with recent reports from clinical trial data, our data support the use of population genotypic tropism testing as a tool for tropism determination before the start of maraviroc.
Keywords: HIV-1 tropism, genotypic tests, phenotypic tests
Introduction
Although more than 25 antiretroviral drugs within six different classes have been approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMEA), current treatment options against HIV-1 remain compromised by broad drug cross-resistance and short- and long-term toxicity.1–3 The finding that several commonly prescribed antiretrovirals, including didanosine, abacavir and ritonavir-boosted protease inhibitors, may be associated with increased cardiovascular risk underscores the need for drugs with an improved safety profile.4,5 Entry of HIV-1 into lymphocytes and monocytes requires binding of the envelope glycoprotein (gp120) to the CD4 receptor, followed by interaction with one of two main co-receptors, CCR5 or CXCR4. The preferential use of CCR5 or CXCR4, called viral tropism, is mainly determined by the amino acid sequence of the V3 region of gp120, although regions outside V3, such as V1/V2, C4 and the bridging sheet, may also be involved.6 Strains that use the CCR5 co-receptor are called CCR5-tropic or R5 viruses, those that use CXCR4 are labelled as CXCR4-tropic or X4 viruses. Dually tropic strains can use both CCR5 and CXCR4 and are known as R5/X4 viruses. As the CD4 cell count starts to drop in patients with late stage HIV-1 infection, the prevalence of X4 or R5/X4 strains rises within the viral quasi-species, and starting from a minority population, these strains can finally emerge as a majority population.7
Maraviroc is the first CCR5 antagonist to receive FDA and EMEA approval. Determination of viral tropism is a prerequisite to the use of these CCR5 antagonists. Several techniques to determine HIV-1 tropism have been developed over the years, including phenotypic and genotypic methods, which show varying degrees of correlation in direct comparisons. A diagnostic gold standard is currently lacking. Among the phenotypic tropism tests (PTTs), the Trofile™ assay (Monogram Biosciences, San Francisco, CA, USA) in its original version was used for the prospective recruitment of patients within clinical trials.8 The original Trofile™ assay (OTA) allowed detection of CXCR4-using strains with a sensitivity approaching 100% at a prevalence of ≥10% of the quasi-species in clonal mixtures.9 A more sensitive assay version, the ‘enhanced sensitivity Trofile™ assay’ (ESTA) increased the sensitivity of detection down to 0.3%, again based on mixtures of viral clones.10 Another PTT has been developed by Virco (Beerse, Belgium) for research purposes only.11 This assay was able to detect CXCR4-using strains in clinical samples when present at ≥10% of the quasi-species.12 It should be noted, however, that the optimal sensitivity of detection of CXCR4-using virus for effectively guiding clinical practice has not been conclusively determined.
Co-receptor use can also be predicted based on the amino acid sequence of the gp120 V3 loop using several publicly available interpretation algorithms.13,14
Among these, Geno2Pheno[coreceptor] uses a support vector machine statistical learning model. Geno2Pheno has been designed by taking mainly genetic and phenotypic information derived from HIV-1 subtype B into consideration. The recent introduction of the 454 technology for ultra-deep sequencing offers a new opportunity for characterizing the viral quasi-species by generating several thousands of sequences from each sample,15–17 thus allowing the fine characterization of drug-resistance patterns.15,18
According to the FDA and EMEA labels, maraviroc can be used only in patients with R5 virus. In the registration trials, the OTA was used for PTTs. The OTA, however, is no longer available, as it was replaced by the ESTA in July 2008. Current consensus from the European, British and German–Austrian guidelines for tropism testing indicate that tropism determination can be performed by a PTT or genotypic tropism test (GTT).19–21 The supporting evidence for the GTT as an acceptable alternative to the PTT, and for ESTA as a valid PTT assay, has been mainly derived from retrospective evaluations of patients enrolled in clinical trials.10,22,23 The goal of this study was to evaluate the usefulness of an academic developed population GTT in comparison with the OTA (assay available at the time of sampling) in an independent dataset in routine clinical practice.
Methods
Study subjects and clinical samples
Plasma samples were collected from 49 patients eligible for maraviroc initiation, for which an OTA was performed. Patients were selected from the maraviroc expanded access programme in three Belgian AIDS Reference Centres (Institute for Tropical Medicine, Antwerp; Centre Hospitalier Universitaire Saint-Pierre, Brussels; and University Hospital, Liege), the Royal Free Hospital HIV Centre and Wharfside Clinic, St Mary's Hospital (London, UK) between 1 October 2007 and 31 December 2008. Patients were triple-class experienced, with advanced HIV disease (median CD4 nadir count 133 cells/mm3) and with a median HIV-1 RNA load of 80 000 copies/mL at the time of tropism testing. Patients were included within the maraviroc expanded access programme (http://clinicaltrials.gov/ct2/show/NCT00426660). Optimized background regimens were guided by drug resistance testing. Informed consent was obtained from all patients enrolled, and the study was approved by the Ethics Committee of the University Hospital, Ghent, Belgium. Samples were stored in aliquots at −80°C until further processing. Plasma HIV-1 RNA was quantified with the Amplicor HIV Monitor v1.5 test (Roche Diagnostics Systems, Basel, Switzerland) with a lower limit of detection of 50 RNA copies/mL.
Definition of the reference tropism result (RTR)
The RTR was defined in two steps. In the first step, the RTR criteria were met if the combination of four different assays showed concordance between at least three of the four assays. The four assays comprised: (i) the Virco population PTT (VpPTT), which uses the NH2-V4 fragment of gp120 on the Antivirogram® platform; (ii) the OTA, which uses the gp160 envelope gene on a PhenoSense platform; (iii) GTT by ultra-deep pyrosequencing on the 454 platform (UDSGTT) (454 Life Sciences, Branford, CT, USA), combined with interpretation provided by the Geno2Pheno bio-informatic tool (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php); and (iv) GTT by population sequencing (PGTT), combined with the Geno2Pheno bio-informatic tool. In the second step, discordant samples that did not reach the criteria of three or more concordant results were analysed using clonal PTT of up to 24 clones in order to allow a final tropism call.
According to the RTR, results were reported as R5 (patients eligible for maraviroc treatment) and non-R5 tropic (patients not eligible for maraviroc treatment according to the FDA/EMEA label). The non-R5 group included the CXCR4 (X4) and the dual and mixed tropic samples.
V3 haplotype
The term V3 haplotype was used to describe a viral population representing individual viruses with an identical V3 sequence fragment.
VpPTT
The assay was performed as described by Van Baelen et al.11 Following RNA extraction, the gp120 NH2-V4 region was amplified in a one-step RT–PCR. The purified NH2-V4 amplicons were recombined with an HIV-1 backbone deleted for gp120 NH2-V4. Recombinant plasmids were transfected into 293T cells. The generated virus stocks were used for infection of U87-CD4-CXCR4 and U87-CD4-CCR5 cells. Infection was evaluated by fluorescence readout. Results were reported as R5, non-R5 or failure.
Virco clonal phenotypic tropism testing
The assay was performed essentially as the VpPTT assay, but the starting material consisted of plasmids containing a clonal amplicon of NH2-V4. Results were reported as R5, X4 or dual tropic (D), or not viable.
OTA
Clinical samples were collected as part of routine patient management and sent to Monogram Biosciences for tropism testing.8 Results were reported as R5, non-R5 or failure.
UDSGTT
The assay was performed as described by Vandenbroucke et al.24 In summary, viral RNA was extracted from 256 µL of plasma and reverse transcribed into cDNA, then the V3 loop was amplified in a nested PCR using bar-coded primers (HXB2 positions: forward primer 6986–7012, reverse primer 7520–7540). To maximize the number of input templates and to minimize variation due to PCR drift, seven parallel RT–PCRs were performed and pooled for each sample.25 Bar-coded amplicons were equimolarly pooled and sequenced on the 454 GS-FLX instrument according to the manufacturer's amplicon sequencing protocol (454 Life Sciences, Roche Diagnostics Systems). Hidden Markov models were applied for alignment of the V3 loop. V3 amino acid sequences were used for tropism prediction. The statistics on the sequence-read numbers from the combined 49 clinical isolates were as follows: average ± SD = 11 836 ± 341 (range 2279–21 220). Prediction of the co-receptor tropism was performed using the clonal Geno2Pheno prediction algorithm (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php) with a false positive rate (FPR) of 5%.26 The following calls were generated: with <5% of the virus population labelled X4 (expressed by the total number of sequencing reads), the sample was considered R5; with >5% of the virus population as a mixed tropic, the sample was considered non-R5.
PGTT
RNA was extracted from 200 to 500 µL of plasma using the High Pure Viral RNA Kit (Roche Diagnostics Systems). Elution was performed with 50 µL of elution buffer. Nested-PCR amplification of a fragment spanning the V1 to V4 region of the env gene was performed (HXB2 primer positions: outer PCR, forward primer 6540–6560 and reverse primer 7701–7721; and inner PCR, forward primer 6561–6580 and reverse primer 7645–7667). Sequencing reactions were prepared using the BigDye® Terminator Cycle Sequencing kit v3.1 (Applied Biosystems, Foster City, CA, USA) with degenerate internal primers: sense 5′-AGYRCAGTACAATGYACACATGG-3′; sense 5′-TCAACHCAAYTRCTGTTAAATGG-3′; and antisense 5′-ATTTCTGGRTCYCCKCCTG-3′. Sequencing products were run on an ABI3130xl automated sequencer. Sequence editing and contig assembly were performed using the Smartgene™ HIV software package (Integrated Database Network System, Smartgene, Zug, Switzerland). V3 nucleic acid sequences were used for tropism prediction with the clonal Geno2Pheno prediction algorithm using an FPR of 5%.
Results
Results of four tropism assays
Results of the four assays are summarized in Table 1. Samples were divided into subgroups according to the individual test results (in the following order: VPPTT/OTA/UDSGTT/PGTT): A = R5/R5/R5/R5; B = non-R5/non-R5/non-R5/non-R5; C = R5/non-R5/R5/R5; D = R5/non-R5/non-R5/non-R5; E = non-R5/R5/non-R5/non-R5; F = non-R5/non-R5/non-R5/fail; G = R5/fail/R5/R5; H = non-R5/fail/non-R5/non-R5; I = fail/R5/R5/R5 (samples were categorized as showing concordance in at least three assays); J = R5/R5/non-R5/fail; K = R5/R5/non-R5/non-R5; L = non-R5/non-R5/R5/R5; M = R5/fail/non-R5/R5; and N = non-R5/fail/non-R5/R5 (samples were categorized as lacking concordance in at least three assays).
Table 1.
Overview of tropism assignment according to the two-step approach and concordance of (i) VpPTT; (ii) OTA; (iii) UDSGTT; and (iv) PGTT
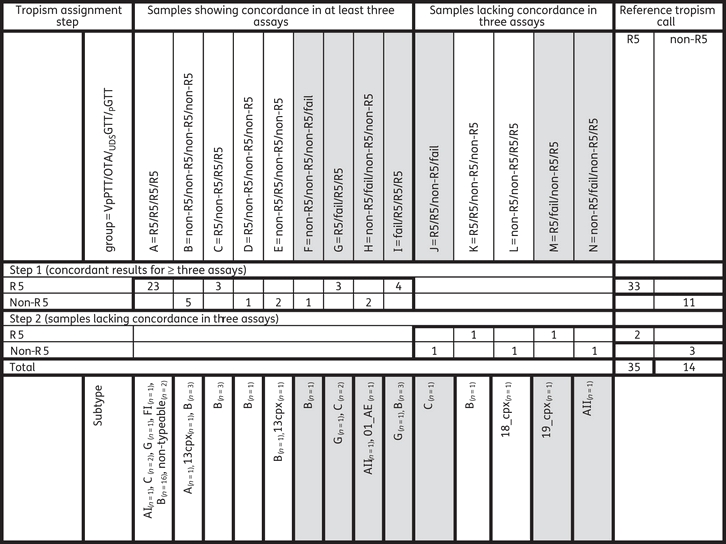 |
Grey shading indicates samples for which at least one assay failed.
Samples showing concordance in at least three assays
In the groups A–I, at least three assay test results could be obtained. For these samples, a detailed analysis was performed of the discordant call. In group C, three samples were non-R5 by OTA but R5 by the other three assays. The ultra-deep sequencing assay did not detect X4 strains in two of the three samples, and detected a low prevalence (1.6%; but because the cut-off of 5% labelled as R5 only) of X4 strains in the third sample. Thus, the final call for group C favoured R5. In group D, one sample was R5 by VPPTT but non-R5 by the other three tests. The ultra-deep sequencing assay identified 46% X4 strains. Thus, the final call for group D favoured non-R5. In group E, two samples were R5 by OTA but non-R5 by the other three tests. The ultra-deep sequencing assay identified 24% and 65% X4 strains. Thus, the final call for group E strains favoured non-R5.
Samples lacking concordance in at least three assays
With samples lacking concordance in three or more assays, clonal phenotyping was performed to gain further insights into the discrepancies (see Table 2). In group J, one sample was R5 by VPPTT and OTA, whereas UDSGTT detected 16% X4 strains, while no result was obtained by PGTT. By UDSGTT, three different populations (present >5%; haplotypes 1–3) could be distinguished. In one population representing 11% X4 virus of the total population by UDSGTT, clonal phenotyping determined three clones as dual tropic, classifying the sample as non-R5. The subgroup K sample was classified R5 by both VPPTT and OTA, but X4 by UDSGTT (77% X4 strains) and PGTT. Of the three populations, two (haplotypes 1 and 2; both representing ±36% of the population) were classified X4 by the G2P prediction tool. However, both populations harboured one or more R5 clones determined by clonal phenotyping and no X4 or dual tropic clones. These data suggest that the prediction tool is making an error both on a population as well as on a UDSGTT level. The final call for this sample is R5. The group L sample was non-R5 by both VPPTT and OTA, but R5 by both UDSGTT and PGTT. The two major populations representing haplotype 1 (14.9%) and haplotype 2 (82.3%) were classified R5 by G2P both on a UDSGTT and population genotype level. The clonal phenotyping determined 20 clones as dual tropic from the major population representing 82.3%. Again these data suggest that the prediction tool is making an error both on a population as well as on an UDSGTT level, which might be due to the relatively uncommon subtype (18_cpx). The final call for this sample is non-R5. In group M, one sample was R5 by VPPTT and PGTT, but non-R5 (5.2% X4) by UDSGTT. In clonal phenotyping, 20 clones were classified R5, and no X4 clones were detected. The final call for this sample was R5. Of the three major populations, one population representing 5.2% was not picked up by the clonal phenotyping. Although 20 viable clones were analysed, the clonal phenotyping has a detection limit of around 5% and it cannot be ruled out that the 5.2% virus population was therefore not picked up by the random sampling (see Table 2). In group N, one sample was non-R5 by VPPTT and UDSGTT (10.2% X4), but was R5 by PGTT. By UDSGTT, three different major populations (>5%) could be distinguished. In one population (haplotype 1) representing 9.3% X4 virus of the total population by UDSGTT, clonal phenotyping confirmed this population as dual tropic, classifying the sample as non-R5. The final conclusion of the combination of first-step and second-step analysis was that 35 samples were classified as R5 and 14 samples as non-R5 (Table 1).
Table 2.
Details of the step 2 tropism identification
 |
D, dual tropic virus; NR, non-reportable.
aPopulation present at the detection limit of the clonal phenotyping.
Parameters of phenotyping and genotyping methods using RTR
By using the RTR, sensitivity and specificity of the VPPTT, OTA, UDSGTT and PGTT could be calculated, as shown in Table 3. The higher failure rate of the OTA affected the negative predictive value. In general, the sensitivity of the OTA, UDSGTT and PGTT was within the same range.
Table 3.
Characteristics of the genotypic and phenotypic assays
| OTA |
VpPTT |
UDSGTT |
PGTT |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTR | N | R5 | non-R5 | F | R5 | non-R5 | F | R5 | non-R5 | F | R5 | non-R5 | F |
| R5 | 35 | 28 | 3 | 4 | 31 | 0 | 4 | 33 | 2 | 0 | 34 | 1 | 0 |
| non-5 | 14 | 3 | 8 | 3 | 2 | 12 | 0 | 1 | 13 | 0 | 2 | 10 | 2 |
| Total | 49 | 31 | 11 | 7 | 33 | 12 | 4 | 34 | 15 | 0 | 36 | 11 | 2 |
| PPV | 90.3 | 93.7 | 97.1 | 94.4 | |||||||||
| NPV | 72.7 | 100 | 86.7 | 90.9 | |||||||||
| Sensitivity | 90.3 | 100 | 94.3 | 97.1 | |||||||||
| Specificity | 72.7 | 85.7 | 92.9 | 83.3 | |||||||||
| Availability | replaced by ESTA | not available | in core facilities | local, home brew protocol | |||||||||
| Regulatory level | CAP/CLIA | research only | research only | dependent on local settings | |||||||||
| TAT | >3 weeks | not relevant | 2 weeks | days | |||||||||
| Cost | moderate to high | not relevant | moderate to high | low | |||||||||
TAT, turnaround time; CAP/CLIA, College of American Pathologists/Clinical Laboratory Improvement Act; PPV, positive predictive value; NPV, negative predictive value; RTR, reference tropism result.
We evaluated the approximate limit of detection for minority variants using PGTT. Only a few samples harboured a population present between 10% and 25%, which is the estimated detection cut-off for minority variants for population genotyping methods. Therefore, it is hard to come up with a well-defined cut-off from this study. If the minority population drops below 10% the PGTT is not picking up this population, as is shown for sample N (X4 present as 9.3% minority), whereas if the population is above 10% the PGTT is picking up the population (group H, sample with 01_AE subtype, X4 present at 12.5% of the total population).
Subsequently the impact of the HIV subtype on the OTA and PGTT was evaluated. In general, discordances for both the OTA and PGTT are evenly distributed between B and non-B subtypes, but the number of patient samples included was too small to draw conclusions (see Table 1). The concordance with the RTR was 31/36 (86%) for OTA and 34/36 (94%) for PGTT. Of the five discordant OTA samples (C and E), four were subtype B. Of the two discordant GTT samples (K and L), one was subtype B.
Clinical decisions based on OTA
In order to better understand the potential value of the different tropism tests, we performed a theoretical evaluation of the impact of using OTA versus PGTT for guiding clinical decisions (see Figure 1). OTA classified 31/49 samples as R5, allowing 31 patients to start a chemokine receptor antagonist as part of their treatment regimen. According to the definition of reference result described previously, 28 of the 31 samples were correctly identified as R5. Assuming use of PGTT instead of OTA, a total of 36 patients would have been returned as R5, leading to maraviroc use, and of these, 34 could be considered as correctly identified as R5. The net result of adopting PGTT as a diagnostic tool would be to allow an additional five patients to receive maraviroc. Conversely, two patients would have started maraviroc in the presence of X4.
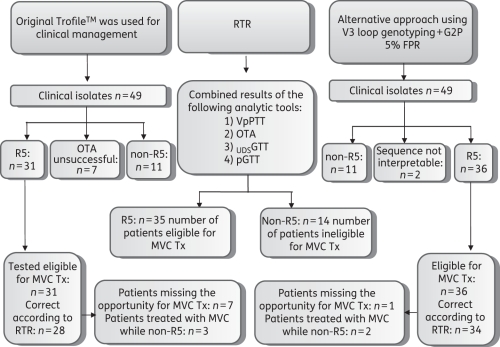
Figure 1.
Overview of the study. RTR, reference tropism result; MVC, maraviroc; Tx, treatment.
According to the reference tropism test, a total of 35 patients would have been eligible for maraviroc therapy. These 35 eligible individuals were distributed in the OTA results as follows: 28 in the R5 cohort, 4 from the OTA failures and 3 from the non-R5 cohort. Hence, a total of seven patients missed the opportunity to be treated with a co-receptor antagonist, while another three were labelled as candidates for such treatment while non-R5 virus was present.
Similarly, the 35 eligible individuals were distributed in the PGTT results as follows: 34 in the R5 cohort and 1 from the non-R5 cohort. This would result in one patient missing the opportunity to be treated with a co-receptor antagonist, while another two were labelled as candidates for such treatment while X4 virus was present (Table 1).
Discussion
In our study, 35 of the 49 plasma samples were classified R5 according to PGTT and 31 samples were classified as R5 according to OTA. If samples for which one assay failed (groups F, G, H, I, J, M and N) were ignored, concordance for all four assays was achieved in 28/36 samples (77%). The concordance with the RTR was 31/36 (86%) for OTA and 34/36 (94%) for pGTT. Discordant results can be attributed to several reasons. In group C, three plasma samples were R5 by PGTT and VPPTT, but non-R5 by OTA. In ultra-deep sequencing, G2P scored 0% of the reads as X4 in two samples. Thus, the FPR of OTA seems to be higher than 0%. Indeed, the only reported FPR was measured using plasma samples of other non-retroviruses.8 Another factor that might have driven discordances is that OTA was running as a routine diagnostic test and the other tests were performed retrospectively in a research setting. In other cases, variability of detection at the lower end of assay sensitivity may explain the discrepancy. One sample in group C was R5 by PGTT and VPPTT, but non-R5 by OTA, and ultra-deep sequencing detected 1.6% X4 reads, not accounted for because of the cut-off of 5%. Although it is generally accepted that the detection of minorities is superior for ultra-deep sequencing, the clonal phenotyping was used to generate the final call. By doing this we allowed for the pick up of potential errors in the interpretation tool that would classify both the population and ultra-deep V3 loop sequences identical. In both samples K and L the G2P interpretation algorithm is making a different interpretation than the clonal phenotype. In these two samples, it is not a question of whether or not the ultra-deep sequencing has a better sensitivity for the detection of minorities than the clonal phenotyping, as the populations for which the G2P differs are not present as a minority, but as a majority. The high sensitivity for minority detection of X4 variant by UDSGTT could be of value for the tropism determination of sample M, now classified as R5, as the 5.2% UDSGTT determined X4 could easily be missed by the clonal phenotyping of 24 clones.
A further item arises from the interpretation of sequencing results analysed, especially for rare and complex sequences. In our analysis, if we excluded the samples where one assay failed, the results of the PGTT analysis could not be confirmed by RTR for samples K (subtype B) and L (18_cpx). The results of the OTA could not be confirmed by the RTR for samples C and E (subtype B, n = 4; 13_cpx, n = 1). Although the ESTA is now performed with new primers that improve tropism prediction on non-B viral strains, the impact of introducing this new assay with adapted primers might have had a minor influence on the concordance with the RTR, as most samples were subtype B.27
In this study we evaluated the potential value of population genotyping in clinical practice. Importantly, 36 patients could have started and benefited from maraviroc. Detailed analysis suggests that 33 of these 36 patient samples did indeed harbour <5% dual or mixed tropic virus, which is associated with a good response to therapy, shown in a previous analysis by McGovern et al.22 Our data support the findings reported earlier from the International HIV Drug Resistance Meeting 2010 indicating that, in treatment-experienced patients, population genotyping is comparable in predicting clinical outcome and response to chemokine receptor antagonist independent of the use of the G2P or position-specific scoring matrix (PSSM) interpretation algorithm with the OTA.28 Our data represent the first dataset in clinical practice where V3 loop tropism determination has been assessed outside the context of a clinical trial with subsequent detailed 454 analysis. In line with the recent European, British and German guidelines, we provide further data to support that population V3 loop genotyping is comparable to OTA in predicting tropism in clinical practice.19–21 Population V3 loop genotyping can serve as a tropism determination tool, but full standardization of the sequence method and interpretation remains a requirement. Although our study evaluated two phenotypic assays, population sequencing and ultra-deep V3 loop sequencing, and for some discordant samples also clonal phenotyping, results should be interpreted with caution, as this is a retrospective evaluation of PGTT on a limited number of samples. Prospective evaluation of PGTT in clinical practice remains a topic of further research.
Funding
This work was supported by internal funding without pharmaceutical sponsoring. L. V. is supported by the Flemish Fund for Scientific Research.
Transparency declarations
A. M. G. receives consultancy fees and research funding from and is on the speaker's bureau for Pfizer, GSK, ViiV, Monogram Biosciences, MSD, Tibotec, J&J, Roche Diagnostics, Gilead, BI, BMS, Virco and Abbott Diagnostics. L. V. receives consultancy fees and research funding from and is on the speaker's bureau for Pfizer, GSK, ViiV, Gilead, BMS, Virco, Tibotec, J&J and MSD. N. M. receives consultancy fees from and is on the speaker's bureau for Tibotec, BMS, ViiV, Merck and Abbott. S. D. W. receives consultancy fees from and is on the speaker's bureau for Pfizer, GSK, BMS and Abbott. E. F. received consultancy fees from Abbott, BMS, Gilead, MSD, J&J and ViiV Healthcare. W. M., K. V. B., I. V., V. V. E., H. V. M. and L. J. S. are employees of Tibotec Virco Virology BVBA. All other authors: none to declare. While Tibotec is commercializing antiviral compounds for HIV-1 treatment, the current work does not represent or influence any of these compound-related commercial activities. Virco has developed and commercialized vicroTYPE for reverse transcriptase and protease resistance testing. Tropism testing tools and algorithms were developed for research purposes only, and no activities for commercialization are planned or intended.
Acknowledgements
We would like to thank Martin Fisher, Margaret Johnson, Nicky Perry and Angela Strang for their contributions.
References
- 1.Llibre JM, Schapiro JM, Clotet B. Clinical implications of genotypic resistance to the newer antiretroviral drugs in HIV-1-infected patients with virological failure. Clin Infect Dis. 2010;50:872–81. doi: 10.1086/650732. doi:10.1086/650732. [DOI] [PubMed] [Google Scholar]
- 2.Feeney Eoin R, Mallon Patrick WG. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr Pharm Des. 2010 doi: 10.2174/138161210793563482. in press. [DOI] [PubMed] [Google Scholar]
- 3.Lang S, Mary-Krause M, Cotte L, et al. Impact of individual antiretroviral drugs on the risk of myocardial infarction in human immunodeficiency virus-infected patients: a case-control study nested within the French Hospital Database on HIV ANRS cohort CO4. Arch Intern Med. 2010;170:1228–38. doi: 10.1001/archinternmed.2010.197. doi:10.1001/archinternmed.2010.197. [DOI] [PubMed] [Google Scholar]
- 4.Friis-Moller N, Reiss P, Sabin CA, et al. Class of antiretroviral drugs and the risk of myocardial infarction. N Engl J Med. 2007;356:1723–35. doi: 10.1056/NEJMoa062744. doi:10.1056/NEJMoa062744. [DOI] [PubMed] [Google Scholar]
- 5.Sabin CA, Worm SW, Weber R, et al. Use of nucleoside reverse transcriptase inhibitors and risk of myocardial infarction in HIV-infected patients enrolled in the D:A:D study: a multi-cohort collaboration. Lancet. 2008;371:1417–26. doi: 10.1016/S0140-6736(08)60423-7. doi:10.1016/S0140-6736(08)60423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. doi:10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsibris AM, Kuritzkes DR. Chemokine antagonists as therapeutics: focus on HIV-1. Annu Rev Med. 2007;58:445–59. doi: 10.1146/annurev.med.58.080105.102908. doi:10.1146/annurev.med.58.080105.102908. [DOI] [PubMed] [Google Scholar]
- 8.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–75. doi: 10.1128/AAC.00853-06. doi:10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petropoulos C, Limoli K, Whitcomb J, et al. Validation studies defining the performance of Monogram's HIV coreceptor tropism assay. Abstracts of the Fifth European HIV Drug Resistance Workshop, Cascais, Portugal, 2007; Abstract 57. [Google Scholar]
- 10.Saag M, Heera J, Goodrich J. Reanalysis of the MERIT study with the enhanced Trofile assay (MERIT-ES). Abstracts of the Forty-eighth Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 2008; Washington, DC, USA: American Society for Microbiology; Abstract H-1232a. [Google Scholar]
- 11.Van Baelen K, Vandenbroucke I, Rondelez E, et al. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays. J Virol Methods. 2007;146:61–73. doi: 10.1016/j.jviromet.2007.06.003. doi:10.1016/j.jviromet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Vandekerckhove L, Verhofstede C, Vandenbroucke I, et al. on behalf of the Tropism Study Group. Comparison of population and 454 genotyping and Trofile phenotyping for tropism determination in patients selected for maraviroc initiation. Abstracts of the Eighteenth International HIV Drug Resistance Workshop, Fort Myers, FL, 2009; Abstract 57. [Google Scholar]
- 13.Chan SY, Speck RF, Power C, et al. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–8. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen MA, van ’t Wout AB. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 2003;5:104–12. [PubMed] [Google Scholar]
- 15.O'Meara D, Wilbe K, Leitner T, et al. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J Clin Microbiol. 2001;39:464–73. doi: 10.1128/JCM.39.2.464-473.2001. doi:10.1128/JCM.39.2.464-473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. doi:10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 17.Shi MM. Enabling large-scale pharmacogenetic studies by high-throughput mutation detection and genotyping technologies. Clin Chem. 2001;47:164–72. [PubMed] [Google Scholar]
- 18.Mitsuya Y, Varghese V, Wang C, et al. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol. 2008;82:10747–55. doi: 10.1128/JVI.01827-07. doi:10.1128/JVI.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandekerckhove LPR, Wensing AMJ, Kaiser R, et al. Consensus statement of the European guidelines on clinical management of HIV-1 tropism testing. Abstracts of the Tenth International Congress on Drug Therapy in HIV Infection, Glasgow, Scotland, 2010; Abstract O121. [Google Scholar]
- 20.Geretti A, Mackie N. British HIV Association Guidelines on Determining HIV-1 Tropism in Routine Clinical Practice. http://www.bhiva.org/Tropism.aspx. (October 2010, date last accessed) [Google Scholar]
- 21.German Recommendations for Determining HIV-1 Coreceptor Usage. http://www.viro.med.uni-erlangen.de/nrz/recommendation080324.pdf. (September 2010, date last accessed) [Google Scholar]
- 22.McGovern R, Dong W, Mo T, et al. Optimization of clinically relevant cutpoints for the determination of HIV co-receptor usage to predict maraviroc responses in treatment experienced (TE) patients using population V3 genotyping. Abstracts of the EACS Conference, Cologne, 2009; Abstract PE3.4/8. [Google Scholar]
- 23.Su Z, Gulick RM, Krambrink A, et al. Response to vicriviroc in treatment-experienced subjects, as determined by an enhanced-sensitivity coreceptor tropism assay: reanalysis of AIDS clinical trials group A5211. J Infect Dis. 2009;200:1724–8. doi: 10.1086/648090. doi:10.1086/648090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandenbroucke I, Van Marck H, Mostmans W, et al. HIV-1 V3 envelope deep sequencing for clinical plasma specimens failing in phenotypic tropism assays. AIDS Res Ther. 2010;7:4. doi: 10.1186/1742-6405-7-4. doi:10.1186/1742-6405-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandenbroucke I, Eygen VV, Rondelez E, et al. Minor variant detection at different template concentrations in HIV-1 phenotypic and genotypic tropism testing. Open Virol J. 2008;2:8–14. doi: 10.2174/1874357900802010008. doi:10.2174/1874357900802010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sing T, Low AJ, Beerenwinkel N, et al. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir Ther. 2007;12:1097–106. [PubMed] [Google Scholar]
- 27.Napolitano L, Tressler T, Coakley E, et al. Incorporation of optimized primers into the Trofile assay substantially improves determination of viral tropism in genetically diverse HIV subtypes. Abstracts of the Forty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, 2009; Washington, DC, USA: American Society for Microbiology; Abstract 908. [Google Scholar]
- 28.Swenson LC, McGovern AR, Dong W, et al. Phenotypic screening for HIV tropism versus both population-based and “deep” sequencing. Abstracts of the Forty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, 2009; Washington, DC, USA: American Society for Microbiology; Abstract H-899/378. [Google Scholar]