Abstract
The standardization of methods for human body fluid protein isolation is a critical initial step for proteomic analyses aimed to discover clinically-relevant biomarkers. Several caveats have hindered pancreatic fluid proteomics, including the heterogeneity of samples and protein degradation. We aim to optimize sample handling of pancreatic fluid that has been collected using a safe and effective endoscopic collection method (ePFT). Using SDS-PAGE protein profiling, we investigate (1) precipitation techniques to maximize protein extraction, (2) auto-digestion of pancreatic fluid following prolonged exposure to a range of temperatures, (3) effects of multiple freeze-thaw cycles on protein stability, and (4) the utility of protease inhibitors. Our experiments revealed that trichloroacetic acid (TCA) precipitation resulted in the most efficient extraction of protein from pancreatic fluid of the eight methods we investigated. In addition, our data reveal that although auto-digestion of proteins is prevalent at 23°C and 37°C, incubation on ice significantly slows such degradation. Similarly, when the sample is maintained on ice, proteolysis is minimal during multiple freeze-thaw cycles. We have also determined the addition of protease inhibitors to be assay-dependent. Our optimized sample preparation strategy can be applied to future proteomic analyses of pancreatic fluid.
Keywords: body fluid, mass spectrometry, proteomics, biomarker, ePFT
1. Introduction
Diseases of the exocrine pancreas have a significant global impact on the healthcare system [1]. Acute pancreatitis, for instance, accounts for one in every 200 hospital admissions and for over $2 billion in direct annual costs in the United States [2, 3]. Similarly, chronic pancreatitis is often associated with diabetes, pain, malabsorption of fat and protein, and to a lesser degree, the development of pancreatic cancer, which is the fourth leading cause of cancer death in the United States [4]. Improved strategies for the diagnosis and treatment of pancreatic disease are necessary to reduce healthcare and patient burdens.
Application of proteomics to the study of pancreatic disease presents a unique opportunity to accelerate the discovery of physiologically- and clinically-relevant biomarkers. Although blood or its derivatives (i.e., plasma or serum) are commonly utilized in the search for disease-specific markers, such systemic fluids represent the state of the entire body such that the majority of the proteins detected are not necessarily related to the pancreatic disease of interest [5]. The analysis of a proximal body fluid, such as pancreatic fluid, will increase the likelihood of biomarker discovery in the context of particular diseased organs (i.e., the pancreas) [6]. Pancreatic fluid is an excellent clinical specimen for identification of novel biomarkers as its protein composition is of lower complexity compared to serum. In addition, the proteins in pancreatic fluid predominantly originate from the exocrine pancreas.
Biomarkers discovered from pancreatic fluid may provide not only a method for the diagnosis and/or prognosis of pancreatic diseases, but also may generate insights into the pathophysiology of the pancreas. To date, several proteomic investigations of pancreatic fluid have been performed using specimens collected surgically or via endoscopic retrograde cholangiopancreatography (ERCP) [7-15]. Both methods are highly invasive and associated with significant risks for the patients. In contrast, we applied the ePFT (endoscopic pancreatic function test) collection method, which is much less invasive compared to ERCP and surgery, and permits the safe collection of 10-fold larger volumes of pancreatic fluid, making it a well-suited method for comprehensive proteome analysis [16-21].
Establishing clear and consistent sample collection and processing methodologies is an initial stage in the development of clinical proteomics assays. In translational research, there often are insufficient standardized protocols regarding specimen collection, storage and processing. In urine proteomics, for example, several recent reviews have stressed the importance of standardized sample handling in reducing experimental variability [22-28]. For pancreatic fluid, such variations are especially pronounced due to its inherently high concentration of active proteolytic enzymes; however standardized protocols have yet to be established. Significant changes in the proteomic profile may also be introduced during sample preparation if consensus methodology is not in place. Procedural artifacts may mask a potentially fortuitous result or be more likely to identify false positive biomarker candidates. We realize that variability cannot be entirely avoided; however, it may be kept to a minimum by careful and standardized sample handling.
The purpose of this study is to assess various laboratory methods of processing ePFT-collected pancreatic fluid to minimize protein degradation that may occur due to the high concentration of endogenous proteases. Following the example of several other reports that used polyacrylamide gel electrophoresis to optimize the preparation of body fluids [29-31], we used SDS-PAGE analysis to (1) investigate protein isolation techniques to maximize protein yield from pancreatic fluid, (2) evaluate the effects of temperature and time on pancreatic fluid auto-digestion, and (3) examine protein stability following multiple freeze-thaw cycles. In addition, during each experiment we explored the utility of protease inhibitors, as data both for and against their use in pancreatic fluid studies have been reported [12, 27, 32, 33]. In summary, we aimed to optimize the handling of pancreatic fluid for use in future proteomic analyses.
2. Materials and Methods
The protocol for collecting pancreatic fluid was approved by the Institutional Review Board (IRB # 2007-P-002480/1) at Brigham and Women's Hospital and Children's Hospital Boston. The pancreatic fluid utilized for the three optimization experiments outlined below was from excess fluid specimens. This fluid was obtained from a subject with chronic abdominal pain who had undergone an endoscopic pancreas function test (ePFT) and was subsequently determined not to have pancreatic disease. Replicate experiments were performed with pancreatic fluid collected from other patients in a similar manner.
Fluid specimens collected during endoscopy from twelve patients were used to test the reproducibility of our optimized protein precipitation procedure. The samples used in these experiments were regarded as “excess samples” in that they were not specifically collected for this study, but were in excess of what was needed for the clinical diagnostic study and would otherwise have been discarded. Although samples used in our experiments were considered “excess,” care was taken to immediately remove the fraction that was needed for the clinical tests and the remaining sample was maintained on ice.
The samples were then immediately transported to the proteomics laboratory, centrifuged to remove particulates, and used in experiments or frozen in 1 mL aliquots. The pancreatic fluid specimens from these twelve patients included three with chronic abdominal pain (CAP) and nine with pancreatic disease (three each with chronic pancreatitis (CP), acute pancreatitis (AP), and cystic neoplasms (CN)). Furthermore, the pancreatic fluid that was used in these experiments was not contaminated with blood. In general, blood contamination was rarely seen with ePFT collection of pancreatic fluid specimens, which lends support to the safety of this pancreatic fluid collection approach.
Materials
ChiRhoStim® synthetic human secretin was from ChiRhoClin (Burtonsville, MD). Complete Mini protease inhibitor cocktail (11836153001) was from Roche (Mannheim, Germany). SeeBluePlus2 Pre-Stained standard (LC5925), LDS (lithium dodecyl sulfate) sample buffer (NP0008), NuPAGE 4-12% Bis-Tris polyacrylamide gels (NP0335), SimplyBlue Coomassie stain (LC0665), and MES-SDS (2-(N-morpholino)ethanesulfonic acid-sodium dodecyl sulfate) buffer (NP002) were from Invitrogen (Carlsbad, CA). PD-10 desalting columns (17-0851-01) were from GE Healthcare (Piscataway, NJ). Microcon centrifugal filter concentrators (5 kDa molecular weight cut-off; UFC3LCC00) were from Millipore (Bedford, MA). C4 trapping column cartridges (004/25111/01) and column holder (TH1/25111/01) were from Michrom BioResources (Auburn, CA). PANHEHA pH indicator paper strips (10360005) were from Sigma-Aldrich (St. Louis, MO). MaxymumRecovery ™ 1.7 mL microtubes (MCT-175-L-C) were purchased from Axygen (Union City, CA). Other reagents and solvents were from Sigma-Aldrich and Burdick and Jackson, respectively.
Procedural overview
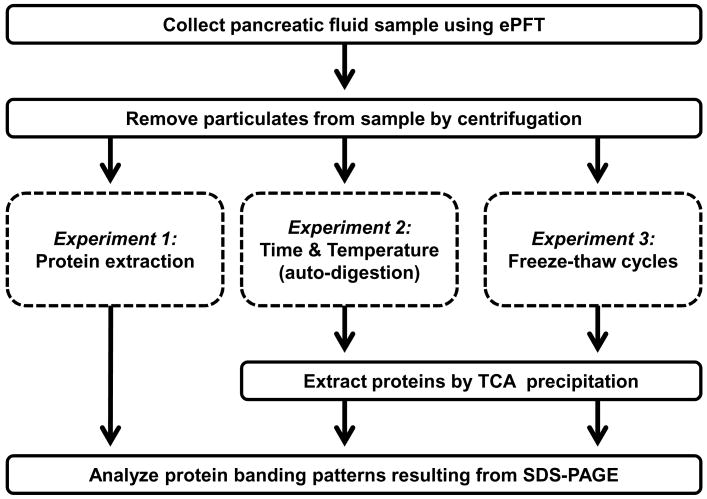
An overview of the procedures used in our investigation was illustrated in Figure 1. Briefly, pancreatic fluid was collected via ePFT and immediately placed on ice, particulates were removed by centrifugation (3,000×g for 15 minutes) at 4°C, and samples were aliquoted on ice for use in the experiments described in detail below. To evaluate whether adding protease inhibitors to pancreatic fluid prevented protein degradation, a protease inhibitor cocktail was added to one aliquot of sample, while an equal volume of water was added to a duplicate aliquot. We used SDS-PAGE analysis to determine the relative amount of protein and the degree of proteolysis for each optimization experiment.
Figure 1. Experimental workflow for the optimization of sample handling of endoscopically (ePFT) collected, secretin-stimulated pancreatic fluid.
Pancreatic fluid collection (ePFT)
Samples were collected by the endoscopic pancreatic function test (ePFT) method following secretin-induced pancreas secretion for chronic abdominal pain (CAP), chronic pancreatitis (CP) and acute pancreatitis (AP) samples [19], or endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) for cystic neoplasms (CN) samples [34, 35].
The ePFT procedure was as follows: A) pre-procedural assessment, B) endoscopic procedure and C) post-procedural assessment/recovery.
A. Pre-procedural assessment
Prior to upper endoscopy, all study subjects underwent a history and physical examination including list of allergies, medications, substance use/abuse, and vital signs.
B. Procedure
Endoscopic collection was performed in a stepwise manner as follows: 1) The patient was placed in left lateral decubitus position with slight head elevation. 2) The posterior pharynx is sprayed with topical cetacaine spray. 3) A sedation and analgesia bolus was administered. 4) Further sedation doses were administered if necessary for patient comfort. 5) After the sedation bolus, a bite-block was placed. 6) Esophagogastroduodenoscopy (EGD) was performed using a standard (10 mm) or thin (6 mm) gastroscope for visualization of the esophagus, stomach, and duodenum (2 to 5 minutes). 7) Gastroduodenal fluid was aspirated (approximately 1 minute) as completely as possible through the gastroscope. 8) A test dose of synthetic human hormone secretin (ChiRhoStim®) was administered and patients were monitored for anaphylaxis or adverse reaction, followed by a standard weight-based intravenous bolus (0.2 μg/kg) given over 1 minute. 9) Pancreatic fluid was aspirated from the descending duodenum at specific timed intervals following hormonal stimulation and stored on ice.
The duodenal aspirates were collected at 0, 5, 10, 15, 20, 30, 45 and 60 minutes after stimulation. Only the 30-minute time point was used for the ensuing analysis. Biopsies of the stomach and duodenum were obtained to rule out microscopic gastrointestinal disease, such as Helicobacter pylori or celiac sprue, as a cause of dyspepsia.
C. Post-procedural Assessment/Recovery
Study participants were discharged from the endoscopy unit based on hospital procedural sedation guidelines assessing level of consciousness, vital signs, oxygen saturation, alertness, gag reflex, degree of nausea, and ability to ambulate.
Immediately following collection, samples were placed on ice, and divided for SDS-PAGE and biochemical analysis [36]. The samples for SDS-PAGE analysis were transported on ice to the proteomics laboratory and centrifuged at 3,000×g for 15 minutes at 4°C to remove particulates. The supernatant was then divided into 1 mL aliquots and frozen at -80°C. For samples collected via the ePFT method, analysis was performed using pancreatic fluid that was collected 45 minutes post-secretin stimulation.
Experiment 1: Comparison of methods to maximize pancreatic fluid protein extraction
To determine the method which maximized protein extraction, 3.2 mL of pancreatic fluid was divided into 200 μL aliquots to ensure equal protein content for each method examined (approximately 250 μg of total protein per aliquot, as determined using the BioRad Protein Assay [37]). As mentioned in the Procedural overview, duplicate samples were processed with and without the addition of protease inhibitors. Protein extraction efficiency was determined based upon SDS-PAGE protein banding patterns and intensities.
Method 1: in vacuo centrifugation
Aliquots of pancreatic fluid (200 μL each) were directly dried at 23°C in a vacuum centrifuge for approximately three hours (SPD1010 Thermo Savant, Waltham, MA).
Method 2: PD-10 desalting column
Samples (200 μL) of pancreatic fluid were diluted in 2.5 mL of HPLC-grade water and added to an equilibrated PD-10 desalting column (GE Healthcare). Samples were processed using the gravity protocol as specified in the manufacturer's instructions. The eluted samples were dried at 23°C in a vacuum centrifuge.
Method 3: Microfiltration
Samples (200 μL each) were loaded onto 5kDa Microcon centrifugal filter concentrators (Millipore, Billerica, MA) and centrifuged at 14,000×g at 4°C until the sample volume was at least one-tenth of the original volume. The retained samples were washed twice with 200 μL of water in the centrifugal filter concentrator and the final 20 μL sample was dried at 23°C in a vacuum centrifuge.
Method 4: C4 column
A final concentration of 0.1% trifluoroacetic acid (TFA) and 2% acetonitrile (ACN) was added to 200 μL of pancreatic fluid. The samples were loaded onto a C4 trapping column (Michrom) which was pre-equilibrated with 0.1% TFA/2% ACN (Buffer A). Samples were washed twice with 500 μL of Buffer A and subsequently eluted with 0.1% TFA/90% ACN. The eluted samples were dried at 23°C in a vacuum centrifuge.
Method 5: TFA/A precipitation
Two sample volumes (400 μL) of ice-cold 0.1% trifluoroacetic acid (TFA) in 99.9% acetonitrile (ACN) were added to 200 μL of pancreatic fluid. The mixture was vortexed and centrifuged at 20,000×g at 4°C for 30 minutes. The supernatants were carefully aspirated and the pellets were allowed to air dry at 23°C.
Method 6: TCA precipitation
Ice-cold 100% trichloroacetic acid (25 μL, TCA) was added to 200 μL of pancreatic fluid, vortexed and incubated at 4°C for 2 hours. The sample was centrifuged at 20,000×g at 4°C for 30 minutes and the supernatant was carefully aspirated. One milliliter of 100% ice-cold acetone was added to the pellets which were briefly vortexed and incubated at -20°C for 1 hour. The sample was centrifuged at 20,000×g at 4°C for 30 minutes and the pellet was washed twice with 100% ice-cold acetone. The final pellets were allowed to air dry at 23°C.
Method 7: Ethanol precipitation
A total of 800 μL of 100% ethanol was added to 200 μL aliquots of pancreatic fluid. The mixtures were vortexed, placed on ice for 30 minutes, and centrifuged (14,000×g at 4°C for 30 minutes). The supernatants were carefully aspirated and the pellets were allowed to air dry at 23°C.
Method 8: Acetone precipitation
A total of four sample volumes (800 μL) of ice-cold 100% acetone was added to 200 μL of pancreatic fluid, vortexed briefly, and incubated at -20°C for 3 hours. Subsequently, the samples were centrifuged at 20,000×g at 4°C for 30 minutes. The supernatants were carefully aspirated and the pellets were allowed to air dry at 23°C.
Acidification of pancreatic fluid prior to ethanol or acetone precipitation
Fourteen aliquots of 200 μL of pancreatic fluid were prepared. To triplicate samples, a final volume of 0.1%, 1%, 5% and 12.5% of 100% TCA was added, essentially resulting in three sets of each TCA concentration. To the remaining two tubes, no TCA is added. Samples were vortexed briefly and pH was determined by spotting ∼2 μL of each mixture on each segment of PANHEHA pH indicator paper strips. Two sets of TCA concentrations (including one sample in which TCA was added) were either ethanol (Method 7) or acetone (Method 8) precipitated. The remaining set containing 0.1%, 1%, 5% and 12.5% TCA was subjected to centrifugation and washed with acetone as was performed for the standard 12.5% TCA precipitation, which is outlined in Method 6 above. The protein content from each precipitation was analyzed by SDS-PAGE.
Experiment 2: Evaluation of temperature and time on pancreatic fluid auto-digestion
Ten milliliters of pancreatic fluid were thawed on ice and divided into six aliquots of equal volume. From each aliquot, 200 μL of fluid was removed, labeled as (t = 0), and frozen at -80°C. The remaining samples were incubated on ice, at 23°C, or at 37°C. For each temperature investigated, one sample was supplemented with the protease inhibitor cocktail and an equal volume of HPLC-grade water was added to the duplicate sample. Subsequently, another 200 μL aliquot from both samples was removed after 2 hours (t = 2), 4 hours (t = 4), 8 hours (t = 8), 24 hours (t = 24), and 48 hours (t = 48). The protein content from each sample was extracted by TCA precipitation (see Experiment 1, method 6) and analyzed by SDS-PAGE.
Experiment 3: Evaluation of freeze-thaw (FT) cycles on pancreatic fluid protein stability
Two milliliters of pancreatic fluid were thawed on ice and divided into two aliquots. To one aliquot, the protease inhibitor cocktail was added, and to the second aliquot, an equal volume of HPLC-grade water was added. From each aliquot, 200 μL of sample was removed, labeled as (FT1), and frozen at -80°C. The remaining fluid was frozen at -80°C for 1 hour and thawed on ice, after which a sample (FT2) was removed. A total of 5 freeze-thaw cycles (FT1-FT5) were performed. The protein content from each sample was extracted by TCA precipitation and analyzed by SDS-PAGE.
SDS-PAGE protein profile analysis
For Experiment 1, dried samples were resuspended in 20 μL of 1× LDS sample loading buffer. For Experiments 2 and 3, proteins were extracted first by TCA precipitation prior to resuspension in 20 μL of 1× LDS sample buffer. For all three experiments, 2 μL of 0.1 M DTT were added to each sample, which was then incubated at 56°C for 1 hour. After cooling, samples were alkylated with 2 μL of 40% acrylamide for 30 minutes at 23°C. SDS-PAGE protein separation was performed at 150 volts in MES buffer for 45 minutes. Gels were rinsed in deionized water for 10 minutes, fixed in 45% methanol, 45% water, 10% acetic acid for 30 minutes, and stained with SimplyBlue Coomassie for 1 hour. Gels were subsequently destained overnight in deionized water. Gel densitometry was performed using the publically-available ImageJ software [38].
3. Results
Experiment 1: Comparison of methods to maximize pancreatic fluid protein extraction
Two-hundred microliters of pancreatic fluid, equivalent to approximately 250 μg of protein were precipitated using a variety of techniques. Different protein banding patterns were revealed by SDS-PAGE analysis according to the particular protein extraction method used. As illustrated in Figure 2, the protein extraction methods that do not acidify the solution (i.e., vacuum centrifugation, PD-10 column, ultrafiltration, ethanol, and acetone) resulted in generally lower staining intensity and the presence of more low molecular weight proteins than acidifying methods (i.e., TFA/A, C4 trapping column, and TCA). As pancreatic enzymes, such as trypsin and chymotrypsin, were active in basic pH, but were inactivated at acidic pH, such a result was expected. The blue smears appearing near the bottom of the SDS-PAGE gel were indicative of proteolysis occurring during extended incubation/preparation periods and/or exposure to room temperature.
Figure 2. Experiment 1- Comparison of methods to maximize pancreatic fluid protein extraction.
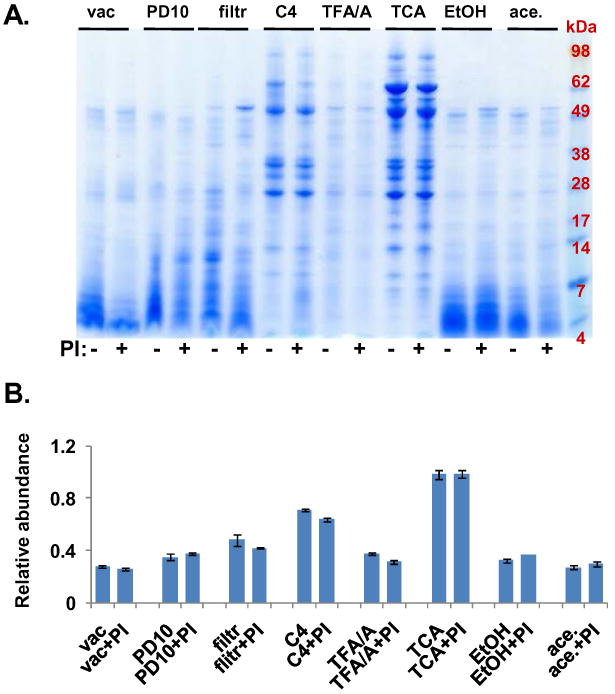
A. SDS-PAGE gel image of pancreatic fluid proteins extracted using the following techniques: vac, vacuum centrifugation; PD10, PD-10 column; filtr, 5 kDa molecular weight cut-off Centricon filtration; C4, C4 trapping column; TFA/A, triflouroacetic acid/acetonitrile precipitation; TCA, trichloroacetic acid precipitation; EtOH, ethanol precipitation; ace, acetone precipitation; PI, protease inhibitors. B. Quantitative gel densitometry measurements of gel lanes (between the 98 kDa and the 7 kDa marker) as determined using ImageJ software. Data points were normalized with respect to the maximum value (TCA plus protease inhibitors).
Of the chemical precipitation techniques that we investigated, adding a final volume of 12.5% TCA to pancreatic fluid resulted in the extraction of the greatest amount of protein, both in the number of sharp, distinct bands and band intensity, while minimizing the lower molecular weight band smears. Quantitatively, ImageJ gel densitometry analysis (of protein bands between approximately 98 and 7 kDa) estimated that protein yields for TCA were ∼30-70% higher than those of other methods tested, as can be approximated by values in Figure 2B.
Acidification of pancreatic fluid prior to ethanol or acetone precipitation
To explore further the effects of acidification of pancreatic fluid on protein extraction, we performed acetone and ethanol precipitations of samples containing increasing concentrations of TCA (0%, 0.1%, 1%, 5%, and 12.5%). Using PANHEHA pH indicator paper strips, we estimated the solution containing no TCA and 0.1% TCA to be pH 8.5, that 1% TCA was pH 6.0, and that 5% and 12.5% TCA was ∼ pH 0. SDS-PAGE gel analysis (Figure 3 A) illustrated that 0.1% TCA did not enhance acetone precipitation. Similarly, ethanol precipitation did not improve protein extraction when using 1% TCA or less. Lower molecular weight smears, as were most apparent in samples to which TCA had not been added, were not apparent when samples were acidified with >1% TCA. Although only a small amount of protein was precipitated with 0.1% TCA, 1% TCA alone was able to precipitate relatively as much protein as acid-supplemented acetone and ethanol precipitations indicating that TCA and not acetone or ethanol was primarily responsible for the successful precipitation. TCA-extraction of protein was maximized at 5%-12.5%, as was supported by gel densitometry analysis (Figure 3 B).
Figure 3. Acidification of pancreatic fluid prior to ethanol or acetone precipitation.
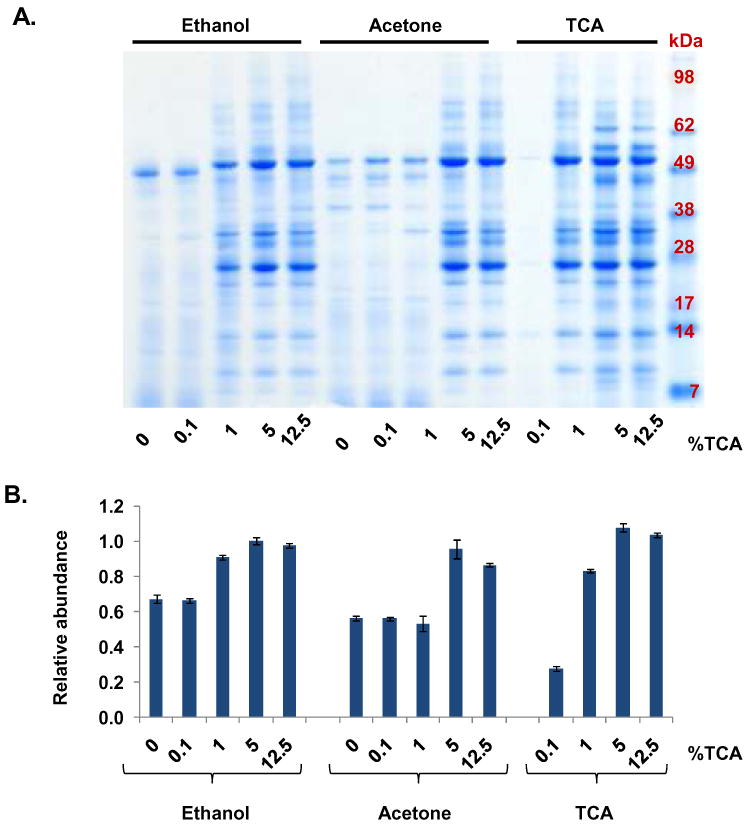
A. SDS-PAGE separation of ethanol, acetone and TCA precipitations of samples containing increasing concentrations of TCA: 0%, 0.1%, 1%, 5%, and 12.5%. B. Quantitative gel densitometry measurements of entire gel lanes as determined using ImageJ software. Data points were normalized with respect to the maximum intensity value.
In summary, TCA precipitation had the advantages of concentrating and desalting the solution, while simultaneously acidifying it, thereby inactivating proteases via denaturation [39]. Thus, for subsequent experiments, TCA precipitation was used to extract proteins from our samples. Furthermore, our SDS-PAGE-based comparison of samples with and without protease inhibitor supplementation revealed that the presence of the inhibitors did not result in major differences in the protein banding patterns for each method investigated.
Experiment 2: Evaluation of temperature and time on pancreatic fluid auto-digestion
We hypothesized that abundant proteases (e.g., trypsin, chymotrypsin, aminopeptidases, carboxypeptidases) in pancreatic fluid remain active following secretin-stimulated ePFT collection, thus pancreatic fluid samples at elevated temperatures were prone to auto-proteolysis (auto-digestion). To test this hypothesis, we incubated pancreatic fluid samples i) on ice, ii) at 23°C, and iii) at 37°C and removed aliquots at different time points (Figure 4 A). Samples, which were prepared with and without the addition of protease inhibitors, were subsequently treated with TCA and analyzed by SDS-PAGE to observe protein degradation (Figure 4 B). It was apparent from Figure 4 B that in the absence of protease inhibitors, protein degradation was evident after 8 hours on ice (particularly in the high molecular weight band region), 2 hours at 23°C, and 0.5 hour at 37°C. This result was supported by gel densitometry quantitation of entire gel lanes with ImageJ, as plotted in (Figure 4 C). As expected, with the addition of the protease inhibitor cocktail, the overall degradation was considerably less.
Figure 4. Experiment 2- Evaluation of temperature and time on pancreatic fluid auto-digestion.
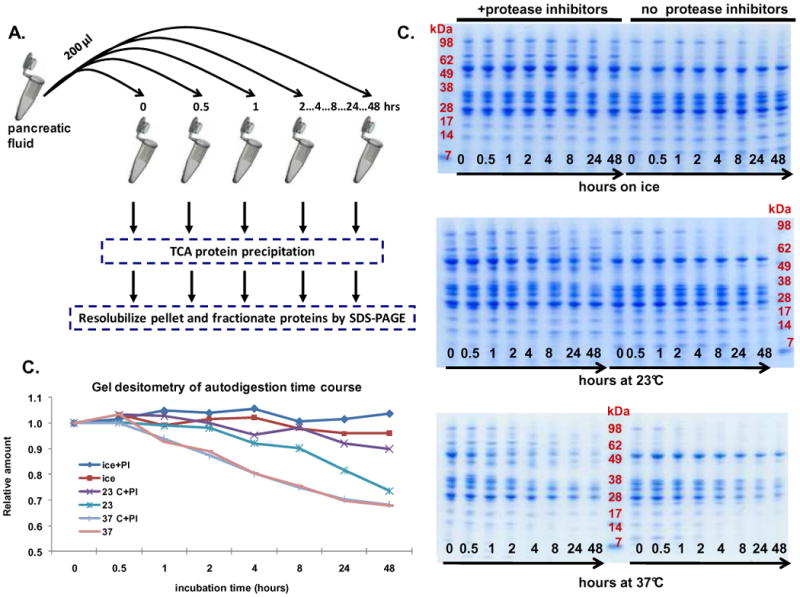
A. Workflow of auto-digestion experiment. B. SDS-PAGE separation of samples incubated on ice (top), at 23°C (middle) and at 37°C (bottom). C. Quantitative gel densitometry measurements of entire gel lanes as determined using ImageJ software. Data points for each curve were normalized using the reading at the t=0 time point.
Experiment 3: Evaluation of freeze-thaw (FT) cycles on pancreatic fluid protein stability
To minimize protein degradation, samples for proteomic analysis are generally stored at -80°C. To validate previously collected proteomics data, it is often necessary to re-analyze samples which have been previously thawed. As a result of the protease-rich nature of pancreatic fluid, we were concerned with protein degradation, which is potentially detrimental to the reproducibility of proteomics experiments. Samples were subjected to repeated cycles of freezing at -80°C and thawing on ice for approximately 1 hour with intermittent gentle agitation. After five freeze/thaw (FT) cycles, SDS-PAGE protein banding pattern analysis (Figure 5 A) revealed only slight differences in the protein bands within the sample groups regardless of protease inhibitor cocktail supplementation. ImageJ gel densitometry analysis of entire gel lanes (Figure 5 B) did not indicate any significant variations and thus also demonstrated the limited degradation resulting from multiple freeze-thaw cycles.
Figure 5. Experiment 3- Evaluation of freeze-thaw (FT) cycles on pancreatic fluid auto-digestion.
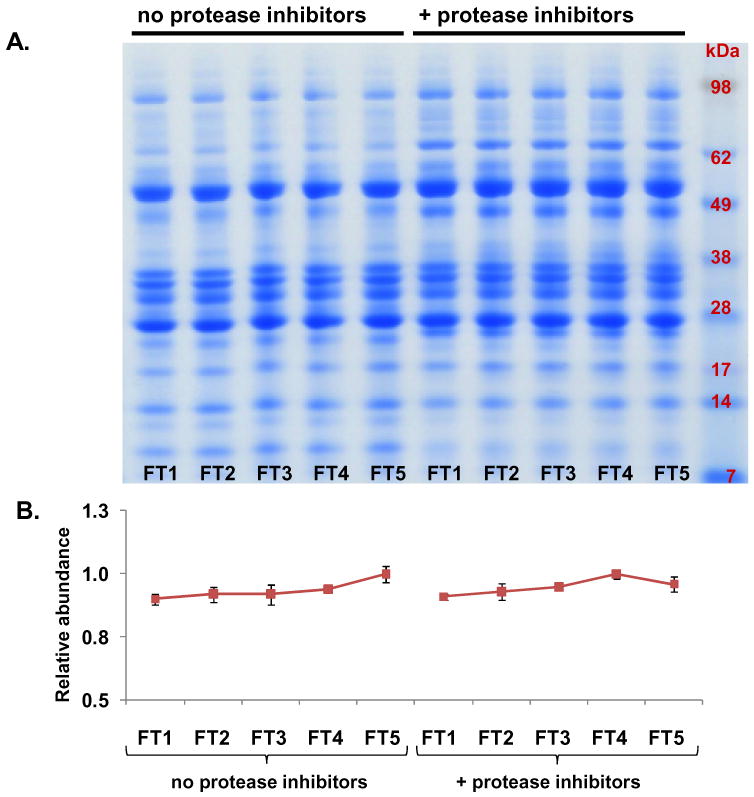
A. SDS-PAGE separation of pancreatic fluid samples undergoing a total of five freeze-thaw cycles. B. Quantitative gel densitometry measurements of entire gel lanes as determined using ImageJ software [38]. Data points for each curve were normalized with respect to the reading of the FT1 time point.
Application of optimized protocol to other pancreatic fluid samples
We assessed the application of our optimized protocol (TCA precipitation, sample handling on ice, no addition of the protease inhibitors) using a range of pancreatic fluid samples. These specimens were collected from 12 different patients diagnosed with either chronic abdominal pain (CAP), acute pancreatitis (AP), chronic pancreatitis (CP), or cystic neoplasms (CN). The specimens were processed with our optimized method, resulting in the SDS-PAGE analysis illustrated in Figure 6. While some patient-to-patient variability was expected, in these results the banding pattern was significantly less variable than previously ERCP-collected pancreatic fluid specimens [27]. Compared to these previous studies, it could be argued that the overall protein banding pattern among all 12 samples was relatively consistent, although not identical. This data demonstrates that our protocol is robust, as relatively reproducible protein patterns were formed by pancreatic fluid specimens collected from patients with various pancreatic diseases.
Figure 6. Reproducibility of our optimized protocol.
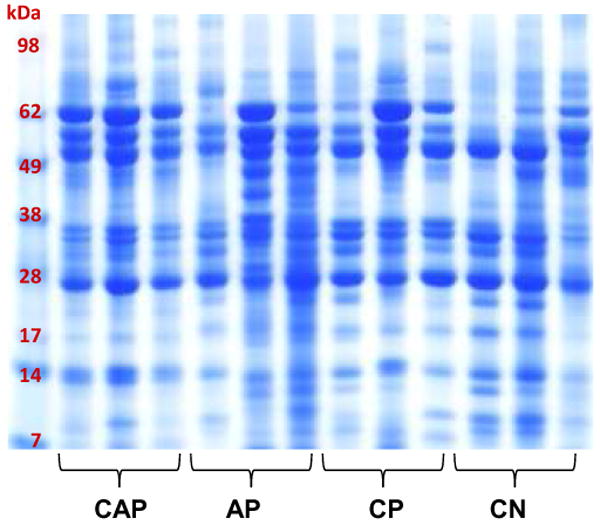
SDS-PAGE separation of TCA precipitated pancreatic fluid samples using our optimized methodology. The samples are from patients with diagnoses as follows: chronic abdominal pain (CAP), acute pancreatitis (AP), chronic pancreatitis (CP), or cystic neoplasms (CN).
4. Discussion
We have optimized the preparation of endoscopically (ePFT) collected pancreatic fluid for SDS-PAGE analysis. This is the first investigation directed toward enhancing the handling and processing of secretin-stimulated pancreatic fluid using ePFT as the collection method. We aim to maximize the integrity of our sample for downstream analyses, as protein degradation is expected in this endogenously protease-rich proximal body fluid. Each of our rigorous experiments examine a crucial aspect of sample preparation in an effort to minimize protein loss and/or degradation, during (1) protein extraction, (2) sample handling, (3) freeze-thaw cycles, and to investigate (4) the utility of protease inhibitors. Furthermore, the application of our methods to pancreatic fluid from various pancreatic diseases is robust and reproducible.
Our approach is novel as we collect our pancreatic fluid samples with the ePFT method and use TCA precipitation for protein extraction. Previous proteomic studies of the pancreas have obtained specimens using ERCP fluid collection [8, 33, 40, 41] or surgery [7, 9, 11-13]. It is not justified to obtain disease-free pancreatic fluid from healthy subjects by ERCP (endoscopic retrograde cholangiopancreatography) because of significant procedure-associated risks [8]. Thus, previously published pancreatic fluid analyses have utilized benign, non-malignant patient specimens as “surrogate” controls. To date, the proteome of pancreatic fluid from a disease-free cohort has not been investigated [42]. The ePFT method enables the collection of pancreatic fluid without cannulation of the pancreatic duct, thus avoiding any risks for procedure-related injury [18, 19, 21]. Moreover, analysis of the electrolyte composition and enzyme activity ePFT-collected fluid supports the notion that this fluid reproduces the classic acinar and duct cell secretory profiles following hormonal stimulation [16-20]. Furthermore, the ePFT is now considered an acceptable alternative method for the assessment of pancreas secretory physiology {Pollack, 2006 #1421; Forsmark, 2008 #1432; Wu, 2009 #221}. Unlike previously published investigations, ePFT significantly enhances fluid collection volume (>10-fold) thereby allowing for more comprehensive proteomic analysis. Moreover, our ePFT method is much less invasive than the aforementioned techniques (ERCP and surgery) and permits the safe collection of pancreatic fluid even from healthy individuals. Thus, the ePFT collection method is a valuable alternative tool to acquire pancreatic fluid including that from subjects without pancreatic disease. We aim to establish this collection technique as the “gold-standard” method to study proteins in pancreatic fluid.
In regards to protein extraction, we have determined that TCA precipitation resulted in the largest amount of extracted protein from pancreatic fluid (Figure 2 B) and that this approach is reproducible (Figures 4 to 5). The various methods were chosen to include common protein extraction techniques, including non-chemical methods (in vacuo centrifugation, PD-10 desalting column, microfiltration), acidification methods (TFA/acetonitrile precipitation, TCA precipitation), and use of organic solvents (ethanol precipitation, acetone precipitation). Although this list is not inclusive of all possible precipitation strategies, as such would be beyond the scope of this work, we aim to utilize those methods commonly in body fluid analyses [27]. Although some publications investigating fluids of pancreatic origin do not specifically mention the protein precipitation method used [7, 8, 11, 12, 33], other publications have cited acetone precipitation for protein extraction without indicating whether or not the samples were acidified prior to precipitation [13, 40]. However, of the methods which we examine, TCA precipitation produces the most intense and well-resolved bands. While the cold temperatures slow enzymatic reactions, TCA precipitation more efficiently and rapidly inactivates enzymes resulting in less overall protein degradation.
Although we conclude that of the approaches that we investigate, TCA is the optimal precipitation technique, we realize that improvements may also be made to the other precipitation strategies investigated herein. We note that there is some band distortion at the lower molecular weight region (below 7 kDa) in the lanes of Figure 2 corresponding to the non-chemical methods, vacuum centrifugation (vac), PD-10, and microfiltration (filtr). For these samples, additional clean-up prior to SDS-PAGE analysis, such as the use of Pierce PAGEprep (Thermo Fisher) or PlusOne (GE Healthcare), may eliminate or reduce interfering salt, lipid, or low molecular weight compounds. Alternatively, attempts to bring the electroconductivity of the samples within the same range of each other may also be beneficial to reduce band distortion. Although these additional steps may improve the banding patterns of the gel, it is unlikely that any other precipitation method would approach the precipitation efficiency of TCA.
In addition, we realize the importance to minimize protein losses due to plastic consumable for the various precipitation strategies which we investigate. From our experience, and that of others, we recognize that the quality of the microfuge tube is important to minimize protein and peptide losses. To prevent protein retention to the walls of the tube, we used low-retention tubes from Axygen, which have been shown to prevent binding to the sides of the microfuge tube [45, 46]. The use of such tubes is particularly important to the vacuum centrifuge approach, as the combined effects of heat and time will promote retention to the tube wall. In addition, we use low retention pipette tips throughout our experiments. Similarly, retention to the plastic and/or filter may also result in additional protein losses. Acidification or supplementation with a minimal volume of organic solvent may improve protein recovery. In addition, sonication may decrease retention to the walls of the tubes in all approaches investigated, including the chemical precipitation methods (e.g. TCA precipitation). Such modifications to our protocols may merit further investigation to determine if the implementation of the additional steps does, in fact, improve the relative protein recoveries of the extraction methods that we investigated.
As mentioned in the results, acidifying conditions demonstrate greater protein extraction than non-acidifying techniques. Such data are expected as pancreatic enzyme activity is inhibited by acidic conditions. We then set out to test if acidification of pancreatic fluid would enhance the frequently used precipitation methods of ethanol and acetone precipitation. We performed ethanol and acetone precipitations of pancreatic fluid samples containing increasing concentrations of TCA (0.1% to 12.5%) (Figure 3). We demonstrate that the greatest amount of protein is extracted with precipitation using 5-12.5% final volume of TCA, which is irrespective of the addition of ethanol or acetone addition. If for certain applications, ethanol or acetone is desired as the precipitation reagent, then the solutions should be acidified prior to precipitation to maximize protein extraction. However, another drawback of either acetone or ethanol precipitation is that the working volume is increased four- to nine-fold, which may be impractical if larger volumes of pancreatic fluid are necessary. An extension of our work may include pre-acidification prior to precipitation using several of the other methods. However, caution should be observed as pre-acidification with high acid concentrations may result in some protein precipitation, which may obstruct the flow of column-based protein extraction methods. In our hands, 12.5% TCA has consistently extracted the most protein from pancreatic fluid.
To test the robustness and reproducibility of TCA precipitation for SDS-PAGE analysis of pancreatic fluid specimens, subsequent experiments are performed using pancreatic fluid from 12 additional individuals, that include those with various pancreatic diseases (chronic pancreatitis, acute pancreatitis, cystic neoplasms), as illustrated in Figure 6. It is noteworthy, that the overall protein banding pattern of our samples is similar even among different pancreatic diseases despite the high concentration of endogenous proteases. Such a result underscores the notion that ePFT-collected pancreatic fluid specimens may be more adequately suited for proteomic analyses than ERCP-collected specimens.
We show that proteolytic enzymes remain active in pancreatic fluid following collection by ePFT and that elevated temperatures enhance protease activity, resulting in a higher degree of proteolysis (Figure 4). Downstream functional investigations of pancreatic enzymes may exploit this retention of activity, however, for proteomic analysis, active proteolytic enzymes are likely to be detrimental to the reproducibility of the protein banding pattern as depicted by SDS-PAGE. In this study, we perform our three experiments with and without supplementation of the protease inhibitor cocktail to investigate the utility of these proteolytic enzyme inhibitors on our sample preparation methodology. SDS-PAGE analysis (Figure 4 B) reveals that protein degradation in pancreatic fluid (prior to precipitation and to which no protease inhibitor cocktail is added) is evident after 2 hours and 30 minutes at 23°C and 37°C, respectively. Specifically, the proteins above 62 kDa and below 17 kDa are degraded more readily in samples without protease inhibitor supplementation, as expected. However, even with the addition of protease inhibitors, some protein degradation can be observed, which may be attributed to the limited half-life and reversibility of the protease inhibitors, as well as the high concentration of endogenous proteases. In particular, we note greater protein degradation in a relatively intense protein band of approximately 50 kDa in the sample supplemented with the protease inhibitor cocktail at incubation temperatures of 23°C and 37°C compared to the sample without the protease inhibitors at these temperatures. This difference may indicate that selective degradation is occurring, i.e., certain proteins/enzymes are more prone to be degraded than others. Further studies investigating the addition of individual protease inhibitors may provide insight into specifically which proteases are active in the pancreatic fluid [47], potentially resulting in the differentiation among various pancreatic disease states according to enzymatic activity.
We also illustrate, using SDS-PAGE analysis (Figures 2 to 6), that auto-digestion is minimized whether or not the sample is supplemented with the protease inhibitors if samples are handled on ice and/or are TCA precipitated. However, there are instances when the presence or absence of protease inhibitors may be detrimental to particular experiments, so that the use of protease inhibitors is assay-specific. For example, certain protease inhibitors may interfere with bottom-up proteomic experiments (particularly for in-solution digests) where specific enzymatic digestion (e.g., with trypsin) is necessary. Also, protease inhibitors should not be added to pancreatic peptidomics studies aiming to investigate the temporal changes of enzyme activity [48, 49]. Conversely, the addition of protease inhibitors would not be detrimental, but might be advantageous for many applications. The addition of protease inhibitors to pancreatic fluid to be processed further by in-gel tryptic digestion would not be likely to have adverse effects, as low molecular weight protease inhibitors are separated from the proteins and/or inactivated during SDS-PAGE. However, supplementation of protease inhibitors may also unintentionally alter the migration of some proteins on SDS-PAGE gels and thus have an effect on the overall protein banding pattern [50]. In addition, protease inhibitor supplementation may be beneficial for the shipment of pancreatic fluid for off-site analysis that may risk changes in temperature during shipment or storage. Although we show that pancreatic fluid proteins are resistant to degradation resulting from multiple freeze-thaw cycles, it may be of additional benefit to store samples for extended periods in smaller aliquots to minimize, or essentially eliminate, the need for multiple sample thaws.
The Roche Complete Protease Inhibitor tablets used in this experiment are of a proprietary formulation. We, however, chose to supplement our samples with the convenient, effective, and commonly-used protease inhibitor cocktail according to the manufacturer's recommended final concentration. Nevertheless, as the major protein components of pancreatic fluid are proteolytic enzymes, future experiments may be designed to investigate the addition of this protease inhibitor cocktail at a higher concentration to assay for enhanced effectiveness of protease inhibition at elevated temperatures. In addition, future studies may utilize known mixtures of protease inhibitors and investigate the effects of each inhibitor on protein degradation and/or migration patterns.
In all samples that were supplemented with the Roche Complete Protease Inhibitor tablets, there is a prominent band that migrates above the 62 kDa protein standard band (Figures 4 and 5). In Figure 4, this band is present in the t = 0 samples, however, it is degraded after 24 hours at 37°C. Coincidently, this band is not degraded during freeze-thaw cycles (Figure 5). Single band-targeted mass spectrometric analysis of the in-gel tryptic digestion of this band identified several proteins including α2-macroglobulin, trypsin, carboxypeptidase, and lipase, all of which have been identified in the more intense band migrating at approximately 50 kDa. It is possible that these proteins have been covalently modified by the protease inhibitors, thus shifting the protein electrophoretic mobility on the SDS-PAGE gel. However, more in-depth analysis will be necessary to determine if this band is significant or simply an artifact of protease inhibitor addition. It is also worth noting a protein band which migrates between the 7 and 14 kDa protein standard markers in the protease inhibitor-deficient samples, but not in the protease inhibitor-containing samples. Single band-targeted in-gel tryptic digestion identified peptides associated with common pancreatic fluid proteins (e.g., albumin, amylase, trypsin, and lipase, which we also identified in other bands in this lane). As these proteins migrate at such a low molecular weight and their intensity increase slightly over time, we believe that the presence of these fragments is due to protease activity in the pancreatic fluid. Such a result indicates that some degree of proteolysis has indeed been prevented by the addition of protease inhibitors, as these bands are not present in the protease inhibitor-deficient sample.
The comprehensive analysis of protein mixtures, i.e. the identification of hundreds or thousands of proteins, is possible for clinical applications with recent developments in high-throughput mass spectrometry [51-54]. Proteomics can facilitate the elucidation the proteins which regulate the pathogenesis of pancreatic disease and facilitate the discovery of clinically-relevant biomarkers. However, the quality of proteomic results depends heavily on the methodology by which samples are prepared. Variations in methods may introduce discrepancies that can impede the progress of pancreatic fluid proteomics. Standardized methods, such as those investigated herein, can minimize the heterogeneity of samples and maximize protein extraction by reducing protein degradation. The establishment and optimization of clear and consistent sample collection, handling, and processing methodologies is paramount to the development of clinical proteomics assays. Our sample optimization experiments were designed to create a platform upon which such assays can be further developed.
In summary, we have shown that (1) enzyme inactivation and protein extraction of ePFT-collected pancreatic fluid is maximized using TCA precipitation, (2) auto-digestion is minimal if samples are efficiently handled or stored on ice for up to 8 hours, and (3) protein degradation remains limited after multiple freeze-thaw cycles. Furthermore, we have shown that the addition of protease inhibitors may be ineffective over extended periods at elevated temperatures and that their addition is assay specific. Finally, application of our optimized methodology to pancreatic fluid from various pancreatic disease states revealed robust and reproducible protein banding patterns via SDS-PAGE analysis. Our optimized sample preparation methods can be utilized in future studies further investigating the proteome of pancreatic fluids.
Acknowledgments
We would like to thank the Burrill family for their generous support through the Burrill Research Grant. In addition, funds were provided by the Harvard Digestive Diseases Center (NIH 5 P30 DK034854-24) and the NIH/NIDDK NRSA Fellowship (NIH NIDDK 1 F32 DK085835-01A1). We would like to thank members of the Steen Lab at Children's Hospital Boston and Harvard Medical School, in particular Damon Anderson, Robert Everley, Yin Yin Lin, Zachary Waldon, and Dominic Winter for their technical assistance and critical reading of the manuscript. We would also like to thank members of the therapeutic endoscopy group at Brigham and Women's Hospital and Harvard Medical School, in particular Christopher Thompson, David Carr-Locke and John Saltzman, for their assistance in specimen collection techniques.
Abbreviations
- ePFT
endoscopic pancreatic function test
- ERCP
endoscopic retrograde cholangiopancreatography
- MES-SDS
2-(N-morpholino)ethanesulfonic acid-sodium dodecyl sulfate
- LDS
lithium dodecyl sulfate
- vac
vacuum centrifugation
- PD10
PD-10 column
- filtr.
5kDa molecular weight cut-off filtration
- C4
C4 trapping column
- TFA/A
triflouroacetic acid/acetonitrile precipitation
- TCA
trichloroacetic acid precipitation
- EtOH
ethanol precipitation
- ace.
acetone precipitation
- PI
protease inhibitors
- EUS-FNA
endoscopic ultrasound-guided fine needle aspiration
- CAP
chronic abdominal pain
- CP
chronic pancreatitis
- AP
acute pancreatitis
- CN
cystic neoplasms
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Everhart JE, Ruhl CE. Gastroenterology. 2009;136:1134–1144. doi: 10.1053/j.gastro.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 2.Fagenholz PJ, Castillo CF, Harris NS, Pelletier AJ, Camargo CA., Jr Annals of epidemiology. 2007;17:491–497. doi: 10.1016/j.annepidem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Fagenholz PJ, Fernandez-del Castillo C, Harris NS, Pelletier AJ, Camargo CA., Jr Pancreas. 2007;35:302–307. doi: 10.1097/MPA.0b013e3180cac24b. [DOI] [PubMed] [Google Scholar]
- 4.Ghaneh P, Costello E, Neoptolemos JP. 2008:478–497. doi: 10.1136/gut.2006.103333. [DOI] [PubMed] [Google Scholar]
- 5.Issaq HJ, Xiao Z, Veenstra TD. Chemical reviews. 2007;107:3601–3620. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 6.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Brentnall TA, Pan S, Cooke K, et al. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Pan S, Cooke K, Moyes KW, et al. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R, Yi EC, Donohoe S, Pan S, et al. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Tian M, Zong M, Teng M, et al. Pancreatology. 2008;9:89–98. doi: 10.1159/000178879. [DOI] [PubMed] [Google Scholar]
- 11.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, et al. Journal of proteome research. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 12.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, et al. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Ke E, Patel BB, Liu T, Li XM, et al. Pancreas. 2009;38:33–42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Simeone DM, Brenner DE, Anderson MA, et al. Journal of proteome research. 2008;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Lee WN, Lim S, Go VL, et al. Analytical chemistry. 2009;81:764–771. doi: 10.1021/ac801905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens T, Conwell DL, Zuccaro G, Van Lente F, et al. Gastrointest Endosc. 2004;60:351–355. doi: 10.1016/s0016-5107(04)01809-7. [DOI] [PubMed] [Google Scholar]
- 17.Stevens T, Conwell D, Zuccaro G, Van Lente F, et al. Dig Dis Sci. 2004;49:1405–1411. doi: 10.1023/b:ddas.0000042238.80040.cc. [DOI] [PubMed] [Google Scholar]
- 18.Conwell DL, Zuccaro G, Jr, Vargo JJ, Morrow JB, et al. Clin Gastroenterol Hepatol. 2003;1:189–194. doi: 10.1053/cgh.2003.50028. [DOI] [PubMed] [Google Scholar]
- 19.Conwell DL, Zuccaro G, Jr, Vargo JJ, Trolli PA, et al. Gastrointest Endosc. 2003;57:37–40. doi: 10.1067/mge.2003.14. [DOI] [PubMed] [Google Scholar]
- 20.Conwell DL, Zuccaro G, Morrow JB, Van Lente F, et al. Pancreas. 2002;25:350–354. doi: 10.1097/00006676-200211000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, Conwell DL. The American journal of gastroenterology. 2009;104:2381–2383. doi: 10.1038/ajg.2008.181. [DOI] [PubMed] [Google Scholar]
- 22.Barratt J, Topham P. Cmaj. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decramer S, Gonzalez de Peredo A, Breuil B, Mischak H, et al. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Hortin GL, Sviridov D. Pharmacogenomics. 2007;8:237–255. doi: 10.2217/14622416.8.3.237. [DOI] [PubMed] [Google Scholar]
- 25.Muller H, Brenner H. Clinical chemistry. 2006;52:562–573. doi: 10.1373/clinchem.2005.062919. [DOI] [PubMed] [Google Scholar]
- 26.Munro NP, Cairns DA, Clarke P, Rogers M, et al. International journal of cancer. 2006;119:2642–2650. doi: 10.1002/ijc.22238. [DOI] [PubMed] [Google Scholar]
- 27.Thongboonkerd V. Proteomics of human body fluids : principles, methods, and applications. Humana Press; Totowa, N.J.: 2007. [Google Scholar]
- 28.Thongboonkerd V. Molecular bioSystems. 2008;4:810–815. doi: 10.1039/b802534g. [DOI] [PubMed] [Google Scholar]
- 29.Havanapan PO, Thongboonkerd V. Journal of proteome research. 2009;8:3109–3117. doi: 10.1021/pr900015q. [DOI] [PubMed] [Google Scholar]
- 30.Mary Jayne K, Angela G, Ruifeng S, Michael M, Jon K. PROTEOMICS - CLINICAL APPLICATIONS. 2009;3:989–999. doi: 10.1002/prca.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thongboonkerd V, Mungdee S, Chiangjong W. Journal of proteome research. 2009;8:3206–3211. doi: 10.1021/pr900127x. [DOI] [PubMed] [Google Scholar]
- 32.Lohr M, Faissner R. Pancreatology. 2004;4:67–75. doi: 10.1159/000077212. [DOI] [PubMed] [Google Scholar]
- 33.Wandschneider S, Fehring V, Jacobs-Emeis S, Thiesen HJ, Lohr M. Electrophoresis. 2001;22:4383–4390. doi: 10.1002/1522-2683(200112)22:20<4383::AID-ELPS4383>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 34.DeWitt J, Jowell P, Leblanc J, McHenry L, et al. Gastrointestinal endoscopy. 2005;61:689–696. doi: 10.1016/s0016-5107(05)00287-7. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez LV, Bhutani MS. Gastrointest Cancer Res. 2008;2:198–202. [PMC free article] [PubMed] [Google Scholar]
- 36.Forrester RL, Wataji LJ, Silverman DA, Pierre KJ. Clin Chem. 1976;22:243–245. [PubMed] [Google Scholar]
- 37.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 38.Girish V, Vijayalakshmi A. Indian J Cancer. 2004;41:47. [PubMed] [Google Scholar]
- 39.Mechin V, Damerval C, Zivy M. Methods in molecular biology. Vol. 355. Clifton, N.J: 2007. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Lu Z, Yang A, Deng R, et al. Proteomics. 2007;7:1345–1355. doi: 10.1002/pmic.200600086. [DOI] [PubMed] [Google Scholar]
- 41.Chen R, Pan S, Yi EC, Donohoe S, et al. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 42.Chen R, Pan S, Brentnall TA, Aebersold R. Mol Cell Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Pollack BJ, Grendell JH. Am J Gastroenterol. 2006;101:356–359. doi: 10.1111/j.1572-0241.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 44.Forsmark CE. Clin Gastroenterol Hepatol. 2008;6:1291–1293. doi: 10.1016/j.cgh.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Bark SJ, Hook V. Journal of proteome research. 2007;6:4511–4516. doi: 10.1021/pr070294o. [DOI] [PubMed] [Google Scholar]
- 46.Gaillard C, Strauss F. American Clinical Laboratory. 2001;20:52–55. [Google Scholar]
- 47.Robinson S, Niles RK, Witkowska HE, Rittenbach KJ, et al. Proteomics. 2008;8:435–445. doi: 10.1002/pmic.200700680. [DOI] [PubMed] [Google Scholar]
- 48.Schulz-Knappe P, Schrader M, Zucht HD. Comb Chem High Throughput Screen. 2005;8:697–704. doi: 10.2174/138620705774962418. [DOI] [PubMed] [Google Scholar]
- 49.Ivanov VT, Yatskin ON. Expert Rev Proteomics. 2005;2:463–473. doi: 10.1586/14789450.2.4.463. [DOI] [PubMed] [Google Scholar]
- 50.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, et al. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 51.Han X, Aslanian A, Yates JR., 3rd Current opinion in chemical biology. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsangaris GT. Expert Rev Proteomics. 2009;6:235–238. doi: 10.1586/epr.09.14. [DOI] [PubMed] [Google Scholar]
- 53.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 54.Latterich M, Abramovitz M, Leyland-Jones B. Eur J Cancer. 2008;44:2737–2741. doi: 10.1016/j.ejca.2008.09.007. [DOI] [PubMed] [Google Scholar]