Abstract
While women are more vulnerable than men to many of the medical consequences of alcohol abuse, the role of sex in the response to ethanol is controversial. Neuroadaptive responses that result in the hyperexcitability associated with withdrawal from chronic ethanol likely reflect gene expression changes. We have examined both genders for the effects of withdrawal on brain gene expression using mice with divergent withdrawal severity that have been selectively bred from a genetically heterogeneous population. A total of 295 genes were identified as ethanol regulated from each gender of each selected line by microarray analyses. Hierarchical cluster analysis of the arrays revealed that the transcriptional response correlated with sex rather than with the selected withdrawal phenotype. Consistent with this, gene ontology category over-representation analysis identified cell death and DNA/RNA binding as targeted classes of genes in females, while in males, protein degradation, and calcium ion binding pathways were more altered by alcohol. Examination of ethanol-regulated genes and these distinct signaling pathways suggested enhanced neurotoxicity in females. Histopathological analysis of brain damage following ethanol withdrawal confirmed elevated cell death in female but not male mice. The sexually dimorphic response was observed irrespective of withdrawal phenotype. Combined, these results indicate a fundamentally distinct neuroadaptive response in females compared to males during chronic ethanol withdrawal and are consistent with observations that female alcoholics may be more vulnerable than males to ethanol-induced brain damage associated with alcohol abuse.
Keywords: alcoholism, microarray, kwdneurotoxicity, animal model
INTRODUCTION
Alcohol (ethanol; EtOH) abuse is a major public health problem; it is now estimated that harm caused by EtOH nearly equals that from smoking (Pearson, 2004). About half of the nearly 20 million alcoholics in the United States develop neuropsychological difficulties (Oscar-Berman and Marinkovic, 2003). Alcoholic dementia, associated with global severe amnesia, and intellectual impairment, is the second-leading cause of adult dementia in the United States and accounts for 10% of all cases (Eckardt and Martin, 1986). It has been observed that, ‘depending on age, the brain of the detoxified alcoholic can appear as ravaged as that of a patient with Alzheimer's disease’ (Sullivan and Pfefferbaum, 2005). The neurotoxicity associated with chronic EtOH abuse is postulated to result from decreased γ-aminobutyric acid and increased glutamate function, resulting in oxidative stress and excitotoxic brain damage (Prendergast et al, 2004). Although smoking and nutritional deficiencies also contribute to disease, regional brain damage and cognitive dysfunction are seen even in uncomplicated alcoholics with no specific neurological or hepatic problems (Gazdzinski et al, 2005; Harper and Matsumoto, 2005).
Long-term chronic EtOH consumption can produce physical dependence as measured by signs of withdrawal, considered a hallmark of alcoholism. This process is hypothesized to be mediated by molecular and structural changes in the central nervous system (CNS) termed neuroadaptation, a homeostatic response to the presence of EtOH, a CNS depressant. The net effect of these responses is increased neuronal excitability, the development of tolerance to the effects of EtOH, and following long-term exposure, physical dependence. EtOH withdrawal (ie removal of EtOH) unmasks this state of neuronal hyper-excitability, with a constellation of symptoms including seizures as one manifestation that is observed in nearly every species studied. Estimates suggest that 4–7% of alcoholics suffer from seizures (Hauser, 1990). In addition, withdrawal likely plays a major role in alcohol-induced neurotoxicity (Favalli et al, 2002). Finally, withdrawal severity has been shown to be inversely genetically correlated with alcohol consumption in animal models (Metten et al, 1998). It is thus important to develop a better understanding of the mechanisms underlying EtOH dependence and withdrawal.
Sex (biological gender) has been implicated as a relevant variable in many neurological diseases in humans including a liability for alcohol abuse (Holden, 2005), consistent with females having a lower incidence of alcoholism (Kessler et al, 1994). Thus, alcoholism has traditionally been considered a male disease because there are more alcoholic males than females. However, increased EtOH use in females is being observed, such that almost half of all women report drinking alcohol to excess (Schmid et al, 2003). This problem will likely increase as young women are drinking more, with consumption by 18 to 24-year-old women across both the United States and Europe rising from 110 per person in 1999 to 148 liters in 2004 (Butler, 2005). Furthermore, the incidence of and the frequency of women seeking help for alcoholism are both increasing, demonstrating that alcoholism is a clinical problem in women as well as men (Breitenfeld et al, 1998).
Nevertheless, alcoholic women have received less research attention than alcoholic men despite good evidence that women may be particularly vulnerable to alcohol's damaging effects on many key organ systems. For example, alcoholic women develop cirrhosis (Loft et al, 1987), alcohol-induced cardiomyopathy (Fernandez-Sola and Nicolas-Arfelis, 2002) and peripheral neuropathy (Ammendola et al, 2000) after fewer years of heavy drinking than do alcoholic men. Although sex differences have been only infrequently addressed with respect to EtOH-induced brain damage, accumulating data indicate that brain damage may also be enhanced in females, with males showing relative neuroprotection (Hommer et al, 2001). It appears that women suffer as much brain damage as men after a shorter period of abuse (so-called telescoping), also suggesting a higher vulnerability to EtOH damage among women (Mann et al, 2005). However, other studies have been contradictory (Hommer, 2003). Thus, both the observation of risk in female alcoholics (Wuethrich, 2001) and the potential mechanism(s) underlying EtOH-induced brain damage remain controversial, and such analysis is difficult to perform in human populations. Notwithstanding the clinical evidence regarding the distinct biological and behavioral responses of females to EtOH, few studies have specifically addressed the response of the CNS to EtOH between females and males.
The purpose of this study was to characterize CNS gene expression differences associated with neuroadaptive responses after chronic EtOH exposure and withdrawal in both sexes. We employed lines of mice with divergent withdrawal severity derived by selective breeding from a genetically heterogeneous population; the Withdrawal Seizure-Prone (WSP) and Withdrawal Seizure-Resistant (WSR) mice (Kosobud and Crabbe, 1986). In addition to extreme differences in EtOH-withdrawal severity, WSP, and WSR mice exhibit many EtOH-related behavioral differences, including dissimilarities in voluntary consumption (Kosobud et al, 1988), conditioned taste aversion (Chester et al, 1998), and anxiety (Atkins et al, 2000), as correlated responses. Given the complex polygenic nature of human alcoholism, dissecting genetic contributions to the disease is difficult. The use of selected lines provides a genetically rich model where the various alleles related to the selection phenotype present in the initial heterogeneous population become segregated in the respective lines. The WSP and WSR lines serve as an animal model of extremes in neuronal hyperexcitability following chronic EtOH exposure, and thus represent an important tool for the identification of underlying neuroadaptive gene expression changes. To address molecular mechanisms underlying neuroadaptive changes associated with alcohol abuse, we used microarray hybridization to characterize gene expression profiles after chronic EtOH exposure at peak withdrawal in male and female WSP and WSR selected lines, with unsupervised hierarchical cluster and gene ontology (GO) and k-means cluster analyses for characterization, and brain histological examination for biological confirmation.
METHODS
Animal Subjects
The WSP and WSR selective breeding protocol was replicated; thus there are two independently derived replicate WSP and WSR lines (Kosobud and Crabbe, 1986). Female and male mice from both replicates were tested in these studies. WSP and WSR mice, derived by selective breeding from the genetically heterogeneous HS/Ibg mice, were provided by the laboratory of Dr John Crabbe in Portland, OR. Drug-naïve adult mice from selected generation 26 (filial generations G77-G90) were used. Ages of the animals at the onset of the experiments ranged from 55–93 days, mean age 77 days; body weights ranged from 21.6–34.6 g, with a mean body weight of 27.6 g. Mice were maintained under a light/dark cycle of 0600–1800 h light with water and Purina Lab Diet chow available ad libitum. Room temperatures were maintained at 22±1°C. All animal procedures were approved by the Portland Oregon VA Medical Center Institutional Animal Care and Use Committee and followed US National Institutes of Health animal welfare guidelines.
EtOH Exposure and Brain Harvest
Mice were made dependent upon EtOH using a method of vapor inhalation in chambers manufactured in-house, with modifications published previously (Beadles-Bohling and Wiren, 2005, 2006). EtOH exposure was initiated between 0730 and 0830 h. Briefly, on day 1 EtOH mice were weighed, injected i.p. with EtOH at 1.5 g/kg for WSP-1, WSR-1, and WSR-2 and 1.75 g/kg for WSP-2 animals (necessary to maintain similar blood EtOH concentrations (BECs) between the selected lines (Terdal and Crabbe, 1994)) and 1.0 mmol/kg pyrazole HCl (Pyr; an alcohol dehydrogenase inhibitor used to maintain constant blood EtOH levels). Control animals were placed into air chambers and received Pyr only; a saline-only control was not included because previous data has shown there is no difference between saline and Pyr-treated animals with respect to broad profiles of gene expression analyzed by mRNA differential display (Schafer et al, 1998 and Wiren KM, unpublished observation). All mice received Pyr boosters (68.1 mg/kg) at 24 and 48 h to reduce fluctuations in BEC. Following 72 h of constant EtOH vapor exposure, all mice were removed from the chambers, weighed, and had blood samples drawn for BEC determinations. BEC was determined by gas chromatography as described previously (Beadles-Bohling and Wiren, 2006). Brain tissue was harvested from animals 8 h after removal from the chambers for RNA analysis, a time point corresponding to peak withdrawal in WSP mice as assessed by handling induced convulsions (HIC) (Finn and Crabbe, 1999). All animals used in these experiments were purposely not scored for HIC to limit the effects of withdrawal seizures per se on measurements of both gene expression and brain damage. Prefrontal cortex (PFC) was carefully isolated by dissection from whole brain by first discarding the olfactory bulb and making a coronal slice 2 mm into the frontal region of the cortex. Lateral regions were removed by cutting the tissue at a 45° angle with the lobes facing upward. Animals used for brain histology were allowed to proceed through withdrawal and were euthanized 10 days after the cessation of EtOH exposure.
Chemicals
Pyrazole HCl (Pyr) was purchased from Sigma-Aldrich Chemical Co. (St Louis, MO). EtOH (ethyl alcohol, absolute, 200 proof) for use in chemical assays was purchased from AAPER Alcohol and Chemical (Shelbyville, KY), and Pharmco Products Inc. (Brookfield, CT) for use in the EtOH vapor chambers and injections. Pyr was dissolved in 0.9% saline (NaCl) and administered via intraperitoneal (ip) injection. EtOH (20% v/v) was mixed with 0.9% saline and injected ip or introduced without mixing as a vapor into the chambers.
RNA Isolation and GeneFilter Microarray Processing
Total RNA was isolated using RNA Stat-60 (Tel-Test, Inc. Friendswood, TX). To remove traces of contaminating genomic DNA, the RNA was digested by RNase-free DNase followed by Zymo-spin column purification following manufacturer's recommendations (Zymo Research, Orange, CA). DNase treated total RNA isolated from the PFC of two animals (2.5 μg each) was pooled for each complex probe labeling reaction. Probe labeling from the total RNA mixture was performed by linear synthesis with 33P-dATP incorporation using the Array Advantage kit (Ambion, Austin, TX). Complex probes were purified by passage through a NucAway spin column (Ambion, Austin, TX). Efficiency of incorporation generally ranged between 15 and 20% using 5 μg total RNA. Microarray hybridization with complex labeled RNA probe was performed overnight at 50°C with 20 ml Ultra-array hybridization buffer (Ambion, Austin, TX). The final post-hybridization wash was at 50°C in 0.5 × SSC, 0.5% SDS similar to standard Northern blot procedures.
Research Genetics GF400 mouse microarrays (Invitrogen, Carlsbad, CA) employed for these studies contained 4300 unique mouse cDNA accessions (both genes and ESTs) that average 1500 bp in length, generally with one spot per cDNA. Several advantages are notable with the use of cDNA arrays. First, the gene sets chosen by Invitrogen are highly enriched in expressed sequences (improving our ability to characterize the biological consequences of expression differences). Furthermore, the longer hybridization sequences vs oligos allow for higher stringency in the hybridization and wash procedures, for greater gene specificity, and better characterization of genes on the array (Dai et al, 2005). Generation of the complex probe is performed by linear reverse transcription, that is, without additional steps or amplification procedures that potentially skew probe populations. Thus, the higher sensitivity of cDNA arrays using radioactive detection (Bertucci et al, 1999) is particularly advantageous in the identification of differences in ethanol-regulated transcription, known to demonstrate only modest regulation (Treadwell and Singh, 2004). Background hybridization values (typically very low) were subtracted from each hybridization value. Some arrays were excluded from analysis based on correlation coefficients values below 90% for ‘replicate’ arrays (ie Female WSP-1 Air Array 1 vs Female WSP-1 Air Array 2).
Microarray Data Analysis
Microarrays were exposed to storage phosphor screens for 2 h and scanned at high resolution with a Cyclone phosphorimager (Packard Bell-Perkin Elmer, Shelton, CT). Data from all samples were log2-transformed, converted into z-scores, multiplied by two and added to eight so that each array had a mean of eight, a variance of four, a SD of two (WebQTL, Wang et al, 2003b). This conversion procedure results in a 1 unit expression difference corresponding to a twofold change in expression. The level of expression of each gene or EST was determined using OptiQuant software and imported into Pathways v4 software (Research Genetics, Carlsbad, CA). Raw expression values were then exported, normalized, and analyzed using Vector Xpression 3.1 (Invitrogen, Carlsbad, CA). Differential regulation was determined by comparing groups in the following manner with duplicate arrays. For male WSR mice, EtOH regulation was determined by comparing four WSR arrays from eight male air-exposed WSR mice to four WSR arrays from eight male EtOH-exposed WSR mice. An uncorrected Student's t-test (n = 4 for each group, two-tailed, α = 0.05) was used to identify a large group of differentially regulated transcripts (Rodd et al, 2006). Similar to that report, we chose to relax the stringency of our analysis to increase the likelihood of identifying broad gene categories that are regulated by EtOH, recognizing the possibility of an increase in false positives.
All significantly regulated transcripts were used in an unsupervised hierarchical cluster analysis (average linkage, Euclidian distance) with 100 resampling iterations to determine Bootstrap values for each node using the TIGR Multiple Experiment Viewer (TMEV) software (The Institute For Genomic Research, see reference Saeed et al, 2003). Multivariate analysis of variance (MANOVA) of microarray data was done using 50–50 MANOVA (Langsrud, 2002). Additional multivariate analysis using k-means clustering was carried out using TMEV with the number of clusters set to 10, average linkage, and Euclidian distance metric.
To identify cellular pathways and biological themes that were affected during EtOH withdrawal, we used EASE (http://david.niaid.nih.gov/david/ease.htm) to assign genes to the categories of the Gene Ontology Consortium (www.geneontology.org) as described in Ashburner et al (2000) and to test statistically (EASE Score, a modified Fisher's exact test) for significant coregulation. This approach identified over-representation of regulated genes (based on spotted clones) within each category. Selected gene expression differences were confirmed by qRT-PCR analysis (see Supplementary Methods in supporting information online for details).
Assessment of EtOH-Induced Brain Damage
For histological analysis, whole brain was fixed and paraffin-embedded. Adjacent 6 μm coronal sections of the forebrain area that included the dorsal hippocampus and parietal cortex were made. Dead or dying neurons were identified in H&E stained sections (for details, see Supplementary Methods in supporting information online). Three contiguous images were captured (904 × 675 μm each) that encompassed the parietal cortex (at co-ordinates Bregma –1.94 to –2.06) of both the right and the left hemispheres using an AxioPlan2e microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY) at × 100 magnification. Counts were determined in the lateral parietal cortex, after visual examination for regions of consistent cell damage. Full thickness of each image was sampled five times counting live, dead, and ambiguous cells using MetaMorph software (Universal Imaging Corporation, Downingtown, PA, USA). All cells were counted because cellularity and vulnerability may be dissimilar in different layers. All counts were conducted with the investigator blind to treatment, selected line, and sex.
Statistical Analyses
Data were analyzed using Prism v4.0 software (GraphPad Software Inc., San Diego, CA), SYSTAT 11 (SYSTAT Software Inc., Point Richmond, CA), and Vector Xpression 3.1 (Invitrogen, Carlsbad, CA). Differences of P<0.05 were considered statistically significant. Results are presented as the mean±SEM. Individual gene expression differences were analyzed by two-tailed t-test between EtOH treated and control samples from each gender and line. Brain damage assessment consisted of three-way ANOVA (sex-treatment × selected line). Weight change during EtOH exposure experiments were analyzed by three-way ANOVA (sex × treatment × selected line). BECs during EtOH exposure were analyzed by two-way ANOVA (sex × selected line), using Bonferonni's test in post hoc analysis.
RESULTS
Chronic EtOH Exposure and Withdrawal
Over the course of the 72 h of EtOH exposure, BECs were monitored daily and EtOH vapor levels were modified to achieve BECs of 2.1–2.8 mg/ml (46–62 mM). Notably, these blood levels are within the range (0.5–5.6 mg/ml; 10–125 mM) typically seen in alcoholic patients (Adachi et al, 1991). Thus, the average BECs during the chronic exposure paradigm were in the highly intoxicated range as follows: female WSP, 2.42±0.12; male WSP, 2.79±0.09; female WSR, 2.14±0.07; male WSR, 2.71±0.06 (mg/ml mean ± SEM, with replicate-1, -2 collapsed). BECs in this range would consistently result in severe withdrawal in WSP, but very modest to no withdrawal in WSR as indexed by HIC, a sensitive measure of neuronal hyperexcitability (see Beadles-Bohling and Wiren, 2006). Two-way ANOVA of BEC levels showed a significant sex effect (F(1, 48)=23.941, P<0.0001) with no interactions of line by sex. Bonferroni post-test analysis showed both WSP and WSR males had higher BECs than female WSP and WSR (P<0.01, P<0.001; respectively). With regard to weight loss, three-way ANOVA of line, sex, and EtOH treatment effects on percent weight change over 72 h of EtOH or control exposure show significant effects of sex (F(1, 99)=12.591, P=0.001) and treatment (F(1,99)=97.447, P=0.0001) and a significant interaction of sex by treatment (F(1, 99)=4.160, P=0.044). Post hoc analysis showed EtOH treated animals had more weight loss during exposure (P<0.0001); males had modestly more weight loss during exposure than females (10 vs 14% respectively, P=0.009), consistent with their higher EtOH levels.
Gene Expression Differences During EtOH Withdrawal
We examined the effect of withdrawal from chronic EtOH exposure on brain gene expression in both sexes of WSP and WSR mice, with the expectation that the use of selected lines would aid in our identification of potential mediators of neuroadaptive responses to chronic EtOH underlying the large behavioral difference in EtOH withdrawal severity between the lines. Expression analysis was performed with samples isolated from PFC, since the PFC is activated by withdrawal (Kerns et al, 2005). Additional rationale for analysis in PFC included findings of expression differences in human alcoholics (Flatscher-Bader et al, 2006; Johansson et al, 2007; Lewohl et al, 2004), reports that PFC is a target of brain damage associated with chronic alcohol abuse (Pfefferbaum et al, 1997; Sullivan and Pfefferbaum, 2005), and that PFC can be dissected reproducibly. We used an uncorrected P-value to decrease the chance of excluding EtOH-regulated transcripts (false negatives). Confirmation of differences by qRT-PCR analysis (Supplementary Methods and Supplementary Figure 2 in supporting information online) with samples derived from animals distinct from those employed for the microarray hybridizations indicated a percent positive that was consistent with reports in the literature using other array platforms and analytical approaches (see Tang et al, 2006; Treadwell and Singh, 2004). We selected genes for confirmation from pathways identified as influenced by withdrawal, particularly those involved in cell death and anti-inflammatory processes. Of the 10 selected significantly regulated sequences from the different conditions identified in the microarray hybridization described below, qRT-PCR analysis confirmed seven in this subset, which is notable in that we did not employ an arbitrary cutoff for the level of regulation for these analyses. We were able to confirm EtOH regulation as low as 0.28 (log 2 ratio; Diablo, female WSP) using qRT-PCR which, along with the 70% confirmation of microarray results with qRT-PCR, gives us a high degree of confidence in the array dataset. Overall in our laboratory, we have confirmed ~80% of regulated genes identified in microarray analysis using this approach (data not shown).
Alcohol-Withdrawal Differential Gene Expression in Male and Female WSP and WSR
A total of 295 EtOH up and downregulated transcripts were identified in the characterization of the transcriptional response to EtOH withdrawal in the four comparisons (male WSP, male WSR, female WSP, female WSR). We identified 59 EtOH-regulated genes in female WSP, 55 EtOH-regulated genes in male WSP, 117 EtOH-regulated genes in female WSR, and 90 EtOH-regulated genes in male WSR. The 10 most up or downregulated named sequences are included in Table 1 (for a complete list of EtOH-regulated genes, see Supplementary Table 1). Several genes involved in protein secretion or degradation/ubiquitination were among the most highly differentially expressed (Table 1), indicating one pathway activated by chronic EtOH exposure and withdrawal also previously identified by us (Schafer et al, 2001) and others (Liu et al, 2005). Many genes identified with significantly different expression levels have been identified previously as regulated by EtOH, or in a variety of models of neurodegenerative diseases (eg adenylate cyclase 7 and Ncr1, also see Supplementary Table 4 in supporting information online for additional characterization of concordance). We also identified 200 genes differentially regulated between naïve WSP and WSR mice (sexes collapsed based on selected line) and 186 genes differentially regulated between naïve male and female mice (lines collapsed based on sex). Seventeen percent of the 295 EtOH-regulated genes were identified as differentially regulated between naïve males and females, while 22% of the EtOH-regulated genes were differentially regulated between naïve WSP and WSR mice (Supplementary Table 3), suggesting that basal differences in expression between sexes or selected lines does not dramatically influence the transcriptional response to EtOH we observed.
Table 1.
Top EtOH Regulated Transcripts
| Symbol | Name | Accession | Regulation | Sex | Line |
|---|---|---|---|---|---|
| EtOH decreased | |||||
| Mterfd3 | MTERF domain containing 3 | AK047778 | –0.941 | Male | WSP |
| Yeats2 | YEATS domain containing 2 | BC057045 | –0.813 | Male | WSR |
| Acvrl1 | Activin A receptor, type II-like 1 | AK134712 | –0.800 | Female | WSP |
| Usp38 | Ubiquitin specific peptidase 38 | NM_027554 | –0.687 | Male | WSR |
| Suv420h2 | Suppressor of variegation 4–20 homolog 2 (Drosophila) | AK154848 | –0.678 | Female | WSP |
| Fndc4 | Fibronectin type III domain containing 4 | NM_022424 | –0.677 | Female | WSP |
| Taz | Tafazzin | NM_181516 | –0.675 | Male | WSP |
| Ctsw | Cathepsin W | NM_009985 | –0.670 | Male | WSP |
| Nck2 | Non-catalytic region of tyrosine kinase adaptor protein | NM_010879 | –0.663 | Male | WSR |
| Ncr1 | Natural cytotoxicity triggering receptor 1 | NM_010746 | –0.625 | Male | WSR |
| EtOH increased | |||||
| Nktr | Natural killer tumor recognition sequence | NM_010918 | 0.554 | Male | WSP |
| Adcy7 | Adenylate cyclase 7 | NM_0010377 | 0.653 | Female | WSR |
| D1Ertd161e | Galactosidase, β-1-like | NM_026187 | 0.676 | Female | WSP |
| Cnot2 | CCR4-NOT transcription complex, subunit 2 | AK138077 | 0.733 | Female | WSP |
| Xrra1 | X-ray radiation resistance associated 1 | AK161464 | 0.741 | Female | WSR |
| Psma1 | Proteasome (prosome, macropain) subunit, α-type 1 | NM_011965 | 0.762 | Male | WSR |
| Smg6 | Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) | BC047279 | 0.762 | Female | WSP |
| Asph | Aspartate-β-hydroxylase | AF289487 | 0.860 | Female | WSR |
| Alg6 | Asparagine-linked glycosylation 6 homolog (yeast, α-1,3,-glucosyltransferase) | AK038890 | 0.999 | Male | WSR |
| Ube3a | Ubiquitin protein ligase E3A | AK141032 | 1.445 | Male | WSR |
The 10 most highly up- or downregulated named sequences were identified from the list of 295 significantly regulated transcripts (for full list, see Supplementary Table 1). Transcripts are listed in ascending order. Regulation represents log 2 fold-change of EtOH vs control.
Confirmation of expression differences by qRT-PCR analysis (Supplementary Figure 1 in supporting information online) showed that although the same broad pathway was influenced in a sex-specific fashion, the identical genes were not necessarily regulated in the same way in both WSP and WSR (for either gender). In some cases, regulation of a specific gene was identified in both lines, but for other genes regulation was noted in only one of the selected lines (demonstrating the potential for gender × phenotype interactions). We confirmed Diablo, and Bclaf1 upregulation by EtOH in female WSP. Upregulation of Hbxip and down-regulation of Cx3cl1 was confirmed in male WSP. Smad3 upregulation was confirmed in both WSP and WSR males. Usp38 upregulation by EtOH in female WSR and down-regulation by EtOH in male WSR was confirmed as was Mapk9 upregulation by EtOH in female WSR mice.
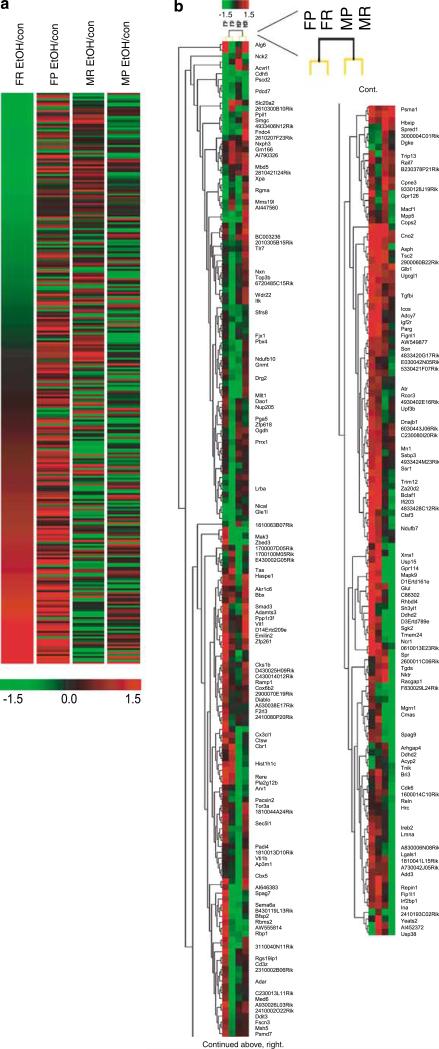
To characterize further patterns of expression differences, false color heat maps were generated for the transcripts that demonstrated significant EtOH regulation in any of the four conditions (Figure 1a). Each column represents combined data from eight arrays for one experimental analysis (four control vs four EtOH, for each sex F, M, and withdrawal phenotype P, R) to visualize the transcriptional response to EtOH withdrawal in each individual condition. Genes are depicted based on the expression ratio for the effect of EtOH vs control with shades of color to indicate EtOH withdrawal upregulation (red), downregulation (green) or no difference (black) for a given gene. Ordering of the genes was established arbitrarily in female WSR. Qualitative examination of the false color heat map suggested that gender, not the WSP or WSR phenotype, correlated best with the transcriptional response to EtOH.
Figure 1.
Heat map and hierarchical cluster analysis of significantly differentially expressed genes. Each column represents data from eight array hybridizations, using male and female WSP and WSR selected lines profiled at peak withdrawal following chronic EtOH exposure. (a) EtOH-regulated genes (in one or more comparison) were sorted based on female WSR EtOH expression ratio. (b) Two-dimensional unsupervised hierarchical clustering analysis was performed on EtOH-regulated genes. Experimental groups (arrays), represented by columns, and genes, represented by rows, were clustered according to their similarities in expression pattern. Top of dendrogram shows analysis of similarity of overall array expression profiles. In the zoomed region, bootstrap analysis using 100 resampling iterations confirmed tree topology, grouping arrays based on sex rather than withdrawal phenotype. FP=female WSP, FR=female WSR, MP=male WSP, MR=male WSR.
Unsupervised hierarchical cluster analysis was then performed to determine the similarity of global expression profiles in significantly regulated genes (Figure 1b). The top dendrogram displays similarity of gene expression for the four experimental groups; relatedness of the arrays is denoted by distance to the node linking the arrays. The two most dissimilar experimental clusters in overall differential gene expression after chronic EtOH withdrawal from all experimental conditions were split primarily upon the sex and not the withdrawal phenotype of the mice (ie female FP and FR were grouped together as related, as were male MP and MR). Thus, clustering analysis identified gender, not selected line phenotype or genotype, as contributing the strongest influence on expression patterns, consistent with the false color heat map qualitative analysis in Figure 1a. Bootstrap analysis (shown in the inset) confirmed the importance of gender, where black nodes indicates 100% support of the tree topology for separation based on sex, and yellow nodes indicates 60–70% support for similarity of the two selected lines (Figure 1b, inset).
Additional statistical characterization of expression profile differences was performed using multivariate analysis of variance (MANOVA) of microarray data with 50–50 MANOVA (Langsrud, 2002), an algorithm designed to adapt MANOVA to situations with a high number of dependent variables (eg gene expression) relative to observations (eg array experiments). In the two-way analysis of gender and line on EtOH-regulated transcripts, we identified significant effects of sex (P<0.00001), line (P<0.0001), and a significant interaction (gender × line, P=0.015). These results suggest the importance of both line and sex in the response to EtOH, but because of the significant interaction, each experimental group was analyzed separately.
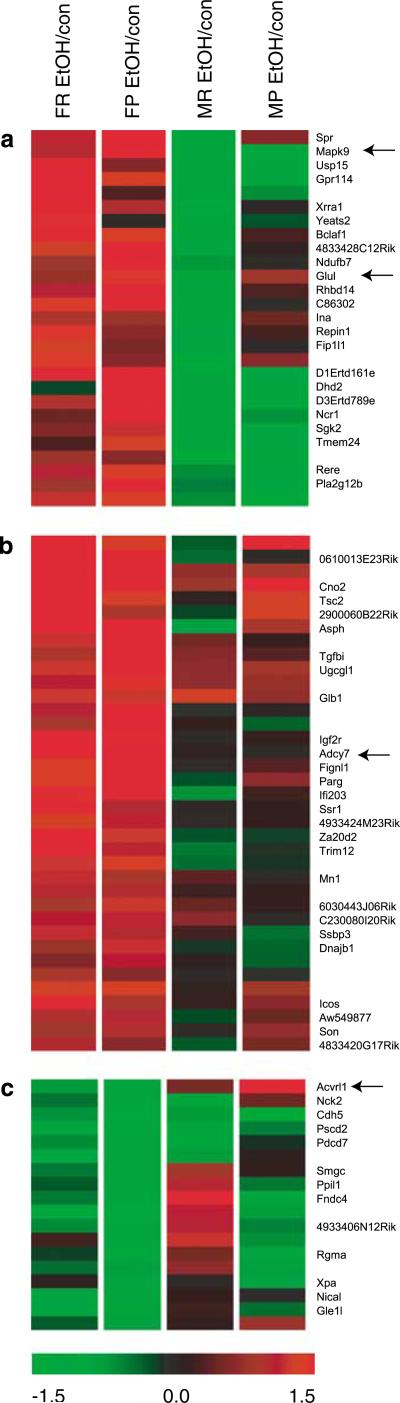
Clustering analysis can also be used to identify groups of genes that demonstrate similar expression profiles. To group similar gene expression patterns, genes (represented by rows in the overview image) were clustered according to a similarity of expression profile as a consequence of treatment for each sex and withdrawal phenotype. The gene-tree shown at the left of the image in Figure 1b corresponds to the degree of similarity of the pattern of expression for specific genes across all experiments. To identify specific groups of genes that demonstrated the strongest correlations in more detail, k-means clustering of genes significantly regulated by EtOH was used. Ten clusters were identified in this analysis; three of ten clusters identified genes with a strong differential response between sexes (Figure 2; the remaining clusters are shown in Supplementary Figure 2 in supporting information online). Genes that showed expression differences that were in opposite directions and thus demonstrating the largest sexually dimorphic response are clustered in Figure 2a. Figure 2b showed a cluster of genes that were upregulated by EtOH in females while the cluster in Figure 2c identified genes downregulated in females. In all clusters identified, gender was more predictive of response to withdrawal than the selected line since none of the groups clustered genes according to selected line differences.
Figure 2.
K-means clustering of differential gene expression. K-means clustering (clusters = 10, average linkage, Euclidian distance) was employed to identify functional relationships between genes. Several clusters were identified with greater similarity between females with different withdrawal phenotype (none were identified showing greater similarity between both genders of a single selected line), suggesting a divergent transcriptional response to EtOH in females vs males. (a) Clustering identified genes with a sexually dimorphic response to withdrawal from chronic EtOH exposure, with upregulation in females, and downregulation in males. (b) Female upregulated genes (upregulation in female WSP and WSR). (c) Female downregulated genes (downregulation in female WSP and WSR). Additional clusters can be seen in Supplementary Figure 3 in supporting information online. No clusters were specifically identified as phenotype-divergent (eg none demonstrated clustering of male and female WSP).
Gene Pathway Enrichment Analysis
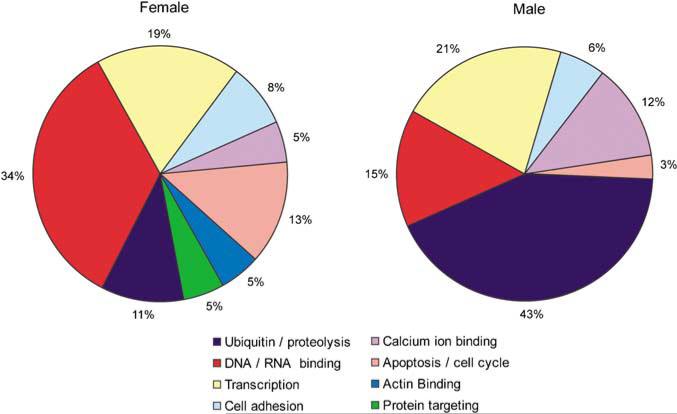
To characterize meaningful signaling pathways that were impacted by EtOH in each sex and selected line in addition to identification of specific genes, computational characterization with GO database analysis was used to group significantly regulated genes into similar biological or molecular functional categories. For this analysis, we employed Expression Analysis Systematic Explorer (EASE) using the list of genes significantly differentially regulated from each EtOH comparison. EASE analysis identified GO groups statistically over-represented in the class of regulated sequences based on presence on the array for each comparison (see Supplementary Table 2 in supporting information online). For visualization, specific signaling categories or redundant pathways identified with EASE analysis were collapsed into related broad classes (Figure 3). Importantly, this process of classification again demonstrated distinct responses between males and females. Examination of the results revealed a notable finding in that apoptotic/cell death signaling was strongly targeted in females compared to males. In this category, representation in females was over fourfold higher in terms of the percentages of genes involved than in males. Females also demonstrated more regulated transcripts relating to DNA and RNA binding and actin binding. In contrast, analysis in males compared to females showed more EtOH-regulated transcripts related to the ubiquitin/protein degradation pathways and calcium ion binding. Genes that were identified as EtOH-regulated in multiple comparisons are presented in Supplementary Table 3 in supporting information online. In addition, characterization of biological pathways influenced by EtOH using Pathway Architect software also showed a sexually dimorphic response irrespective of withdrawal severity phenotype (data not shown), most notably with respect to activation of HNF4 in female WSP and WSR, a transcription factor reflecting oxidative stress and free-radical generation and known to mediate superinduction of iNOS (Guo et al, 2006).
Figure 3.
Summary of compiled GO categories based on sex. The 176 of 295 regulated genes with known functions were classified according to their gene ontology annotation terms for each group. Several GO categories were identified in each comparison as significantly over-represented in the population based on presence on the array platform after EASE analysis. GO categories were collapsed into the broader categories presented here, with genes significantly regulated during EtOH withdrawal placed into eight functional categories.
Enhanced Alcohol-Induced Brain Damage in Females
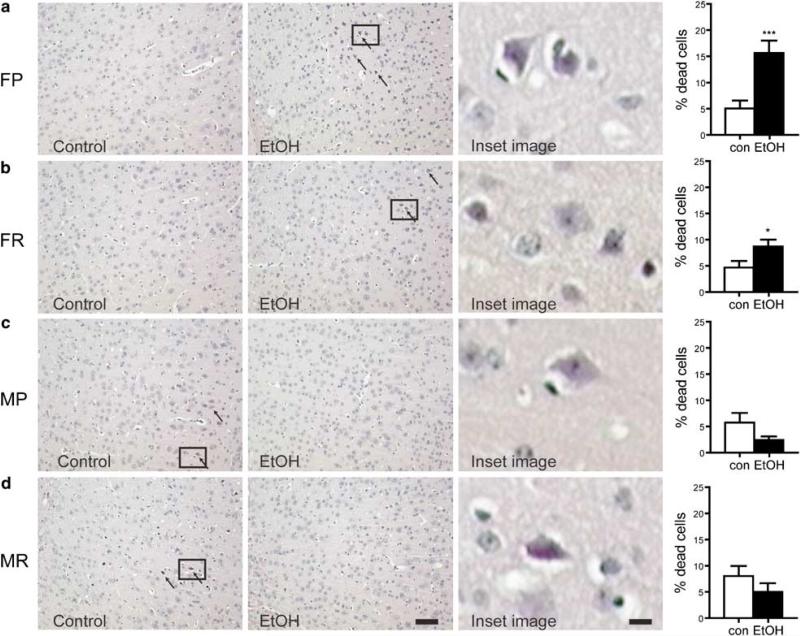
With consideration of the gender-dependent response to withdrawal from chronic EtOH observed in gene expression, the consequence of activation of these functional pathways would indicate a process of EtOH-induced brain damage in females that was distinct from males. As biological confirmation of the identified transcriptional response, we therefore assessed brain damage following chronic EtOH exposure in both sexes. Brains were examined 10 days after withdrawal as the withdrawal process, and not EtOH exposure per se, plays a major role in alcohol-induced toxicity (Favalli et al, 2002). The 10-day time point was based on the time associated with neuronal damage following a prototypical excitotoxic insult (Panegyres, 1998), which is considered an important mediator of alcohol-induced brain damage (Harper and Matsumoto, 2005; Prendergast et al, 2004). To quantify the effect of chronic EtOH withdrawal on brain damage in both sexes, we evaluated histopathology in the lateral parietal cortex. Parietal cortex as a region of interest offered the advantage that, in addition to PFC, it has also been shown to demonstrate damage in human alcoholics (Gazdzinski et al, 2005) and after EtOH withdrawal in rodents (Favalli et al, 2002). In addition, the lateral parietal cortex is part of the network involved in inhibitory control along with the PFC (Watanabe et al, 2002), is a discrete region as opposed to the PFC with seven cortical layers, and was also identified in a visual screen performed to assess brain regions demonstrating neuronal damage. For evaluation of neuro-degeneration following EtOH-induced brain injury, coronal sections were stained with hematoxylin and eosin (H&E) (also see Supplementary Methods). Dead cells were clearly identified as cells with an angular pyknotic nucleus and a hypereosinophilic cytoplasm (Figure 4; note cell morphology in zoomed images as insets). Counting of both dead and live cells was performed as described in Methods. Three-way ANOVA (sex, line, treatment) showed significant effects of sex (F(1, 57)=2.259; P=0.026), interaction of EtOH treatment by sex (F(1, 57)=13.788; P<0.0001), and interaction of sex by line (F(1, 57) = 4.637; P=0.036). Given our a priori hypothesis of divergent cell death between sexes, t-tests were performed for each gender and line. Results showed an increased percentage of dead cells in females that was significant in both WSP (P=0.0003) and WSR (P<0.05) mice. In contrast, both WSP and WSR males exhibited a modest decrease in cell death in EtOH withdrawn mice that did not reach significance.
Figure 4.
Assessment of brain damage in male and female WSP and WSR following chronic EtOH exposure and withdrawal for 10 days. H&E staining was performed and arrows indicate eosinophilic staining characteristic of dead or dying cells (× 100 magnification). Zoomed regions (indicated by square inset region) showing morphology and staining of dead neurons from EtOH females or control males are included. (a) H&E stained sections from control or EtOH withdrawn female WSP mice. The percentage of damaged cells after EtOH withdrawal was significantly elevated in female WSP (FP; ***P<0.0003) mice. (b) Similarly in WSR females (FR), EtOH withdrawal resulted in increased cell death (*P<0.05). (c) In contrast to females, EtOH withdrawal in WSP male (MP) mice resulted in a non-significant decrease in dead or dying cells. (d) Likewise, analysis in male WSR (MR) mice revealed a modest decrease in cell death. Data are mean±SEM. Bar = 100 μm for photomicrographs; 14 μm for zoomed images.
DISCUSSION
We have characterized changes in brain gene expression during peak withdrawal following chronic EtOH exposure in both sexes of WSP and WSR selected lines of mice. We identified sex as a more important modulator of the transcriptional response during EtOH withdrawal than withdrawal severity phenotype. The sexually dimorphic response was noted in both false color heat mapping and with unsupervised hierarchical cluster analysis, with 100% support for separation based on sex in bootstrap analysis. Examination of both significantly regulated individual genes and of the pathways statistically over-represented in EASE and k-means cluster analysis demonstrates a distinct response in males vs females. Among the more note-worthy changes, analysis indicated gene expression differences in females consistent with enhanced cell death and a pro-inflammatory response in the brain. In contrast, males exhibited more EtOH-responsive genes related to the ubiquitin pathway and peptidase/proteolysis, cell proliferation, and anti-inflammatory responses, suggesting the possibility of reduced neurotoxic signaling. We evaluated brain damage after withdrawal from chronic EtOH exposure and have confirmed that females have enhanced cell death vs males.
This sexually dimorphic response was observed in both WSP and WSR selected lines (ie females>males, irrespective of withdrawal severity phenotype or genotype), and was somewhat unanticipated given the large difference in withdrawal severity between the WSP and WSR lines. On further consideration, this result should not have been surprising as there is a large body of evidence in both human and animal models showing profound differences between males and females with respect to a variety of neurological disorders, with gender being one of the strongest predictors of disease severity (Holden, 2005). Sex differences are observed in many EtOH-related behaviors, including EtOH sensitivity and consumption, liability for and the development of EtOH dependence, and differences in withdrawal severity (Brady and Randall, 1999; Carroll et al, 2004; Devaud and Chadda, 2001; Prescott et al, 2005; Sershen et al, 2002; Wang et al, 2003a). Thus, female rodents tend to consume more alcohol yet have reduced withdrawal severity, including in the WSP and WSR mice (Kosobud and Crabbe, 1986). Gender differences have also been reported in terms of alcohol handling; females tend to have increased body fat vs water compared with males, thus altering the volume distribution of alcohol (Addolorato et al, 1999). Females also have reduced levels of alcohol dehydrogenase, a key liver enzyme involved in alcohol metabolism, and removal (Baraona et al, 2001). Notwithstanding these sex differences, EtOH exposure was modestly higher in males in the studies reported here, yet males showed less severe brain damage. Additionally, analysis of percent weight change during EtOH exposure that showed males had modestly more weight loss than females, indicating that the brain damage seen in EtOH exposed females was not due to weight loss.
To confirm the biological relevance of the expression profiling results, we assessed brain damage following 10 days of withdrawal, and show significantly increased cell death in females in both WSP and WSR mice. In contrast, both WSP and WSR males showed a non-significant decrease in the incidence of cell death when compared to controls, suggesting that reduced neurotoxicity might be occurring. These findings provide strong support of the microarray analysis and the expression differences identified, with a functional outcome confirmed by histological assessment. Even though the sexually dimorphic response in overall neuronal death was observed irrespective of the selected line (females always>males), female WSP mice exhibited higher rates of cell death than female WSR mice. Thus, the withdrawal severity phenotype and/or genotype likely plays a modulatory role in the severity of brain damage, and in the overall transcriptional response to EtOH withdrawal. Reports indicate that brain damage observed in settings of excitotoxic damage is not due to seizures per se (Hopkins et al, 2000), and the mice used in these studies were purposely not evaluated for withdrawal severity. Notably, other data suggest that, in the absence of EtOH, males might be more susceptible to excitotoxic brain damage than females, for example during ischemic stroke (Alkayed et al, 1998). It should be noted that the histological analysis reported here was carried out in a region distinct from the region analyzed by microarray (ie lateral parietal cortex vs PFC) and, therefore, may not directly reflect results from the microarray analysis. Also, the time point of the histological analysis of brain damage was selected based on the time course of progression from excitotoxic insult to brain damage (Panegyres, 1998), but other genes regulated during later periods of withdrawal may also account for some of the damage we observed. Nevertheless, these results do support and are consistent with a distinct biological outcome between males and females following chronic EtOH withdrawal.
Potential functional or regulatory relationships between significantly regulated genes were identified by k-means clustering. Genes that showed expression differences that were in opposite directions and thus demonstrating the largest sexually dimorphic response are clustered in Figure 2a. Included in this cluster of genes is Glul (glutamine synthetase), which catalyzes the metabolism of glutamate to glutamine and results in decreased synaptic glutamate levels. Overexpression of Glul has recently been shown to be strongly correlated with resistance to withdrawal seizures in a variety of genetic models (Letwin et al, 2006); here Glul was generally elevated in females and reduced in males after chronic EtOH withdrawal, consistent with the decreased severity of EtOH withdrawal seizures observed in females (Devaud and Chadda, 2001; Kosobud and Crabbe, 1986). Mapk9 (JNK2) is also elevated in females and reduced in males, and is critical for activation of the mitochondrial death pathway in apoptosis and for pro-inflammatory functions in microglia (Waetzig et al, 2005). Figure 2b shows a cluster of genes that were upregulated by EtOH in females. Of interest in this cluster is Adcy7 (adenylyl cyclase type 7), which is associated with alcoholism in humans and sex-specific depression in transgenic and knockout mice (Hines et al, 2006). Acvrl1 (Alk1) is downregulated in females (Figure 2c). Interestingly, Acvrl1 is known to activate Smad signaling through the phosphorylation of Smad1 (Inman et al, 2002). Down-regulation of Smad signaling is of interest here as we have identified Smad3 as upregulated by EtOH in males in both WSP and WSR selected lines.
Expression levels of selected genes were analyzed by qRT-PCR analysis to corroborate microarray results. Confirmation of Diablo upregulation in female WSP mice is interesting because of its role in promoting cell death by binding to inhibitor of apoptosis proteins, promoting the activation of caspase 3 (Chai et al, 2000). Additionally, upregulation of Bclaf1 in female WSP is also pro-apoptotic as overexpression of Bclaf1 in HeLa cells induced apoptosis (Kasof et al, 1999), while upregulation of Mapk9 in female WSR indicates enhanced apoptosis and microglia activation. In contrast, upregulation of Hbxip in male WSP is anti-apoptotic as Hbxip forms a complex with Survivin to prevent caspase-9 activation (Marusawa et al, 2003). Although histological analysis does not clearly identify whether apoptosis or necrosis is more important in the cell death characterized after chronic alcohol exposure, the genes identified in the array analysis suggest a role for apoptotic signaling. Of note, there are few reports identifying genes specifically important in necrotic cell death (and are thus difficult to ascribe this function to); in fact, there is significant overlap in the genes involved in necrotic and apoptotic cell loss (Raza et al, 2004). Thus, the results presented here can be indicative of activation of either or both pathways. The downregulation of Cx3cl1 in male WSP, which encodes a brain chemokine thought to recruit T-cells and monocytes (Bazan et al, 1997), is characteristic of an anti-inflammatory response. Smad3 was confirmed as upregulated in both WSP and WSR males, and upregulation is of interest because Smad3 has been shown to inhibit Class II MHC expression (Dong et al, 2001) resulting in deactivation of microglia (Gordon, 2003), characteristic of an anti-inflammatory response. Smad3 is also known to bind androgen receptor and prevent androgen activated transcription (Rahman et al, 2004), which may provide insight into mechanism(s) for the differential response to EtOH we have observed between males and females. A recent study profiling the gene expression changes of microglia exposed to different activating agents (Thomas et al, 2006) identified several genes that were also regulated by chronic EtOH withdrawal in this study, including Za20d2, Nical, Mef2c, and Cstf3. Combined, these results suggest that there may be sexually dimorphic activation of microglia, contributing to increased neuronal damage in females.
In conclusion, we have demonstrated for the first time that CNS gene expression differences during withdrawal from chronic EtOH exposure are profoundly affected by gender. Examination of individual gene expression differences and the signaling pathways that were influenced indicated the likelihood of enhanced cell death in females. In fact, brain damage was elevated in females but not males, irrespective of genetic background or the withdrawal severity phenotype. Thus, our studies are consistent with some reports demonstrating that in human alcoholics, females have increased susceptibility to brain damage associated with alcohol abuse (Hommer et al, 2001; Mann et al, 2005), and may help to resolve a controversy surrounding the role of gender in brain damage (Wuethrich, 2001). Collectively, the findings reported here indicate a fundamentally distinct neuroadaptive response in females when compared to males during chronic EtOH withdrawal, with females demonstrating enhanced neurotoxicity. Such brain damage might underlie debilitating cognitive dysfunction and motor deficits observed in some alcoholics. In addition, disruption of the PFC normal inhibitory functions (ie inhibition of unnecessary or unwanted behavior) may contribute to excessive drinking and the self-sustaining nature of alcoholism. Thus, the enhanced cell death signaling we observed selectively in females may also have behavioral relevance. Characterization of distinct responses to alcohol between males and females may provide therapeutic insight, and further studies on the effects of alcohol abuse in both sexes are warranted.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the Department of Veterans Affairs Merit Review program (KMW) and the NIH (AA-13783 to KMW). Presented in part at the 28th Annual Meeting of the Research Society on Alcoholism, Santa Barbara, CA, June 25–29, 2005 (Wiren et al, 2006). We thank Drs Deborah A Finn, Patricia Hurn, and Marjorie Grafe for their advice and support, and Michelle Tanchuck for her technical help. We also thank Drs Pamela Metten and John Crabbe for providing the WSP and WSR mice and access to alcohol vapor chambers.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
DISCLOSURE/CONFLICT OF INTEREST The authors declare that, except for income received from their primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
REFERENCES
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52:448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Addolorato G, Capristo E, Caputo F, Greco AV, Ceccanti M, Stefanini GF, et al. Nutritional status and body fluid distribution in chronic alcoholics compared with controls. Alcohol Clin Exp Res. 1999;23:1232–1237. doi: 10.1111/j.1530-0277.1999.tb04283.x. [DOI] [PubMed] [Google Scholar]
- Alkayed NJ, Harukuni I, Kimes AS, London ED, Traystman RJ, Hurn PD. Gender-linked brain injury in experimental stroke. Stroke. 1998;29:159–165. doi: 10.1161/01.str.29.1.159. discussion 166. [DOI] [PubMed] [Google Scholar]
- Ammendola A, Gemini D, Iannaccone S, Argenzio F, Ciccone G, Ammendola E, et al. Gender and peripheral neuropathy in chronic alcoholism: a clinical-electroneurographic study. Alcohol Alcohol. 2000;35:368–371. doi: 10.1093/alcalc/35.4.368. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins AL, Rustay NR, Crabbe JC. Anxiety and sensitivity to ethanol and pentobarbital in alcohol withdrawal seizure-prone and withdrawal seizure-resistant mice. Alcohol Clin Exp Res. 2000;24:1743–1749. [PubMed] [Google Scholar]
- Baraona E, Abittan CS, Dohmen K, Moretti M, Pozzato G, Chayes ZW, et al. Gender differences in pharmacokinetics of alcohol. Alcohol Clin Exp Res. 2001;25:502–507. [PubMed] [Google Scholar]
- Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- Beadles-Bohling A, Wiren K. Alteration of kappa-opioid receptor system expression in distinct brain regions of a genetic model of enhanced ethanol withdrawal severity. Brain Res. 2005;1046:77–89. doi: 10.1016/j.brainres.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Beadles-Bohling A, Wiren K. Anticonvulsive effects of kappa opioid receptor modulation in an animal model of ethanol withdrawal. Genes Brain Behav. 2006;5:483–496. doi: 10.1111/j.1601-183X.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- Bertucci F, Bernard K, Loriod B, Chang YC, Granjeaud S, Birnbaum D, et al. Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples. Hum Mol Genet. 1999;8:1715–1722. doi: 10.1093/hmg/8.9.1715. [DOI] [PubMed] [Google Scholar]
- Brady K, Randall C. Gender differences in substance use disorders. Psychiatr Clin North Am. 1999;22:241–252. doi: 10.1016/s0193-953x(05)70074-5. [DOI] [PubMed] [Google Scholar]
- Breitenfeld D, Lang B, Thaller V, Breitenfeld T, De Syo D, Jagetic N. Psycho-social characteristics of female alcoholics. Coll Antropol. 1998;22:613–618. [PubMed] [Google Scholar]
- Butler D. Alcohol may hit women's brains harder. Nature (news@nature.com) 2005 2005 May 16; doi:10.1038/news050509-15. [Google Scholar]
- Carroll M, Lynch W, Roth M, Morgan A, Cosgrove K. Sex and estrogen influence drug abuse. Trends Pharmacol Sci. 2004;25:273–279. doi: 10.1016/j.tips.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- Chester JA, Risinger FO, Cunningham CL. Ethanol reward and aversion in mice bred for sensitivity to ethanol withdrawal. Alcohol Clin Exp Res. 1998;22:468–473. [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaud L, Chadda R. Sex differences in rats in the development of and recovery from ethanol dependence assessed by changes in seizure susceptibility. Alcohol Clin Exp Res. 2001;25:1689–1696. [PubMed] [Google Scholar]
- Dong Y, Tang L, Letterio J, Benveniste E. The Smad3 protein is involved in TGF-beta inhibition of class II trans-activator and class II MHC expression. J Immunol. 2001;167:311–319. doi: 10.4049/jimmunol.167.1.311. [DOI] [PubMed] [Google Scholar]
- Eckardt M, Martin P. Clinical assessment of cognition in alcoholism. Alcohol Clin Exp Res. 1986;10:123–127. doi: 10.1111/j.1530-0277.1986.tb05058.x. [DOI] [PubMed] [Google Scholar]
- Favalli L, Rozza A, Frattini P, Masoero E, Scelsi R, Pascale A, et al. Ischemia-induced glutamate release in rat front-oparietal cortex after chronic alcohol and withdrawal. Neurosci Lett. 2002;326:183–186. doi: 10.1016/s0304-3940(02)00352-x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sola J, Nicolas-Arfelis J. Gender differences in alcoholic cardiomyopathy. J Gend Specif Med. 2002;5:41–47. [PubMed] [Google Scholar]
- Finn D, Crabbe J. Chronic ethanol differentially alters susceptibility to chemically induced convulsions in Withdrawal Seizure-Prone and -Resistant mice. J Pharmacol Exp Ther. 1999;288:782–790. [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Landis N, Hwang J, Harrison E, Wilce P. Comparative gene expression in brain regions of human alcoholics. Genes Brain Behav. 2006;5(Suppl 1):78–84. doi: 10.1111/j.1601-183X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Gazdzinski S, Durazzo T, Studholme C, Song E, Banys P, Meyerhoff D. Quantitative brain MRI in alcohol dependence: preliminary evidence for effects of concurrent chronic cigarette smoking on regional brain volumes. Alcohol Clin Exp Res. 2005;29:1484–1495. doi: 10.1097/01.alc.0000175018.72488.61. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Guo H, Gao C, Mi Z, Wai PY, Kuo PC. Phosphorylation of Ser158 regulates inflammatory redox-dependent hepatocyte nuclear factor-4alpha transcriptional activity. Biochem J. 2006;394:379–387. doi: 10.1042/BJ20051730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Harper C, Matsumoto I. Ethanol and brain damage. Curr Opin Pharmacol. 2005;5:73–78. doi: 10.1016/j.coph.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Hauser T. Epidemiology of alcohol use and of epilepsy: the magnitude of the problem. In: Mattson R, Porter RJ, Cramer J, Diamond I, editors. Alcohol and Seizures. F.A. Davis Co.; Philadelphia: 1990. pp. 12–21. [Google Scholar]
- Hines LM, Hoffman PL, Bhave S, Saba L, Kaiser A, Snell L, et al. A sex-specific role of type VII adenylyl cyclase in depression. J Neurosci. 2006;26:12609–12619. doi: 10.1523/JNEUROSCI.1040-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden C. Sex and the suffering brain. Science. 2005;308:1574. doi: 10.1126/science.308.5728.1574. [DOI] [PubMed] [Google Scholar]
- Hommer D. Male and female sensitivity to alcohol-induced brain damage. Alcohol Res Health. 2003;27:181–185. [PMC free article] [PubMed] [Google Scholar]
- Hommer D, Momenan R, Kaiser E, Rawlings R. Evidence for a gender-related effect of alcoholism on brain volumes. Am J Psychiatry. 2001;158:198–204. doi: 10.1176/appi.ajp.158.2.198. [DOI] [PubMed] [Google Scholar]
- Hopkins K, Wang G, Schmued L. Temporal progression of kainic acid induced neuronal and myelin degeneration in the rat forebrain. Brain Res. 2000;864:69–80. doi: 10.1016/s0006-8993(00)02137-5. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, et al. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–28. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Kasof G, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999;19:4390–4404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns R, Ravindranathan A, Hassan S, Cage M, York T, Sikela J, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, et al. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry. 1994;51:8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Bodor A, Crabbe J. Voluntary consumption of ethanol in WSP, WSC and WSR selectively bred mouse lines. Pharmacol Biochem Behav. 1988;29:601–607. doi: 10.1016/0091-3057(88)90026-3. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Crabbe J. Ethanol withdrawal in mice bred to be genetically prone or resistant to ethanol withdrawal seizures. J Pharmacol Exp Ther. 1986;238:170–177. [PubMed] [Google Scholar]
- Langsrud O. 50–50 multivariate analysis of variance for collinear responses. The Statistician. 2002;51:305–317. [Google Scholar]
- Letwin NE, Kafkafi N, Benjamini Y, Mayo C, Frank BC, Luu T, et al. Combined application of behavior genetics and micro-array analysis to identify regional expression themes and gene-behavior associations. J Neurosci. 2006;26:5277–5287. doi: 10.1523/JNEUROSCI.4602-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G, et al. The application of proteomics to the human alcoholic brain. Ann N Y Acad Sci. 2004;1025:14–26. doi: 10.1196/annals.1316.002. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2005;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Loft S, Olesen K, Dossing M. Increased susceptibility to liver disease in relation to alcohol consumption in women. Scand J Gastroenterol. 1987;22:1251–1256. doi: 10.3109/00365528708996472. [DOI] [PubMed] [Google Scholar]
- Mann K, Ackermann K, Croissant B, Mundle G, Nakovics H, Diehl A. Neuroimaging of gender differences in alcohol dependence: are women more vulnerable? Alcohol Clin Exp Res. 2005;29:896–901. doi: 10.1097/01.alc.0000164376.69978.6b. [DOI] [PubMed] [Google Scholar]
- Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. Embo J. 2003;22:2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metten P, Phillips T, Crabbe J, Tarantino L, McClearn G, Plomin R, et al. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Oscar-Berman M, Marinkovic K. Alcoholism and the brain: an overview. Alcohol Res Health. 2003;27:125–133. [PMC free article] [PubMed] [Google Scholar]
- Panegyres P. The effects of excitotoxicity on the expression of the amyloid precursor protein gene in the brain and its modulation by neuroprotective agents. J Neural Transm. 1998;105:463–478. doi: 10.1007/s007020050070. [DOI] [PubMed] [Google Scholar]
- Pearson H. Public health: the demon drink. Nature. 2004;428:598–600. doi: 10.1038/428598a. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan E, Mathalon D, Lim K. Frontal lobe volume loss observed with magnetic resonance imaging in older chronic alcoholics. Alcohol Clin Exp Res. 1997;21:521–529. doi: 10.1111/j.1530-0277.1997.tb03798.x. [DOI] [PubMed] [Google Scholar]
- Prendergast M, Harris B, Mullholland P, Blanchard J, II, Gibson D, Holley R, et al. Hippocampal CA1 region neurodegeneration produced by ethanol withdrawal requires activation of intrinsic polysynaptic hippocampal pathways and function of N-methyl-D-aspartate receptors. Neuroscience. 2004;124:869–877. doi: 10.1016/j.neuroscience.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Prescott C, Caldwell C, Carey G, Vogler G, Trumbetta S, Gottesman I. The Washington University Twin Study of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2005;134:48–55. doi: 10.1002/ajmg.b.30124. [DOI] [PubMed] [Google Scholar]
- Rahman M, Miyamoto H, Chang C. Androgen receptor coregulators in prostate cancer: mechanisms and clinical implications. Clin Cancer Res. 2004;10:2208–2219. doi: 10.1158/1078-0432.ccr-0746-3. [DOI] [PubMed] [Google Scholar]
- Raza SM, Fuller GN, Rhee CH, Huang S, Hess K, Zhang W, et al. Identification of necrosis-associated genes in glioblastoma by cDNA microarray analysis. Clin Cancer Res. 2004;10:212–221. doi: 10.1158/1078-0432.ccr-0155-3. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2006 2006 October 10; doi: 10.1038/sj.tpj.6500420. E-pub ahead of print; doi:10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schafer G, Crabbe J, Wiren K. Ethanol-regulated gene expression of neuroendocrine specific protein in mice: brain region and genotype specificity. Brain Res. 2001;897:139–149. doi: 10.1016/s0006-8993(01)02110-2. [DOI] [PubMed] [Google Scholar]
- Schafer GL, Crabbe JC, Wiren KM. Identification of neuroendocrine-specific protein as an ethanol-regulated gene with mRNA differential display. Mamm Genome. 1998;9:979–982. doi: 10.1007/s003359900910. [DOI] [PubMed] [Google Scholar]
- Schmid H, Ter Bogt T, Godeau E, Hublet A, Dias S, Fotiou A. Drunkenness among young people: a cross-national comparison. J Stud Alcohol. 2003;64:650–661. doi: 10.15288/jsa.2003.64.650. [DOI] [PubMed] [Google Scholar]
- Sershen H, Hashim A, Vadasz C. Strain and sex differences in repeated ethanol treatment-induced motor activity in quasi-congenic mice. Genes Brain Behav. 2002;1:156–165. doi: 10.1034/j.1601-183x.2002.10303.x. [DOI] [PubMed] [Google Scholar]
- Sullivan E, Pfefferbaum A. Neurocircuitry in alcoholism: a substrate of disruption and repair. Psychopharmacology (Berl) 2005;180:583–594. doi: 10.1007/s00213-005-2267-6. [DOI] [PubMed] [Google Scholar]
- Tang R, Zhu Z, Qu Y, Li J, Ji Y, Cai Q, et al. The impact of hyperthermic chemotherapy on human gastric cancer cell lines: preliminary results. Oncol Rep. 2006;16:631–641. [PubMed] [Google Scholar]
- Terdal E, Crabbe J. Indexing withdrawal in mice: matching genotypes for exposure in studies using ethanol vapor inhalation. Alcohol Clin Exp Res. 1994;18:542–547. doi: 10.1111/j.1530-0277.1994.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Thomas DM, Francescutti-Verbeem DM, Kuhn DM. Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J. 2006;20:515–517. doi: 10.1096/fj.05-4873fje. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29:357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Waetzig V, Czeloth K, Hidding U, Mielke K, Kanzow M, Brecht S, et al. c-Jun N-terminal kinases (JNKs) mediate pro-inflammatory actions of microglia. Glia. 2005;50:235–246. doi: 10.1002/glia.20173. [DOI] [PubMed] [Google Scholar]
- Wang G, Volkow N, Fowler J, Franceschi D, Wong C, Pappas N, et al. Alcohol intoxication induces greater reductions in brain metabolism in male than in female subjects. Alcohol Clin Exp Res. 2003a;27:909–917. doi: 10.1097/01.ALC.0000071740.56375.BA. [DOI] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003b;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Sugiura M, Sato K, Sato Y, Maeda Y, Matsue Y, et al. The human prefrontal and parietal association cortices are involved in NO-GO performances: an event-related fMRI study. Neuroimage. 2002;17:1207–1216. doi: 10.1006/nimg.2002.1198. [DOI] [PubMed] [Google Scholar]
- Wiren K, Hashimoto J, Alele P, Devaud L, Price K, Middaugh L, et al. Impact of sex: determination of alcohol neuro-adaptation and reinforcement. Alcohol Clin Exp Res. 2006;30:233–242. doi: 10.1111/j.1530-0277.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- Wuethrich B. Neurobiology. Does alcohol damage female brains more? Science. 2001;291:2077–2079. doi: 10.1126/science.291.5511.2077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.