Abstract
The botulinum neurotoxins (BoNTs) produced by different strains of the bacterium Clostridium botulinum are responsible for the disease botulism and include a group of immunologically distinct serotypes (A, B, E, and F) that are considered to be the most lethal natural proteins known for humans. Two BoNT serotypes, C and D, while rarely associated with human infection, are responsible for deadly botulism outbreaks afflicting animals. Also associated with animal infections is the BoNT C-D mosaic protein (BoNT/CD), a BoNT subtype that is essentially a hybrid of the BoNT/C (~two-thirds) and BoNT/D (~one-third) serotypes. While the amino acid sequence of the heavy chain receptor binding (HCR) domain of BoNT/CD (BoNT/CD-HCR) is very similar to the corresponding amino acid sequence of BoNT/D, BoNT/CD-HCR binds synaptosome membranes better than BoNT/D-HCR. To obtain structural insights for the different membrane binding properties, the crystal structure of BoNT/CD-HCR (S867-E1280) was determined at 1.56 Å resolution and compared to previously reported structures for BoNT/D-HCR. Overall, the BoNT/CD-HCR structure is similar to the two sub-domain organization observed for other BoNT HCRs: an N-terminal jellyroll barrel motif and a C-terminal β-trefoil fold. Comparison of the structure of BoNT/CD-HCR with BoNT/D-HCR indicates that K1118 has a similar structural role as the equivalent residue, E1114, in BoNT/D-HCR, while K1136 has a structurally different role than the equivalent residue, G1132, in BoNT/D-HCR. Lysine-1118 forms a salt bridge with E1247 and may enhance membrane interactions by stabilizing the putative membrane binding loop (K1240-N1248). Lysine-1136 is observed on the surface of the protein. A sulfate ion bound to K1136 may mimic a natural interaction with the negatively changed phospholipid membrane surface. Liposome-binding experiments demonstrate that BoNT/CD-HCR binds phosphatidylethanolamine liposomes more tightly than BoNT/D-HCR.
Keywords: botulinum neurotoxin, C-D mosaic, botulism, phosphatidylethanolamine, membrane recognition
1. Introduction
The anaerobic, gram-positive bacterium, Clostridium botulinum, is a NIAID category A pathogen responsible for botulism and produces the most toxic biological substances known, botulinum neurotoxins (BoNTs) [1]. The extreme potency of BoNTs is due to their neurospecific interactions with unmyelinated regions of nerve terminals where they inhibit the release of the neurotransmitter, acetylcholine, leading to flaccid muscular paralysis [2]. While the frequency of natural botulism is low, the high mortality rate of the untreated disease makes it an important human health issue and there is concern for the potential use of BoNTs as agents of bioterrorism [3]. Botulism is also extremely lethal to livestock and responsible for significant economic losses worldwide [4].
Seven BoNT serotypes (A-G) have been identified based on neutralization characteristics of antiserum. Serotypes A, B, E, and F are associated with human botulism while serotype C and D are predominately associated with animal botulism [5,6,7]. In addition to the seven basic serotypes, numerous sub-serotypes have also been identified. Two such sub-serotypes are BoNT mosaic proteins C-D and D-C isolated from C. botulinum strains causing animal botulinum in African and Japan, respectively [4,7], that are essentially hybrids of BoNT/C and BoNT/D. The C-D mosaic (BoNT/CD) is approximately two-thirds BoNT/C and one-third BoNT/D while the D-C mosaic is approximately two-thirds BoNT/D and one-third BoNT/C. All BoNTs are initially translated as ~ 150 kDa single chain polypeptides that are proteolytically activated into a re-associated molecule consisting of an N-terminal ~ 50 kDa light chain (Lc) and the C-terminal ~ 100 kDa heavy chain (Hc) held together with a single disulfide bond [8]. The Lc polypeptide is a zinc-dependent endopeptidase while the Hc polypeptide is divided into an N-terminal translocation domain and C-terminal receptor binding domain (HCR) of approximately equal length. The HCR domain may be further divided into two subdomains: a C-terminal Hcc sub-domain involved in cell surface recognition and an N-terminal Hcn sub-domain with an unclear function, although it has been suggested that it may interact with phosphoinositides [9]. Following endocytosis into neuronal cells, a conformational change triggered by the acidic pH environment inside the endosomes releases the Lc domain to the cytosol [10,11] where it inhibits the function of acetylcholine by hydrolyzing SNARE (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptors) proteins.
The internalization of BoNTs into the host neuronal cells, via endocytosis after the HCR domain binds to the surface of neuronal cells, is crucial for the progression of the disease. Uptake of serotype A, B, E, F, and G occur via a dual receptor mechanism [12] involving a synaptic vesicle protein (Synaptotagmin or synaptic vesicle protein 2) and a ganglioside (reviewed by [13]). The mechanism by which BoNT serotypes C and D enter neuronal cells is not clear. Neurotoxicity is dependent upon ganglioside binding for both C and D [14,15], although a ganglioside binding motif SxWY does not exist in the BoNT/D amino acid sequence. Additionally, protein receptors have not been identified for either BoNT/C or BoNT/D serotype [16]. Furthermore, BoNT/CD-HCR bound to synaptosomes more tightly than BoNT/D-HCR [16,17], and K1118 and K1136 of BoNT/CD were identified as key residues critical for the binding activity [17]. These two lysine residues are absent in BoNT/D. To obtain insights into the entry mechanism for BoNT/D and BoNT/CD, the crystal structure for the BoNT/CD-HCR was determined at 1.56 Å resolution. The structure was compared to previously reported structures for BoNT/D-HCR [15,18,19] and potential roles of residues K1118 and K1136 of BoNT/CD in membrane interactions were analyzed.
2. Materials and methods
2.1. Protein expression and purification
The DNA sequence encoding the receptor binding domain of C. botulinum BoNT/CD (S867-E1280) was codon-optimized for expression in Escherichia coli with a C-terminal 6×his tag (DNA 2.0) and cloned into the expression vector pJexpress411 (see Fig. S1 for the sequence of the optimized cDNA). This recombinant vector was then transformed into the host E. coli bacterial strain BL21 (DE3). Greater than 98% pure BoNT/CD-HCR was obtained following the protocols previously described in detail for BoNT/D-HCR (Y Zhang, X Gao, L Qin, GW Buchko, H Robinson, and SM Varnum, Acta Crystallographica Section F, in press; and [18]) using the antibiotic kanamycin during cell growth in LB media, isopropyl-β-D-1-thiogalactopyranoside induction at 12 °C, initial purification on a Ni-NTA agarose column (Qiagen), and final purification on a HiTrap Q ion exchange column (GE Healthcare).
2.2. Liposome-binding assay
Phosphatidylcholine (PC) (Avanti Polar Lipids) and a 1:1 mixture of PC and bovine brain phosphatidylethanolamine (PE) (Sigma, P7693), were first dissolved in chloroform and then dried under a stream of nitrogen. Following resuspension in TBS buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.0), the lipids were incubated at 30 °C for 1 h, vortexed for 5 min, and centrifuged at 20,000×g (4 °C) for 10 min. One hundred μg liposome pellets were resuspended in 50 μl binding buffer (TBS with 2 mM MgCl2). The binding reaction was then performed by adding 2 μg of purified BoNT/CD-HCR to the resuspended liposomes. Following incubation at 30 °C for 30 min, the solution was centrifuged at 20,000×g (4 °C) and the distribution of BoNT/D-HCR in the pellet and supernatant fractions analyzed by SDS-PAGE [18].
2.3. Crystallization and structure determination
Crystallization screening was performed using the hanging-drop vapor diffusion method. The best crystals grew with mother liquor composed of 20% (w/v) PEG 8000, 10% (w/v) isopropanol, 0.2 M (NH4)2SO4, and 0.1 M Hepes, pH 7.5. Crystals were harvested by stepwise transfer into cryoprotectant solutions containing increasing concentrations of glycerol for cryoprotection. X-ray diffraction data was collected at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory on beamline X29A. Data was processed using DENZO, and integrated intensities scaled using the SCALEPACK from the HKL-2000 program package [20]. Original phasing for the BoNT/CD-HCR crystal was obtained by molecular replacement using the CCP4 suite of programs [21] and the crystal structure of the HCR domain of BoNT/D (PDB ID 3OGG) for the molecular search template [18]. The model was further refined using Coot [22] and PHENIX [23]. Structural validation was performed using Molprobity [24]. Coordinates and structure factors for BoNT/CD-HCR have been deposited in the Protein Data Bank (PDB) with accession numbers 3PME.
3. Results and discussion
3.1. Overall structure
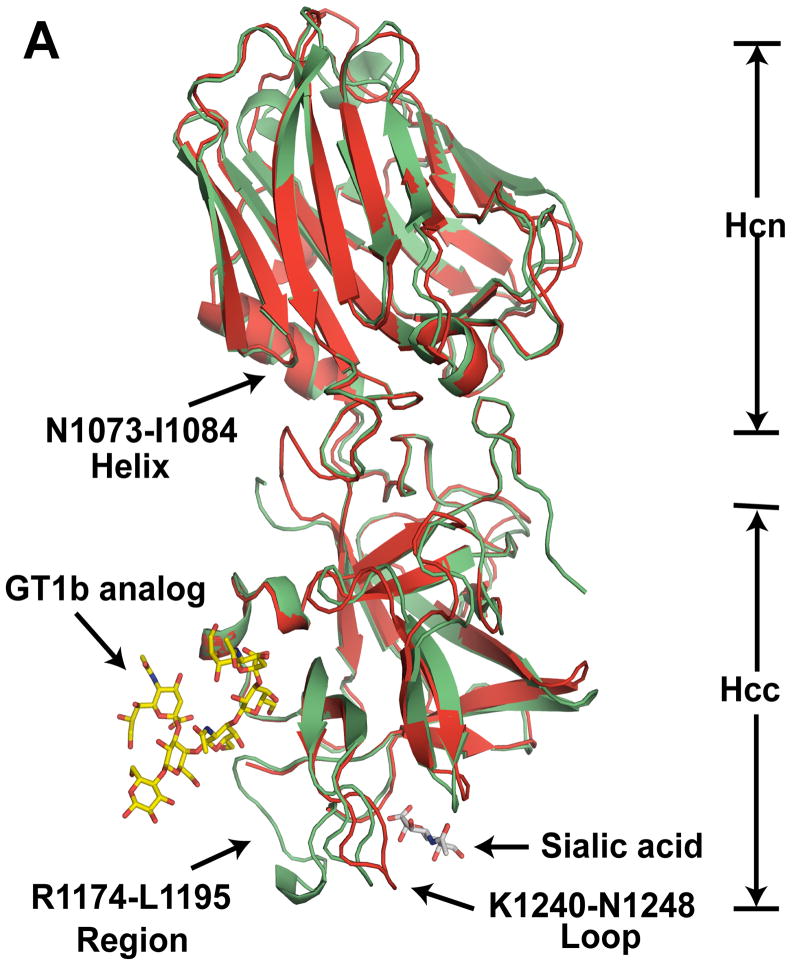
The crystal structure of BoNT/CD-HCR (S867-E1280) at 1.56 Å resolution was solved using a molecular replacement method. The asymmetric unit of the crystal contained a single protein unit, two sulfate ions, a glycerol molecule, and 374 water molecules. The data collection and refinement statistics, summarized in Table S1, show that the final model was a quality representation of the BoNT/CD-HCR structure. Electron density could be modeled into the entire protein except for the final two histidine residues on the C-terminal poly-histidine tag and residues S867, N1218, and K1219. The overall structure of BoNT/CD-HCR, shown in Figure 1, is similar with that of BoNT/D-HCR and the other serotype HCR domains. It contains two sub-domains, Hcn (S867-S1072) and Hcc (L1085-E1280), connected by a helix (N1073-I1084). The Hcn sub-domain folds into a jelly roll barrel motif and the Hcc sub-domain adopts a β-trefoil fold.
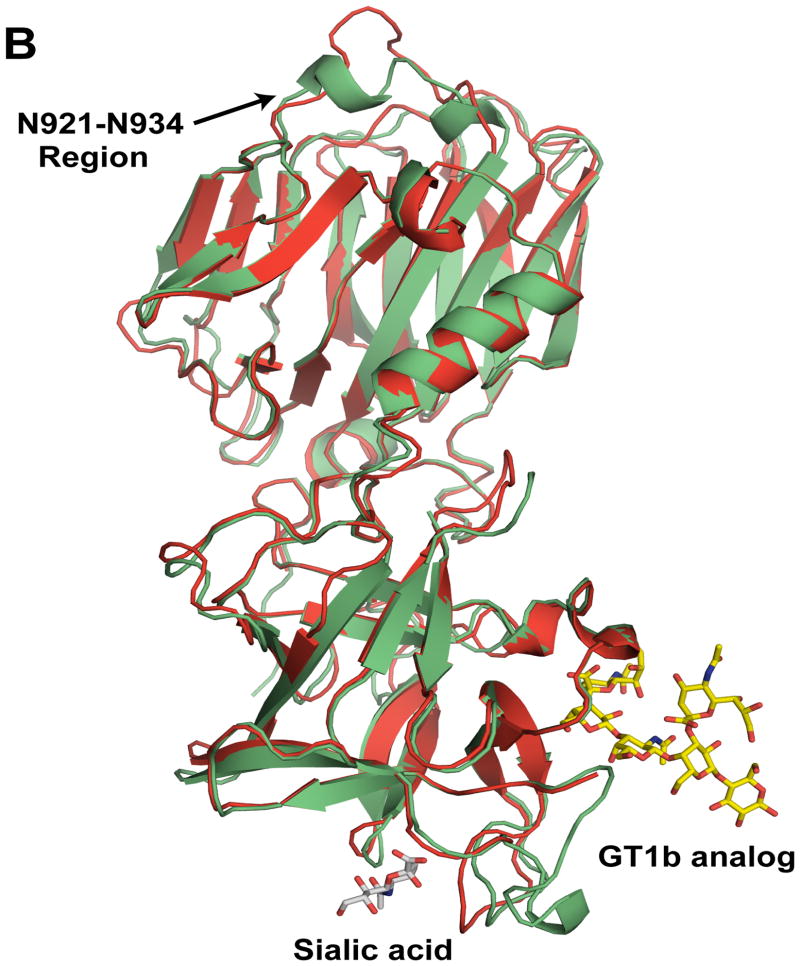
Fig. 1.
Comparison of the crystal structures of BoNT/CD-HCR and BoNT/D-HCR. (A) Cartoon representation of the superposition of the BoNT/CD-HCR crystal structure (green, 3PME) onto the crystal structure of BoNT/D-HCR (red, 3OGG). Also superimposed on the structure of BoNT/CD-HCR are the protein backbones of the structures of the BoNT/A-HCR-GT1b analog complex (2VU9) and the BoNT/D-HCR-sialic acid complex (3OBT) with only the position of the ganglioside co-receptor, GT1b (analog) and sialic acid shown. (B) An approximately 180° rotation of Figure 1A about the y-axis exposes the structural differences in the N921-N934 region. The Hcn and Hcc sub-domains are indicated and important regions are labeled with arrows. Labels for residue numbers in all figures are according to the BoNT/CD-HCR sequence unless specifically stated.
3.2. Structural comparison to BoNT/D-HCR
The amino acid sequence of BoNT/CD-HCR is highly similar with that of BoNT/D-HCR. While the overall tertiary structure for the two proteins is consequently very similar (Fig. 1A and 1B), the two structures contain some differences as the backbone RMSD of the ordered regions, when superimposed by the program SuperPose [25], is 1.05 Å. Many of these differences are due to minor variations in the length of corresponding β-strands and α-helices that are likely without functional consequences. However, a few of the structural differences located on the surface of the protein may be responsible for some of the different synaptosome binding properties observed between BoNT/CD-HCR and BoNT/D-HCR.
Perhaps the most functionally significant structural difference between the BoNT/CD-HCR and BoNT/D-HCR structures is with regard to the N921-N934 region of the Hcn sub-domain (Fig. 1B). Although this region is similar in length between the two proteins, in BoNT/D-HCR the equivalent region (N917-S931) adopts a loop structure that protrudes away from the interior of the molecule [18], thereby potentially providing opportunities for intra- or extra-molecular interactions. In contrast, the N921-N934 region of BoNT/CD-HCR folds toward the interior of the molecule and two short helices form in the middle of the region (N924-L927 and A930-E933). Such an orientation of the N921-N934 region in BoNT/CD-HCR, while different than the corresponding region in BoNT/D-HCR, is similar to the structure adopted by analogous regions in the other serotype HCR domains.
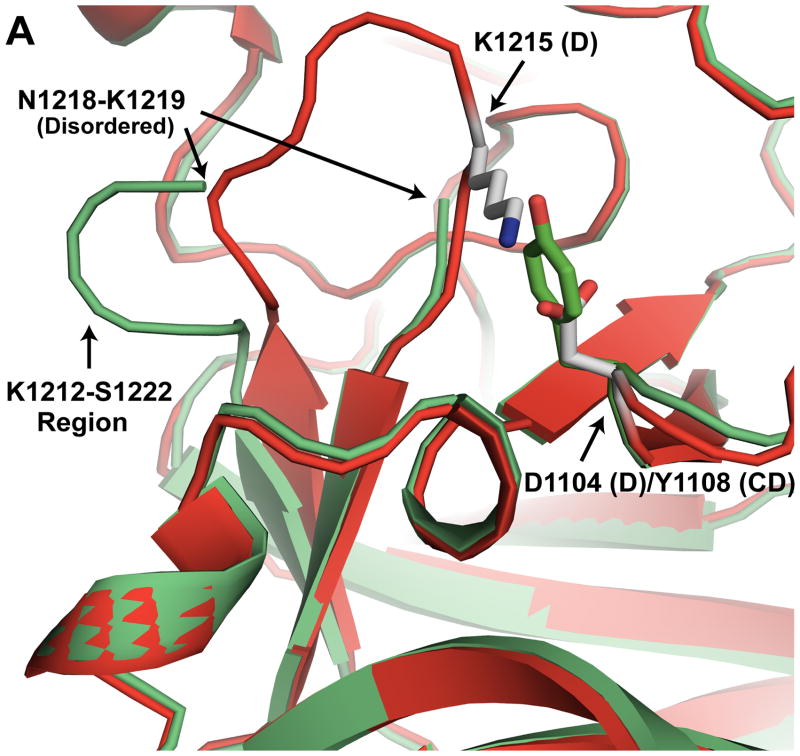
Another potentially significant structural difference between BoNT/CD-HCR and BoNT/D-HCR is in the K1212-S1222 loop (Fig. 2A). In BoNT/CD-HCR, this region is disordered and residues N1218 and K1219 could not be modeled into the structure. Conversely, the similar region in BoNT/D-HCR (N1209-S1218) is a well ordered loop. The reason for the structural difference may be due to a salt bridge in BoNT/D that is absent in BoNT/CD. The potential salt bridge in BoNT/D-HCR is formed between residues D1104 and K1215. On the other hand, in BoNT/CD, residue D1104 is replaced by the equivalent residue, Y1108, a bulkier residue than aspartic acid that is less amiable to salt bridge formation with the corresponding equivalent partner, K1219.
Fig. 2.
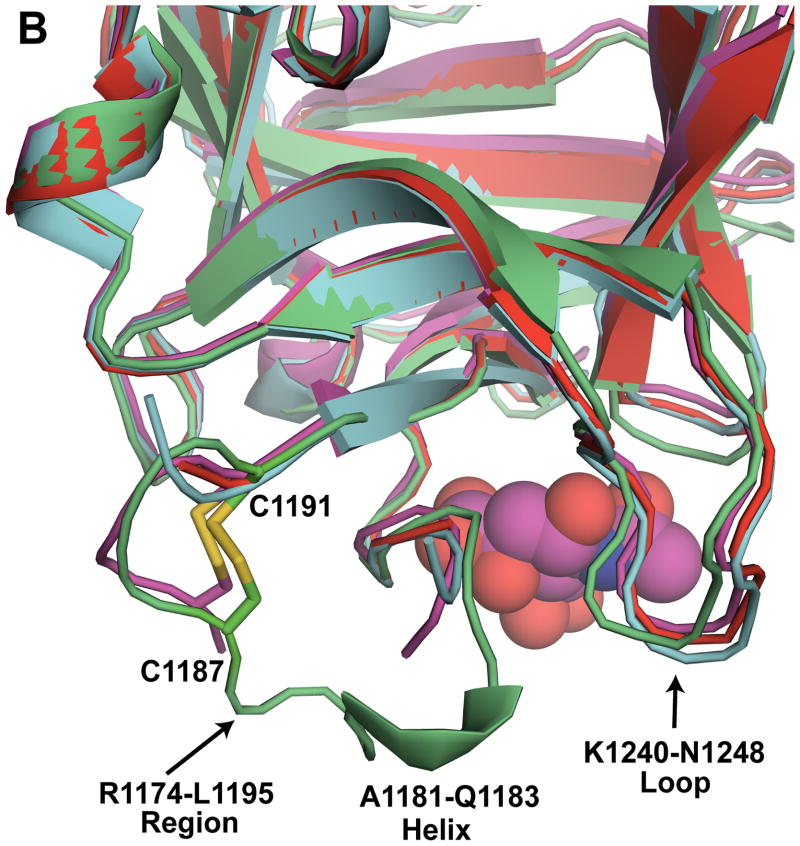
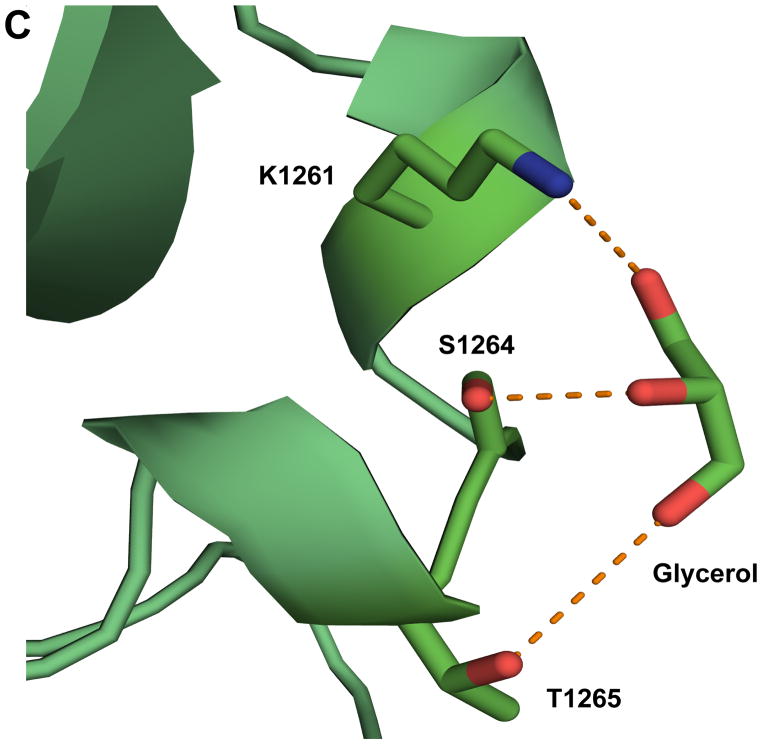
Detailed structural analysis of specific regions of BoNT/CD-HCR. (A) Expansion of the K1212-S1222 region (BoNT/CD-HCR (green) superposed on BoNT/D-HCR (red)) illustrating a potential salt bridge between D1104 and K1215 in BoNT/D that is absent in BoNT/CD where D1104 is replaced by Y1108. The absence of this salt bridge in BoNT/CD may be responsible for the disorder present in the electron density maps in the K1212-S1222 region of BoNT/CD. (B) Expansion of the R1174-L1195 region of BoNT/CD-HCR (green) superposed on the three BoNT/D-HCRs (3OGG, red; 3OBT, pink; 3N7J, cyan). The sialic acid from the BoNT/D-HCR-sialic acid complex (3OBT) structure is shown as a spherical representation. (C) Detailed expansion of the region near a putative ganglioside binding site containing a bound glycerol molecule. Potential hydrogen bonds between glycerol and the side chains of residues K1261, S1264, and T1265 are shown as dashed lines.
As observed in the structure of BoNT/D-HCR, BoNT/CD-HCR contains a region, R1174-L1195, which is longer than the equivalent region in the other serotypes. There is no known function for this region in BoNT-HCRs, however, its proximity to the putative membrane binding loop and receptor binding sites makes it attractive to hypothesize that it is involved in coordinating membrane interactions [18]. In the three recently published BoNT/D-HCR structures, interpretable electron density was missing for Y1176-Q1185 (3OGG), G1180-G1181 (3OBR), and T1178-S1184 (3N7J) [15,18,19]. In contrast, the equivalent region in BoNT/CD-HCR was well ordered with a disulfide bond between C1187 and C1191 likely responsible (Fig. 2B). Such a disulfide bond was only observed in one of the three BoNT/D-HCR structures, 3OBR [15], the BoNT/D-HCR structure with the fewest (two) missing residues. Furthermore, in the middle of the BoNT/CD-HCR R1174-L1195 region there is a short helix, A1181-Q1183, not observed in the equivalent region in the other BoNT/D-HCR structures (Fig. 2B). If the R1174-L1195 region of BoNT/CD-HCR is involved in membrane binding, perhaps these structural differences between this region in BoNT/CD-HCR and BoNT/D-HCR may be responsible for the different synaptosome binding properties of these two proteins.
In one of the three recently determined crystal structures for BoNT/D-HCR, glycerol, an agent used as a cryoprotectant for X-ray diffraction data collection, was observed bound in the putative ganglioside binding site [15]. In the crystal structure of BoNT/CD-HCR, a glycerol molecule was also observed bound to the protein, however, it was located near, not at, the ganglioside binding pocket described for several other BoNT serotypes (Fig. 2C). The three hydroxyl groups of the glycerol are in position to form hydrogen bonds with the side chain hydroxyl group of S1264 (2.8 A), the ε-amino group of K1261 (3.1 Å), and possibly the side chain hydroxyl group of T1265 (3.8 Å). The difference in position of the glycerol bound to BoNT/CD-HCR and BoNT/D-HCR may be related to the different synaptosome binding properties of those two proteins.
3.3. BoNT/CD-HCR binds PE liposomes with higher capacity than BoNT/D-HCR
The HCR domains of BoNT/D and BoNT/CD were both previously shown to bind immobilized PE by a thin layer chromatography assay [16,17]. Compared with immobilized lipids, liposomes provide a better approximation to membrane structures and environments and binding of proteins to liposomes better reflects the in vivo protein-lipid interactions. Using a liposome-binding assay, ~ 80% of BoNT/CD-HCR was observed distributed in the centrifugal pellet fraction after incubation with PE liposomes (Fig. 3), indicating a strong association of BoNT/CD-HCR with PE liposomes. This liposome binding capacity of BoNT/CD-HCR is much greater than the ~ 30% distribution previously reported for BoNT/D-HCR under the same conditions [18], and consistent with the observation that BoNT/CD-HCR binds synaptosomes stronger than BoNT/D-HCR [16]. Although PE is found primarily on the inner leaflet of membranes [26], it can be transferred to the outside of membranes by lipid transporters [27]. Therefore, it is possible that binding of BoNT/D and BoNT/CD to synaptosomes is partially mediated by interactions with PE.
Fig. 3.
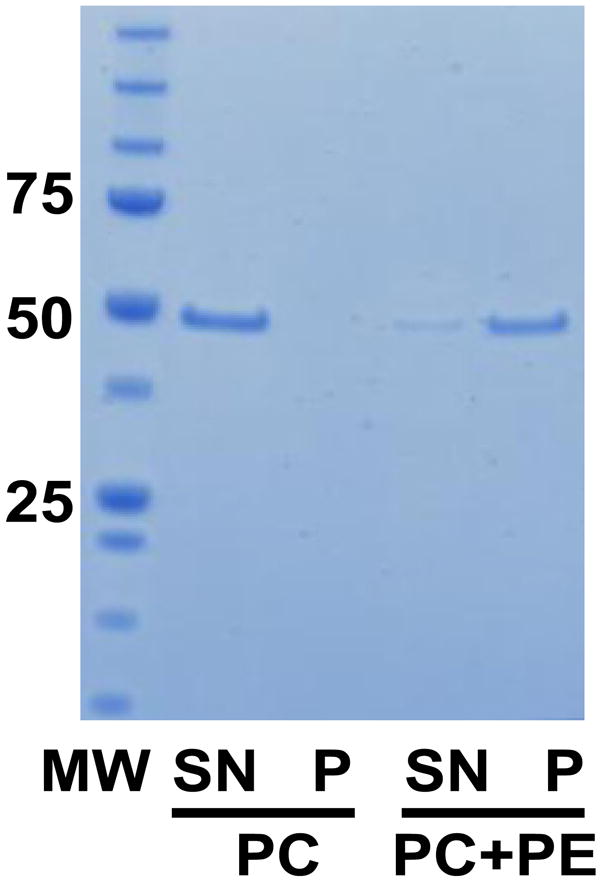
Analysis of PE liposome-binding by BoNT/CD-HCR. SDS-PAGE gel showing the results of the liposome-binding assay. MW, protein molecular weight standard (in kDa); PC, 100% phosphatidylcholine; PC+PE, 1:1 ratio of PC and PE; SN, supernatant fraction showing non-bound protein; P, pellet fraction showing bound protein.
3.4. Potential roles of K1118 and K1136 in membrane recognition
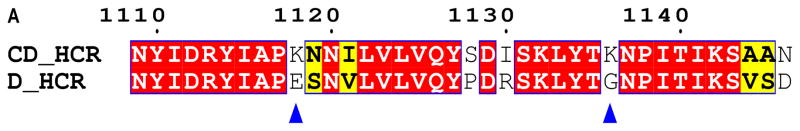
While BoNT/CD does not have an SxWY ganglioside binding motif, BoNT/CD-HCR binds to synaptosomes [16] and PE liposomes. Lysine-1118 and -1136 likely play some role in the lipid binding properties of BoNT/CD-HCR because mutation of these two residues were previously shown to dramatically decrease the binding affinity of BoNT/CD-HCR for synaptosomes [17]. These two residues are not present in BoNT/D-HCR with K1118 and K1136 replaced by E1114 and G1132, respectively (Fig. 4A).
Fig. 4.
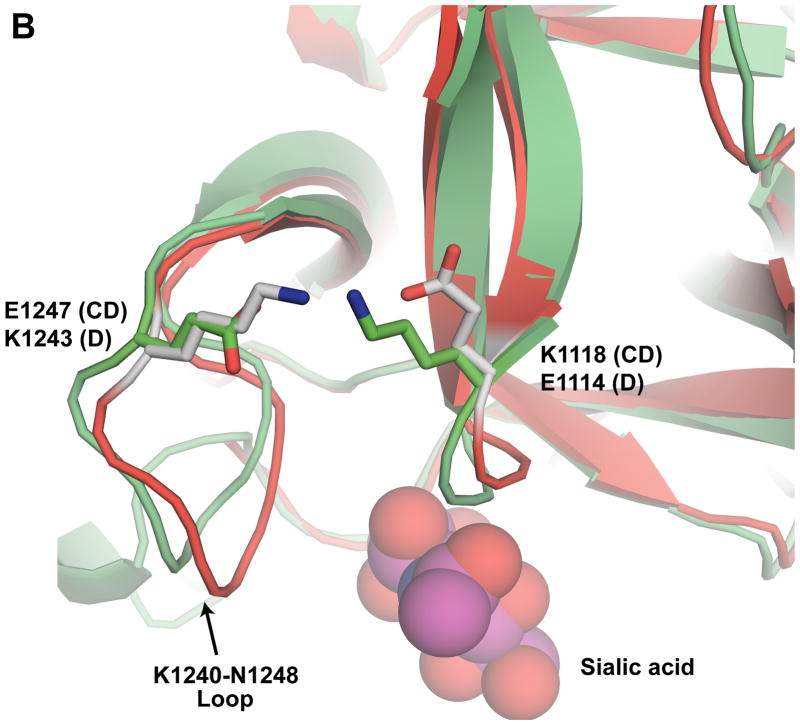
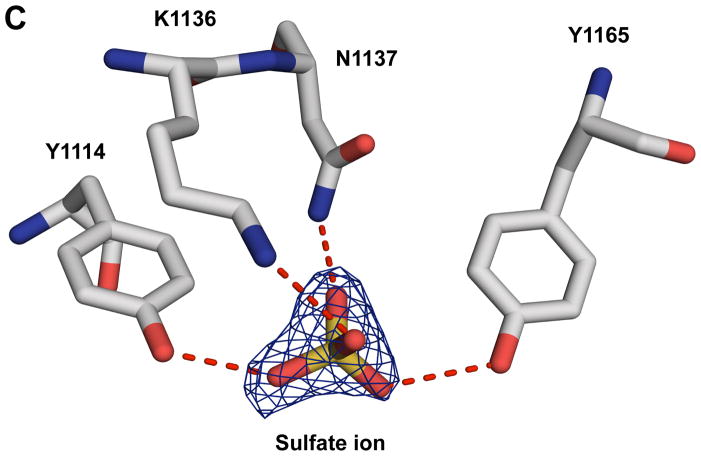
Amino acid sequence and structural details around K1118 and K1136. (A) Sequence alignment of the HCR domains of BoNT/CD and BoNT/D. Blue triangles indicate the location of K1118 and K1136 in BoNT/CD-HCR. Identical and conserved residues are indicated with boxes shaded red and yellow, respectively. The numbering along the top corresponds to the BoNT/CD sequence. The figure was prepared using the online ESPript server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). (B) Expansion of the region around the K1240-N1248 loop shown in Figure 1 with carbon atoms of the BoNT/CD-HCR and BoNT/D-HCR side chains colored green and gray, respectively. The position adopted by the sialic acid molecule when the structure of the BoNT/D-HCR-sialic acid complex (3OBT) is superimposed on the BoNT/CD-HCR structure is shown as a spherical representation. The termini of the K1118 and E1247 side chains of BoNT/CD-HCR (green) are in a position to form a salt bridge similar to the one formed by the equivalent side chains of residues E1114 and K1243 in BoNT/D-HCR (grey). (C) Interactions of the sulfate ion, illustrated with the 2Fo−Fc electron-density map contoured at 1.0 σ, with the side chains of K1136 and surrounded residues. Potential hydrogen bonds between these side chains and the sulfate ion are shown as red dashed lines.
Lysine-1118 is located on the C-terminal end of a β-strand near the putative protein receptor binding site and is in a position to form a salt bridge with E1247 (Fig. 4B) in the K1240-N1248 loop. Based on comparison to the analogous HCR regions in BoNT/B and G, the K1240-N1248 loop in BoNT/CD-HCR shifts > 10 Å into the putative protein receptor-binding groove. The organization of the equivalent region in BoNT/D-HCR was essentially the same: a salt bridge may form between E1114 and K1243 in the K1236-N1244 loop (Fig. 4B) and the loop shifts ~ 13 Å into the putative protein receptor binding groove [18]. Modeling studies with BoNT/D-HCR showed that such an organization sterically blocked the putative protein receptor binding groove from binding to a protein receptor [18]. As a consequence, in BoNT/D-HCR it was proposed that the K1236-N1244 loop might be involved in membrane, instead of protein, interactions [18], a hypothesis supported by the identification of a sialic acid molecule bound near the K1236-N1244 loop in a crystal structure of BoNT/D-HCR [15]. Using the same arguments, the K1240-N1248 loop in BoNT/CD-HCR may also be involved in membrane recognition. A salt bridge might be important for stabilizing this membrane-binding region because it is conserved in the crystal structures of BoNT/CD-HCR and D-HCR. Such a conclusion is further supported by the reduced synaptosome binding activity of the K1118E BoNT/CD-HCR mutant observed by Tsukamoto et al. [17], a mutation that would destroy the salt bridge and subsequently destabilize the K1240-N1248 loop.
Lysine-1136 is located in the middle of the Q1126-I1139 loop on the surface of BoNT/CD-HCR and is exposed to solvent. The ε-amino group is in position to hydrogen bond with a sulfate ion (3.0 Å, Fig. 4C) that was observed bound to the crystal structure of BoNT/CD-HCR. This sulfate ion originates from the crystallization mother liquor solution and is also in position to hydrogen bond with hydroxyl groups of Y1114 (2.7 Å) and Y1165 (2.9 Å), and the side chain amino group of N1137 (2.7 Å) (Fig. 4C). Because the sulfate is similar to a phosphate group on a phospholipid membrane surface (and to a lesser extent, a hydroxyl group of a glycolipid sugar moiety), the observed binding of the sulfate ion to BoNT/CD-HCR may mimic a natural protein-membrane interaction. Moreover, the side chains observed associated with the interactions of the sulfate ion, aromatic residues Y1114 and Y1165, would strengthen hydrophobic interactions with lipid. Lysine-1136 may be central to the protein-membrane interactions since the K1136G mutation reduces the ability of BoNT/CD-HCR to bind synaptosomes [17].
3.5 Summary
By determining the structure of BoNT/CD-HCR and comparing it with the structure of BoNT/D-HCR, subtle structural differences were observed between the two proteins that may be related to biological differences. In addition, analysis of the structure of BoNT/CD-HCR provided structural insights to potentially explain the observation that mutations to K1118 and K1136 in BoNT/CD-HCR dramatically decrease the binding affinity for synaptosomes [17]. Further biochemical analyses, including attempts to co-crystallize BoNT/CD-HCR and BoNT/D-HCR with lipid molecules, will potentially facilitate an understanding of the membrane-binding interactions essential for neuronal cell recognition.
Research highlights.
Crystal structure of botulinum C-D mosaic neurotoxin at 1.56 Å resolution
BoNT/CD-HCR binds PE liposomes more tightly than BoNT/D-HCR
Lysine-1118 may enhance BoNT/CD-HCR synaptosome binding by stabilizing a putative membrane interaction loop
Binding of a sulfate ion to K1136 may reflect natural interactions of BoNT/CD-HCR with membrane phospholipids or glycolipids
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Allergy and Infectious Diseases (NIAID) through award number U01AI081895 and Federal Contract No. HHSN272200700057C. The structure of BoNT/CD-HCR was a community request made to the Seattle Structural Genomics Center for Infectious Disease (SSGCID) and was given the internal identification code ClboA.17807.a.AV2. Portions of the research was performed at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. Department of Energy (DOE)’s Office of Biological and Environmental Research (OBER) program located at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the US DOE under contract (AC06-76RLO 1830). Data for this study were collected on beamline X29A at the National Synchrotron Light Source (NSLS) located at Brookhaven National Laboratory. Financial support for NSLS comes principally from OBER and the Basic Energy Sciences of the US DOE, and from the National Center for Research Resources of the NIH.
Abbreviations
- BoNT
botulinum neurotoxin
- BoNT/CD-HCR
the HCR domain of BoNT C-D mosaic serotype
- BoNT/D-HCR
the HCR domain of BoNT serotype D
- Hc
BoNT C-terminal 100 kDa heavy chain
- Hcc
the C-terminal sub-domain of HCR
- Hcn
the N-terminal sub-domain of HCR
- HCR
the receptor binding domain
- Lc
BoNT N-terminal 50 kDa light chain
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lamanna C. The most poisonous poison. Science. 1959;130:763–772. doi: 10.1126/science.130.3378.763. [DOI] [PubMed] [Google Scholar]
- 2.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 3.Shukla HD, Sharma SK. Clostridium botulinum: a bug with beauty and weapon. Crit Rev Microbiol. 2005;31:11–18. doi: 10.1080/10408410590912952. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura K, Kohda T, Umeda K, Yamamoto H, Mukamoto M, Kozaki S. Characterization of the D/C mosaic neurotoxin produced by Clostridium botulinum associated with bovine botulism in Japan. Vet Microbiol. 2010;140:147–154. doi: 10.1016/j.vetmic.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Lindstrom M, Nevas M, Kurki J, Sauna-aho R, Latvala-Kiesila A, Polonen I, Korkeala H. Type C botulism due to toxic feed affecting 52,000 farmed foxes and minks in Finland. J Clin Microbiol. 2004;42:4718–4725. doi: 10.1128/JCM.42.10.4718-4725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins MD, East AK. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J Appl Microbiol. 1998;84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- 7.Takeda M, Tsukamoto K, Kohda T, Matsui M, Mukamoto M, Kozaki S. Characterization of the neurotoxin produced by isolates associated with avian botulism. Avian Dis. 2005;49:376–381. doi: 10.1637/7347-022305R1.1. [DOI] [PubMed] [Google Scholar]
- 8.DasGupta BR, Sugiyama H. A common subunit structure in Clostridium botulinum type A, B and E toxins. Biochem Biophys Res Commun. 1972;48:108–112. doi: 10.1016/0006-291x(72)90350-6. [DOI] [PubMed] [Google Scholar]
- 9.Muraro L, Tosatto S, Motterlini L, Rossetto O, Montecucco C. The N-terminal half of the receptor domain of botulinum neurotoxin A binds to microdomains of the plasma membrane. Biochemical and Biophysical Research Communications. 2009;380:76–80. doi: 10.1016/j.bbrc.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 10.Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat Struct Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- 11.Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc Natl Acad Sci U S A. 2007;104:10447–10452. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montecucco C. Double Receptors for Tetanus and Botulinum Toxins. European Journal of Cell Biology. 1986;42:13–13. [Google Scholar]
- 13.Montal M. Botulinum neurotoxin: a marvel of protein design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- 14.Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- 15.Strotmeier J, Lee K, Volker AK, Mahrhold S, Zong Y, Zeiser J, Zhou J, Pich A, Bigalke H, Binz T, Rummel A, Jin R. Botulinum neurotoxin serotype D attacks neurons via two carbohydrate binding sites in a ganglioside dependent manner. Biochem J. 2010 doi: 10.1042/BJ20101042. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto K, Kohda T, Mukamoto M, Takeuchi K, Ihara H, Saito M, Kozaki S. Binding of Clostridium botulinum type C and D neurotoxins to ganglioside and phospholipid - Novel insights into the receptor for clostridial neurotoxins. Journal of Biological Chemistry. 2005;280:35164–35171. doi: 10.1074/jbc.M507596200. [DOI] [PubMed] [Google Scholar]
- 17.Tsukamoto K, Kozai Y, Ihara H, Kohda T, Mukamoto M, Tsuji T, Kozaki S. Identification of the receptor-binding sites in the carboxyl-terminal half of the heavy chain of botulinum neurotoxin types C and D. Microb Pathog. 2008;44:484–493. doi: 10.1016/j.micpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Buchko GW, Qin L, Robinson H, Varnum SM. Structural analysis of the receptor binding domain of botulinum neurotoxin serotype D. Biochem Biophys Res Commun. 2010;401:498–503. doi: 10.1016/j.bbrc.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karalewitz AP, Kroken AR, Fu Z, Baldwin MR, Kim JJ, Barbieri JT. Identification of a unique ganglioside binding loop within botulinum neurotoxins C and D-SA. Biochemistry. 2010;49:8117–8126. doi: 10.1021/bi100865f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Research. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwaal RF, Roelofsen B, Colley CM. Localization of red cell membrane constituents. Biochim Biophys Acta. 1973;300:159–182. doi: 10.1016/0304-4157(73)90003-8. [DOI] [PubMed] [Google Scholar]
- 27.Williamson P, Schlegel RA. Transbilayer phospholipid movement and the clearance of apoptotic cells. Biochim Biophys Acta. 2002;1585:53–63. doi: 10.1016/s1388-1981(02)00324-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.