Abstract
Transglutaminase 2 (TG2) is a ubiquitously expressed protein that catalyzes protein/protein crosslinking. Because extracellular TG2 crosslinks components of the extracellular matrix, TG2 is thought to function as a suppressor of cellular invasion. We have recently uncovered that the TG2 gene (TGM2) is a target for epigenetic silencing in breast cancer, highlighting a molecular mechanism that drives reduced TG2 expression, and this aberrant molecular event may contribute to invasiveness in this tumor type. Because tumor invasiveness is a primary determinant of brain tumor aggressiveness, we sought to determine if TGM2 is targeted for epigenetic silencing in glioma. Analysis of TGM2 gene methylation in a panel of cultured human glioma cells indicated that the 5′ flanking region of the TGM2 gene is hypermethylated and that this feature is associated with reduced TG2 expression as judged by immunoblotting. Further, culturing glioma cells in the presence of the global DNA demethylating agent 5-aza-2′-deoxycytidine and the histone deacetylase inhibitor Trichostatin A resulted in re-expression of TG2 in these lines. In primary brain tumors we observed that the TGM2 promoter is commonly hypermethylated and that this feature is a cancer-associated phenomenon. Using publically available databases, TG2 expression in gliomas was found to vary widely, with many tumors showing overexpression or underexpression of this gene. Since overexpression of TG2 leads to resistance to doxorubicin through the ectopic activation of NFκB, we sought to examine the effects of recombinant TG2 expression in glioma cells treated with commonly used brain tumor therapeutics. We observed that in addition to doxorubicin, TG2 expression drove resistance to CCNU; however, TG2 expression did not alter sensitivity to other drugs tested. Finally, a catalytically null mutant of TG2 was also able to support doxorubicin resistance in glioma cells indicating that transglutaminase activity is not necessary for the resistance phenotype.
Keywords: Glioma, Epigenetic silencing, Extra cellular matrix, Tumor invasiveness, Drug resistance
Introduction
Tumor cells arise from normal cells that have acquired abnormal proliferative capability stemming from a multistep process where oncogene activity is increased and tumor suppressor gene (TSG) activity is constrained [1]. Numerous TSGs have been shown to undergo de novo methylation at clustered 5′-CG-3′ dinucleotides (referred to as CpG islands) generally present within their 5′ flank [2]. This cancer-associated DNA hypermethylation is part of a complex set of epigenetic events that result in the transformation of chromatin from a transcriptionally-active to an inactive state [3]. This process, termed epigenetic gene silencing, is now widely recognized as either a causative or correlative event in tumor development.
Our group recently showed that the gene encoding transglutaminase 2 (TGM2) is a target for epigenetic silencing in breast cancer. This gene product, referred to as TG2, is a ubiquitously expressed protein comprised of a single ~ 76 kD polypeptide that, like other transglutaminases, catalyzes the formation of a covalent bond between a free epsilon-amino group of a lysine residue and the gamma-carboxyl group of a glutamine residue [4]. Extracellular pools of TG2 bind to and cross-link numerous components of the extracellular matrix (ECM) such as fibronectin, vitronectin, collagen, fibrin, laminin, osteonectin, and osteopontin [5]. This has led to the view that TG2 cross-linking renders the ECM resistant to mechanical and proteolytic degradation, stabilizes cell-ECM interaction [6], and promotes wound healing [7]. Moreover, this role in ECM stabilization has lead to speculation that TG2 functions as a TSG by inhibiting tumor spread [8, 9].
In addition to this hypothesized TSG activity, TG2 expression also exerts a clear effect on cellular sensitivity to chemotherapeutic drugs. Mehta [10] first observed that a doxorubicin-resistant subclone of the human breast tumor cell line MCF-7 displayed significantly higher levels of TG2 expression activity than doxorubicin-sensitive MCF-7 cells. Since this seminal finding, several studies firmly support the conclusion that TG2 expression modulates sensitivity to doxorubicin in a variety of tumor types [11–14]. While the mechanism responsible for this effect remains controversial, several groups have observed heightened NFkB activity in cells expressing aberrantly high levels of TG2 [15, 16]. Through its pro-survival activity, dysregulated NFkB activity is likely important in conferring the chemoresistant property of tumor cells overexpressing TG2. Moreover, because of its ability to ectopically activate NFkB, it has been proposed that, when overexpressed, TG2 functions as an oncogene [17, 18].
Brain tumors, although a rare malignancy across age groups, are the most common solid tumor in children [19, 20]. Despite advances in treatment for malignant glioma, the poor prognosis associated with this disease has remained relatively unchanged. Morbidity and mortality from malignant glioma is directly correlated with the ability of tumor cells to infiltrate surrounding tissue and malignant cells are found well beyond the gross radiographic margins of the tumor [21]. The infiltrative nature of gliomas makes it impossible to obtain a clear surgical resection margin, therefore, following surgical debulking and/or radiation therapy, clandestine infiltrative tumor cells eventually lead to relapse [22]. In contrast, poorly invasive tumors (i.e., pylocytic astrocytomas and gangliomas) respond well to surgery leading to the widely held view that the invasive nature of brain tumors is a primary prognostic indicator [23].
Glioma cell invasion is likely an early event in disease progression and is already observable in low-grade infiltrative astrocytomas [21]. Tumor invasion is believed to be a multi-step process, involving, in part, alterations in the interaction of the tumor cell with extracellular matrix (ECM), and a generalized loss of ECM integrity often through extracellular protease dysregulation [24, 25]. Studies have shown that at the interface of glioma and normal tissue, the ECM undergoes remodeling with vitronectin, osteopontin, osteonectin, tenacin-C, SPARC and BEHAB becoming prominent components of the ECM in such locations [21]. Moreover, several of these components, as well as the down-regulation of some ECM components, likely promote tumor cell invasiveness [26–29]. Such findings clearly underscore the widely-held view that the composition and tumor-associated changes in the ECM have a profound effect on modulating glioma cell invasion [30, 31].
Although combination therapy of surgery, radiotherapy and chemotherapy are used in the treatment of nearly all brain tumors, the response and regimen of chemotherapy is based on tumor type. At present, carboplatin and vincristine are considered first line agents along with radiation for the treatment of pediatric gliomas [19], whereas, in adults, temozolomide and radiation are emerging as the standard of care for glioblastoma [20]. Prognostic biomarkers of response to different chemotherapy regimens will allow rapid stratification of patients into low and high risk cohorts and allow us to tailor future therapies to maximize curability and minimize treatment associated morbidity.
In light of the view that TG2 modulates ECM stability and tumor cell invasiveness, and that dysregulation of these properties may impact tumor behavior, we sought to determine if the TGM2 gene is targeted for epigenetic silencing in brain tumors. We document that several cultured glioma lines display hypermethylation of TGM2 and that this is associated with reduced TG2 expression and reversal of TGM2 methylation results in re-expression of TG2. Primary brain tumors also commonly display aberrant TGM2 methylation and this likely contributes to the heterogeneous nature of TG2 expression in glioma. Finally, by expressing TG2 in the U118MG glioma line that has silenced TGM2, we document that ectopic expression of TG2 confers resistance to some, but not all, commonly used brain tumor therapeutics.
Materials and methods
Cell culture and drug treatment
All cell lines were obtained from American Type Culture Collection (Manassas, VA) unless otherwise specified and cultured in McCoys-5A media (Cellgrow, Manassas, VA) supplemented with 10% fetal calf serum (Hyclone, ThermoFisher Inc., Rockford, IL), 100 units/ml penicillin and 100 mg/ml streptomycin. Stably transfected cells were grown in the same media supplemented with 200 μg/ml neomycin sulfate (G418). All cells were maintained at 37°C in a humidified 5% CO2 atmosphere.
Epigenetic alteration of genomic DNA in vivo was induced by culturing cells with the global demethylating agent 5-aza-2′-deoxycytidine (5-azadC) and Trichostatin A (TSA) using previously published protocols [32]. Briefly, every 24 h for five consecutive days used medium was removed from cultures of cells, plates were subsequently rinsed with PBS, and fresh culture media containing 5 μM 5-azadC was replaced. On the sixth day, cells were rinsed with PBS, re-fed with fresh culture media containing 100 μM TSA and incubated for an additional 16 h prior to harvesting. Both 5-azadC and TSA were purchased from Sigma-Aldrich (St. Louis, MO).
SDS-PAGE and immunoblotting
Cells were harvested by centrifugation following trypsinization and following several PBS washes, protein extracts were formed by addition of an appropriate volume of SDS lysis buffer (100 mM Tris–HCl, pH 7.5/5 mM EDTA/1% SDS) and placed in a 95°C hot block for 5 min. Lysates were then sonicated and centrifuged at 3000×g for 3 min. Protein concentrations were determined by the BCA method (Pierce), and lysates stored at −80°C.
Prior to electrophoresis, an appropriate volume of cell lysate was diluted in 3× SDS sample buffer (150 mM Tris–HCl, pH 6.8; 10% β-mercaptoethanol; 20% glycerol; 3% SDS; 0.01% bromophenol blue; 0.01% pyronin-Y) and boiled for 5 min. Proteins were resolved on SDS-polyacrylamide gels and electrotransferred onto nitrocellulose membranes O/N at 12V. Immunoblotting was conducted with mouse monoclonal mouse anti-TG2 Ab-3 (Lab Vision) or polyclonal rabbit anti-tubulin Ab15568-500 (Abcam) followed by HRP-conjugated secondary antibody. Signals were recorded on X-ray film using SuperSignal West Pico Chemoluminescent Substrate (Pierce) per manufacturer’s instructions.
RT-PCR
Total RNA was extracted from cultured cells using TriZol reagent (Invitrogen) per manufacturers instructions. RNA was then used for first strand cDNA synthesis reactions using AffinityScript (Stratagene) and random hexamer primers per manufacturer’s protocol. PCR reactions were conducted using Qiagen HotStar Taq polymerase with supplied buffer and MgCl2. Human TG2 PCR primers are 5′-TAAGAGATGCTGTGGAGGAG-3′ (forward) and 5′-CGAGCCCTGGTAGATAAA-3′ (reverse) and were used in the following thermocycling conditions: 1 cycle (95°C, 3 min); 30 cycles (94°C, 45 s/58°C, 45 s/72°C, 60 s); final extension at 72°C for 10 min. Human GAPDH was amplified using the primers 5′-ACCACAGTCCAT GCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTT GCTGA-3′ (reverse) and thermocycling conditions were 1 cycle (95°C, 3 min); 26 cycles (94°C, 45 s; 58°C, 45 s; 72°C, 60 s); final extension at 72°C for 10 min. PCR products were visualized by ethidium bromide staining following electrophoretic separation on 2% agarose gels.
DNA extraction and sodium bisulfite conversion
Genomic DNA (gDNA) was isolated from harvested glioma cell lines by adding 100 μL of proteinase K lysis buffer (10 mM Tris–HCl, 1 mM EDTA, 1% SDS, 0.4 mg/ml proteinase K, and 0.1 mg/ml RNAse A) to pelleted cells. Cells were subsequently incubated at 50°C for 24 h, and DNA obtained by ethanol precipitation.
gDNA was isolated from formaldehyde-fixed, paraffin-embedded tissues as previously reported [33]. Multiple 5 μM tissue sections were collected into microfuge tubes and de-paraffinized with xylene. De-paraffinized samples were sequentially washed in 100% then 75% ethanol, and finally air dried prior to lysis. Lysis was performed as indicated above by resuspension in proteinase K lysis buffer and O/N incubation at 50°C. Following this, lysates were cleared by centrifugation and supernatants used in DNA analyses outlined below.
DNA was bisulfite converted using the EZ DNA methylation kit (ZYMO Research Co., Orange, CA) according to manufacturer’s instructions. Briefly, 1.5 μg of denatured genomic DNA was treated with sodium bisulfite in the dark at 50°C overnight. Following this, samples were washed in the supplied columns, desulphonated, and washed again. DNA was eluted with 20 μl of the provided elution buffer and 1.0 μl of bisulfite-converted DNA was subsequently used in PCR reactions.
Pyrosequencing
Quantitative DNA methylation analysis was conducted by pyrosequencing as previously described [34]. A 201 bp segment of the TGM2 gene was amplified from bisulfite converted DNA using a 5′ biotinylated reverse primer. The primers used are 5′-GTTTTTTGGGTGAGTTTTAG-3′ (forward) and 5′-ATAACTCCTTCCACTAAC-3′ (reverse) and thermocycling conditions used are 95°C, 15 m/52 cycles (94°C, 30 s/53°C, 30 s/72°C, 45 s), final extension at 72°C for 10 min. Following amplification, 5–20 μl of PCR product was mixed with streptavidin-conjugated Sepharose beads (GE Healthcare) in the supplied binding buffer (Biotage AP, Uppsala, Sweden) and the sample was diluted to 60–80 μl total volume with dH2O. The beads were subsequently collected using a vacuum preparation workstation, the sequencing primer 5′-GGGTAATG GGTGGTTTTTTAGG-3′ was added to the beads, the mixture heated to 80°C for 2 min and annealing of the sequencing primer to the biotinylated DNA strand is allowed to occur during cooling to room temperature. Pyrosequencing was subsequently conducted using a PyroMark MD system (Biotage) and methylation analysis was conducted using PyroMark CpG software.
Site-directed mutagenesis and cell transfection
A full-length human TG2 cDNA subcloned into the eukaryotic expression vector pcDNA3.1 was a gift from Dr. H-G Wang (Moffitt Cancer Center, Tampa, FL). Site directed mutagenesis of this TG2 cDNA construct was conducted using the Stratagene (La Jolla, CA) Quick-Change system per manufacturer’s instructions. The oligonucleotide 5′-GGTGTGCTGCTGGGACGCcaGGACA ACAACTACGGG-3′ was used to mutagenize the wildtype tryptophan at TG2 residue 241 to glutamine (mutagenic nucleotides in lower case). The resultant W241Q mutation was confirmed by sequencing.
Cell transfection was performed using Lipofectamine (Invitrogen, Life Technologies, Carlsbad, CA) per manufacturer’s instructions. Briefly, cells were plated in 60 mm tissue culture dishes at high density (>90% confluency) 24 h prior to transfection. The following day, 8 μg purified plasmid DNA and 20 μl of Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, CA) were diluted in 1.0 ml of Opti-MEM I medium (Invitrogen), and this mixture was incubated for 20 min at room temperature. Following this, DNA/lipid complexes were added to cells and then returned to the incubator for 24 h. The following day, cells were split into five 100 mm tissue culture dishes, and 24 h later G418 was added to the medium to a final concentration of 400 μg/ml. Cells were incubated for approx. 3 weeks under G418 selection, single colonies were picked, expanded, and cells screened by immunoblotting using anti-TG2.
Viability (MTT) assay
2000–6000 cells were plated in each well of a 96 well plate in complete growth medium and incubated at 37°C for 24 h. Following this, media was aspirated and 100 μl of fresh media added to each well. Chemotherapeutics were diluted in fresh media and added to the cells at indicated concentrations. (O6 benzylguanine was added to a final concentration of 50 μM in cultures of cells 1 h prior to treatment with temozolomide.) Following the addition of drug, media was added to bring the final volume/well to 200 μl. Plates were subsequently incubated for 72 h at 37°C. MTT reagent (Sigma) was added to each well to a final concentration of 1.0 μg/ml and plates returned to the incubator for 2–4 h. Following this, media was aspirated and cells solubilized in 100 μl/well of DMSO. After a 5 min incubation at 37°C, absorbance at 570 nM was measured using a BioRad plate reader. Relative cell viability vs. untreated cells was subsequently calculated. As a control, doxorubicin treated cells were always run in parallel to assure maintenance of the resistance phenotype.
Results
The TGM2 gene is epigenetically silenced in cultured human glioma cells
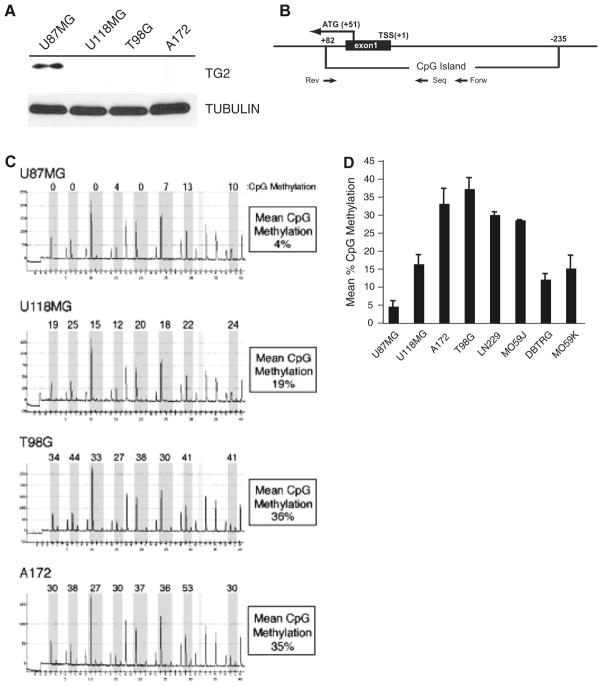
We have recently documented that the gene encoding transglutaminase 2 (TG2) is a target for epigenetic silencing in breast cancer [32]. Whereas the function of TG2 is to covalently link proteins and TG2 has been shown to crosslink components of the ECM, it has been proposed that this protein functions to suppress tumor invasiveness [18]. Because of the invasive nature of glioma, we set out to determine if TG2 is silenced in this tumor type as well. When several glioblastoma cell lines were assayed for expression of TG2 by immunoblotting, we observed abundant expression of this protein in the cell line U87MG (Fig. 1a). Other lines investigated (U118MG, T98G, and A172) displayed no detectable TG2 expression under these conditions.
Fig. 1.
TGM2 is aberrantly hypermethylated in many cultured brain tumor lines. a Protein extracts were formed from cultures of human glioma lines U87MG, U118, T98G and A172. Extracts were immunoblotted with anti-TG2 (top panel) or anti-tubulin (bottom). b The structure of the 5′ flank of the human TGM2 gene is shown. Depicted is the first transcribed exon (exon 1), the boundaries of the CpG island located within this locus, the translational (ATG) and transcriptional start sites (TSS), and the location of oligonucleotide primers used for amplification (Forw, Rev) and sequencing (Seq). c Genomic DNA was isolated from the glioblastoma cell lines U87MG, U118, T98G and A172. After bisulfite conversion, the methylation status of the TGM2 gene was analyzed by pyrosequencing. Shown are resultant pyrograms and mean CpG methylation for the region of the TGM2 gene analyzed. d TGM2 methylation was measured by pyrosequencing for each of the indicated glioma cell lines, graphed is the mean of at least 5 independent pyrosequencing assays
We next conducted methylation analysis of the proximal promoter region of the TG2 gene (termed TGM2) in these lines to determine if reduced expression of TG2 was attributable to aberrant DNA methylation. To conduct this analysis we used pyrosequencing, an established methodology that permits quantitative analysis of CpG methylation within the region of the genome investigated. The pyrosequencing assay that we developed to examine TGM2 examines a 40 bp region of this gene close to the translational start site that contains 8 CpG dinucleotides (Fig. 1b). Pyrosequencing analysis of U87MG determined that this line had a low level of CpG methylation (4%) within the region of the TGM2 gene (Fig. 1c). In contrast, A172 (35%), U118MG (19%), and T98G (36%) each showed high levels of CpG methylation within the TGM2 gene. We conducted pyrosequencing analysis on a number of cultured glioma cell lines and the data from this analysis is shown graphically in Fig. 1d. While the lowest value for average TGM2 CpG methylation was measured in U87MG cells, the other seven brain tumor cell lines all displayed higher levels of CpG methylation within the TGM2 gene ranging from 36% in T98G to 13% in MO59K. Taken together, these findings indicate that methylation of the TGM2 promoter is common in cultured glioma cells.
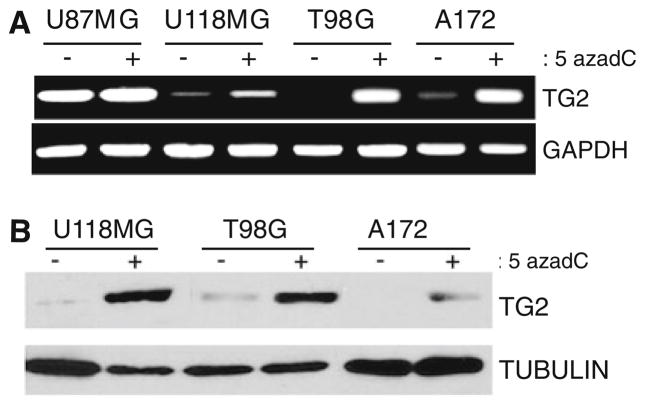
We next sought to determine if CpG methylation within the TGM2 gene was resulting in gene silencing. U87MG, U118MG, T98G and A172 cells were treated with the global DNA demethylating agent 5-aza-2′-deoxycytidine (5-aza-dC) and the histone deacetylase inhibitor Trichostatin A (TSA) to reverse epigenetic gene repression. Following 5-azadC/TSA treatment, total RNA was isolated from treated and untreated cells, and subsequently used in RT-PCR reactions using TG2-specific primers. Consistent with immunoblotting results, we observed that the cell line U87MG expresses high levels of TG2 mRNA prior to treatment with 5-azadC/TSA and that expression does not increase following treatment (Fig. 2a). In contrast, 5-azadC/TSA treatment of U118MG, T98G and A172 cells resulted in a clear increase in TG2 transcript abundance in each line. We conducted parallel immunoblot experiments on U118MG, T98G and A172 and found that 5-azadC/TSA treatment also resulted in increased TG2 protein expression (Fig. 2b). Since pyrosequencing analysis confirmed that 5-azadC/TSA treatment resulted in decreased CpG methylation within the TGM2 gene (data not shown), we conclude that hypermethylation of the TGM2 promoter occurs in glioma cell lines and that this epigenetic event results in silencing of this gene.
Fig. 2.
Global DNA demethylation results in TG2 re-expression in cultured glioma cell lines. a The glioma lines U87MG, U118, T98G and A172 were cultured in the presence or absence of the global DNA demethylating agent 5-azadC as indicated. Total RNA was isolated, cDNA synthesized, and PCR conducted using TG2 specific primers (top) or GAPDH specific primers (bottom). b. Protein extracts were obtained from the glioblastoma cell lines U118, T98G and A172 cultured in the presence or absence of 5-azadC. Cell extracts were immunoblotted with anti-TG2 (top) or anti-tubulin (bottom)
Aberrant hypermethylation of TGM2 occurs in primary brain tumors
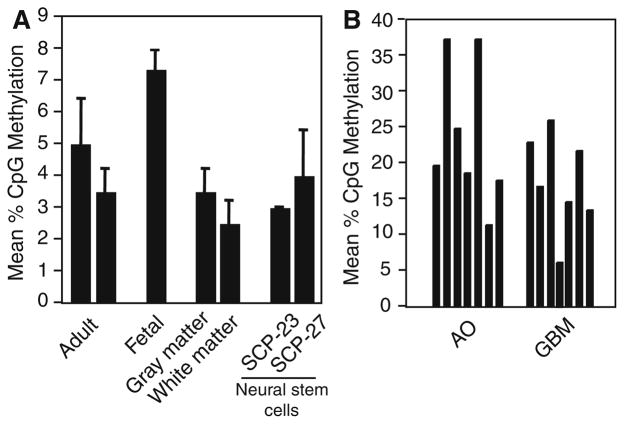
We next sought to determine the methylation status of the TGM2 gene in normal human brain and primary brain tumor specimens. We measured very low levels (≤7%) methylation in DNA samples taken from various normal sources, readings consistent with background methylation scored by pyrosequencing (Fig. 3a). Amongst these are samples taken from two independent normal adult white and grey matter specimens, and one fetal and two adult cortex samples. Additionally, we assayed two neuroprogenitor cell lines SC-23 and SC-27 [35], and found low levels of TGM2 methylation in these lines as well.
Fig. 3.
TGM2 is aberrantly hypermethylated in primary human brain tumors. a Analysis of TGM2 methylation was conducted by pyrosequencing on genomic DNA isolated from various normal human brain specimens including two samples of adult cortex, one from fetal cortex, and two independent samples of dissected white and gray matter. Additionally, genomic DNA harvested from two neural progenitor cell lines (SC-23 and SC-27) was analyzed for methylation of the TGM2 gene. b DNA was isolated from archival specimens of glioblastoma multiformae (GBM) and anaplastic oligodendroglioma (AO) tumors and TGM2 methylation assayed by pyrosequencing. The %CpG methylation graphed for each sample is at least the mean of two independent experiments
TGM2 methylation was measured in genomic DNA samples from diagnosed cases of anaplastic oligodendroglioma (AO, grade III) and glioblastoma multiforme (GBM, grade IV). In these tumor samples we observed, in general, quantitatively higher levels of TGM2 gene methylation than observed in normal brain samples (Fig. 3b). Specifically, we measured the highest level of TGM2 methylation in two AO samples (36%) and the lowest in a GBM (6%). We have not been able to quantify TG2 expression in primary glioma samples, thus we cannot conclude this gene is transcriptionally downregulated in brain tumors. However, our results do indicate that the TGM2 gene is a target for aberrant hypermethylation in primary brain neoplasms and, when combined with the data obtained from cultured glioma lines, strongly suggests that this gene is subject to epigenetic silencing in glioma.
TG2 expression in glioma is heterogeneous
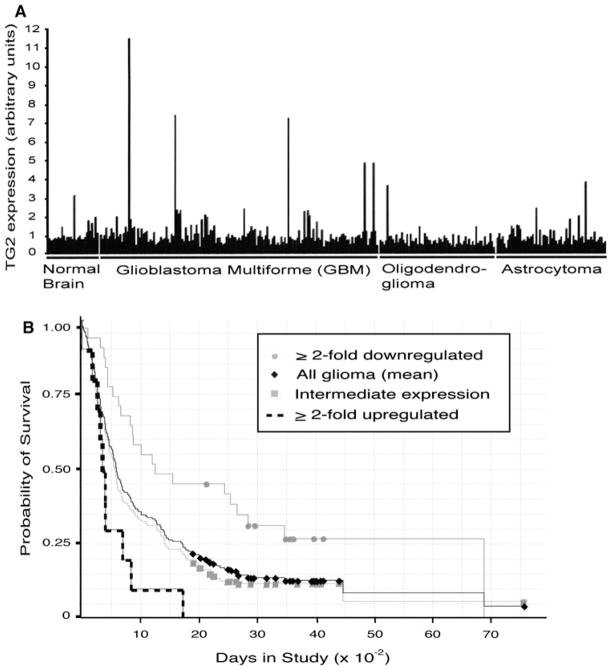
Others have reported high transglutaminase activity occurs in human glioblastoma tumors [36] and our data suggests that TG2 expression may be epigenetically repressed in some brain tumors. To obtain a comprehensive understanding of TG2 expression in brain tumors we queried the publically available Rembrandt dataset maintained by NCI (http://rembrandt.nci.nih.gov; Accessed October 13, 2009). Relative TG2 expression, as scored by hybridization-based microarray analysis using the Affymetrix 201042_at probe, was obtained on 28 non-tumor brain tissues, and 152 GBM, 65 oligodendroglioma (grade II and III), and 64 astrocytoma (grade II and III) tumor samples. Initial inspection of this dataset (Fig. 4a) indicates that TG2 expression is quite heterogeneous in brain tumors. While most tumors show TG2 expression levels comparable (less than two-fold up or downregulated) to non-tumor tissue, 9.6% (27/281) tumors show ≥2-fold downregulation and 5% (14/281) display ≥2-fold upregulation of TG2 expression.
Fig. 4.
TG2 expression is heterogeneous in human brain tumors. a Relative TG2 expression on normal brain tissue, glioblastoma multiforme (stage IV), oligodendroglioma (stage II, III) or astrocytoma (stage II, III) specimens was obtained by query of the Rembrandt database. b Brain tumor samples were stratified into tumor groups that show ≥ 2-fold upregulation (dashed line), ≤ 2-fold downregulation (gray circles), and intermediate levels (gray squares) of TG2 expression. Kaplan–Meier analysis was subsequently conducted with these groupings, as well as mean TG2 expression among all glioma samples (black diamonds), to compare the association of TG2 expression with patient survival
A subset of these tumors have associated patient survival data, thus, conducted a Kaplan–Meier analysis comparing TG2 expression (also using the 201042_at probe) with patient survival (Fig. 4b). By comparing TG2 expression with all glioma samples, we observed a striking difference in survival between patients showing ≥2-fold upregulated vs ≥2-fold downregulated TG2 expression. Specifically, we uncovered that downregulation of TG2 expression is associated with greater survival (P = 0.00016, logrank test). The difference in survival between ≥2-fold upregulated and intermediate TG2 expression (defined as <2-fold up or downregulated from the mean expression measured in all glioma samples) failed to reach significance (P = 0.08); however, comparison of downregulated vs intermediate expression samples was significant (P = 0.04).
Further inspection of this survival data indicated that of the 10 samples displaying upregulated TG2 expression 8 of them were GBM specimens. Conversely, of the 31 samples displaying downregulated TG2 expression, only 9 specimens were from diagnosed GBM tumors, while the remainder were predominantly lower grade oligodendrogliomas and astrocytomas. This result suggested that high grade brain tumors tend to overexpress TG2 while lower grade tumors tend to display reduced expression. To scrutinize this more thoroughly, we examined TG2 expression within grouped GBM, oligodendroglioma, and astrocytoma tumors. We observed that of the 152 GBM tumors in the dataset, 6.6% (10/152) display ≥2-fold upregulation, and 4% (6/152) show ≥2-fold downregulation of TG2 expression compared to the mean expression measured in non--tumor brain tissue. In contrast, only 1.5% (1/65) of oligodendrogliomas display TG2 upregulation but that 20% (13/65) of these tumors display TG2 downregulation. Similarly, 4.6% (3/64) of astrocytomas show upregulation while 14% (9/64) show TG2 downregulation. From this analysis we conclude that upregulation of TG2 expression in brain tumors is associated with high grade tumors and a poorer survival rate, whereas TG2 downregulation is associated with lower grade tumors and a better survival rate. Of course, the number of samples used to conduct this analysis limits statistical power, and more samples need to be included in this analysis to strengthen the validity of these conclusions.
TG2 expression modulates glioma chemosensitivity
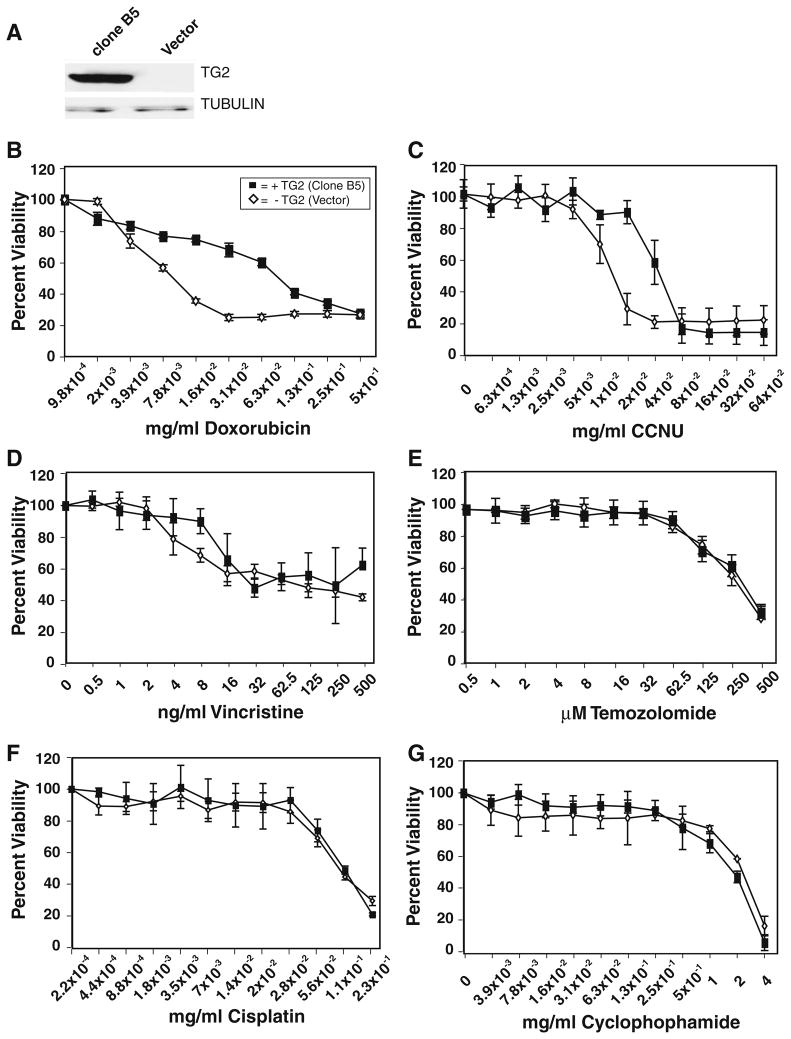
Numerous studies have implicated TG2 expression as a modulator of sensitivity to a variety of different cancer therapeutics in several different tumor cell types [37]. Given this, we set out to determine if TG2 expression could confer chemoresistance in glioma cells to compounds therapeutically relevant to the treatment of brain tumors. To conduct this study we engineered U118MG glioma cells, which have epigenetically silenced its endogenous TGM2 gene and does not express TG2 protein (Figs. 1, 2), to express a transgenic copy of full-length human TG2. Figure 5a shows immunoblot analysis of U118MG cell clones stably transfected with TG2 (clone B5) and empty pcDNA3.1 vector (Vector). Figure 5b–g show the dose response curves for doxorubicin, lomustine (CCNU), vincristine, temozolomide, cisplatin, and cyclophosphamide, respectively, for both clone B5 and the vector control line. Consistent with published reports on lines from other tumor types, we observed that TG2 protein expression induces significant resistance to doxorubicin in U118MG cells over a broad range of drug doses (Fig. 5b).
Fig. 5.
TG2 expression in the glioblastoma cell line U118MG induces chemoresistance to doxorubicin and CCNU but not to other glioma therapeutics. a The glioma cell line U118MG was stably transfected with an expression vector (pcDNA3.1) containing a full-length human TG2 cDNA or empty vector. Immunoblot analysis of extracts U118MG clone expressing TG2 (clone B5) or vector only (Vector) with anti-TG2 (top) or anti-tubulin (bottom) is shown. b U118MG Clone B5 (closed squares) or vector only (open diamonds) were treated with the indicated dose (0–0.5 mg/ml) of doxorubicin and 72 h after drug assayed for viability by MTT analysis. c. Clone B5 and vector only cells were also subject to viability analysis with a range of CCNU (0–0.64 mg/ml). Chemosensitivity of clone B5 and vector cells were also assayed following treatment with Vincristine (0–500 ng/ml) (d); Temozolomide (0.5–500 μM) (e); Cisplatin (0–0.23 mg/ml) (f); Cyclophosphamide (0–4 mg/ml) (g). Error bars = ±1.0 SD
Similar to response to doxorubicin, we observed that TG2 expression conferred resistance to the bifunctional alkylator CCNU (Fig. 5c). However, the dose range where CCNU resistance was observed was found to be more narrow than that measured in doxorubicin treated cells. Nevertheless, a significant difference in sensitivity was measured between B5 and vector cells at the calculated LD50 (12 μg/ml) of CCNU for vector cells (P = 2.2 × 10−7). In cells treated with a range of the microtubule-disrupting drug vincristine (Fig. 5d), we measured significant difference (P = 0.018) in sensitivity at a single concentration (7.8 ng/ml) after averaging five independent experiments. This resistance occurs at a concentration higher than the LD50 for vector cells, and while statistically significant, these results do not allow us to conclude that TG2 expression has a general effect on the sensitivity of U118MG cells to vincristine.
We observed no measurable difference in sensitivity to the monofunctional methylating agent temozolomide in U118MG cells expressing and not expressing TG2 (Fig. 5e). Similarly, we observed that TG2 does not confer resistance to cisplatin (Fig. 5f) nor to cyclophosphamide (Fig. 5g) in this glioma line. In sum, this work indicates that, although TG2 is not a global modulator of chemotherapeutic resistance, it does confer a resistance phenotype in glioma cells against CCNU, a commonly used brain tumor therapeutic.
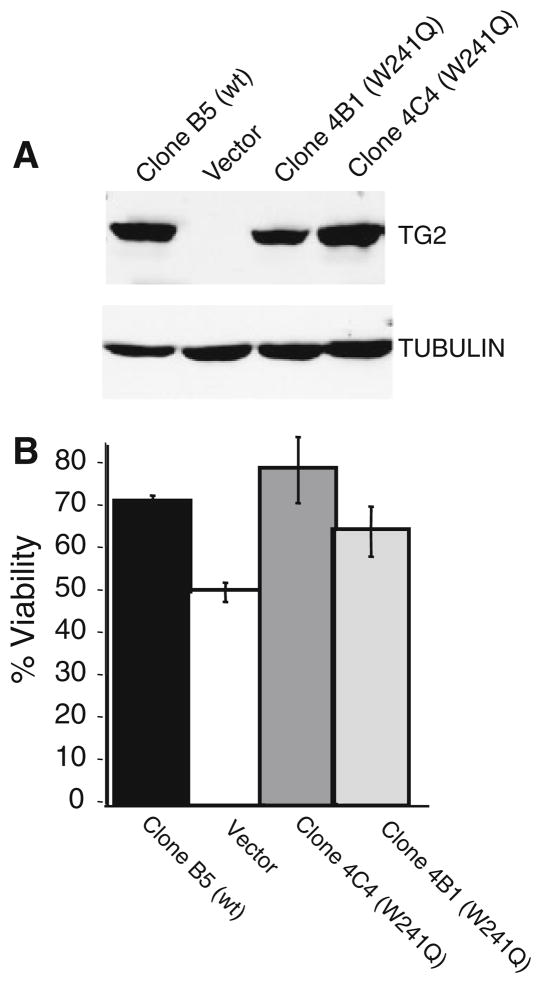
Finally, we sought to determine if the transglutaminase activity of TG2 was required to confer doxorubicin resistance in U118MG glioma cells. Site directed mutagenesis of TG2 cDNA was performed to mutate the tryptophan at residue 241 to glutamine (W241Q) disrupting a “tryptophan bridge” within the active site of TG2 that has previously been shown to ablate the transglutaminase activity of TG2 [38]. Immunoblot analysis of U118MG cells expressing wild type TG2 (Clone B5), pcDNA3.1 only (Vector) and two U118MG clones expressing the W241Q mutant (4B1 and 4C4) shows that wildtype and mutant TG2 expression are well matched in this panel of cell lines (Fig. 6a).
Fig. 6.
TGase activity is not necessary for TG2-induced chemoresistance to doxorubicin in glioma cells. a U118MG cells were stably transfected with an expression vector containing human TG2 cDNA encoding the TGase null mutation W241Q. Shown is an immunoblot of extracts from U118MG cells expressing wild type TG2 (Clone B5), empty vector (Vector), or U118MG cells expressing TG2 W241Q mutants (clone 4B1 and clone 4C4). Extracts were probed with anti-TG2 (top) or anti-tubulin (bottom). b Indicated cell lines were treated with 7.8 μg/ml doxorubicin and 72 h after drug addition cell viability was assayed by MTT. Graphed is the mean of 8 independent data points/cell line, error bars = ±1.0 SD
When cell viability was measured at a doxorubicin concentration close to the LD50 for the vector cells (7.8 μg/ml, 54.7% viability, ±3.27 SD), wildtype TG2 expressing B5 cells display increased viability (73.6%, ±1.54) which is significantly higher (P = 7.7 × 10−6) than that displayed by vector cells (Fig. 6b). Further, neither of the U118MG clones expressing the W241Q mutant display significant differences in doxorubicin sensitivity from that measured in B5 cells expressing wildtype TG2 at this dose. Conversely, both the 4C4 and 4B1 W241Q clones were statistically more resistant to doxorubicin than vector cells (P = 0.002 and 0.017, respectively) when treated with 7.8 μg/ml doxorubicin. Similar results were obtained when these cells were treated with CCNU (data not shown). In sum, these findings indicate that the transglutaminase activity of TG2 is not necessary to confer drug resistance in U118MG glioma cells.
Discussion
Morbidity and mortality from malignant glioma is directly correlated with the ability of tumor cells to infiltrate surrounding tissue as malignant cells are often found well beyond the gross radiographic margins of the tumor [21]. Acquisition of invasion behavior is likely an early event in disease progression [21, 39] and is believed to be a multistep process involving, in part, alterations in the interaction of the tumor cell with ECM, and a generalized loss of ECM integrity often through extracellular protease dysregulation [24, 31]. In accordance with its transglutaminase activity, extracellular pools of TG2 bind to and cross-link numerous components of the ECM, including fibronectin, fibrinogen/fibrin, von Willebrand factor, vitronectin, lipoprotein A, dermatane sulfate proteoglycans, collagen V, osteonectin, laminin, nidogen and osteopontin (for review see [5]). The cross linking of these components is viewed as making the ECM resistant to proteolytic or mechanical damage and in the context of cancer, it has been proposed that TG2 acts as a barrier to tumor spread [8, 9]. Supporting this notion is the finding that expression of recombinant TG2 in hamster fibrosarcoma cells increased adherence to a fibronectin-coated substratum and that subclones of the endothelial cell line ECV304 expressing reduced TG2 showed decreased cross-linking of fibronectin compared to controls [40, 41]. Additionally, Swiss 3T3 fibroblasts expressing TG2 showed decreased migration on fibronectin-coated surfaces [42, 43]. Haroon et al. [8] observed that the injection of purified TG2 into xenografts of the rat mammary tumor line R3230Ac caused a significant tumor growth delay. More recently, it was observed that pre-treatment of Matrigel with purified TG2 resulted in an inhibition of the migration of the breast cancer line MDA-MB-231 through this basement membrane extract [44].
While the findings outlined above are consistent with TG2 acting to suppress of tumor invasiveness, other studies drew opposite conclusions. For example, Mangala et al. [45] found that downregulation of TG2 expression in MDA-MB-231 cells using RNAi resulted in decreased tumor survival and invasion. Others showed that RNAi-induced knockdown or pharmacological inhibition of TG2 resulted in reduced motility of the ovarian tumor cell line SKOV3 in vitro, or tumor dissemination when implanted into the peritoneum of nude mice [46]. Later, Verma et al. [47] showed that downregulation of TG2 in pancreatic adenocarcinoma cells inhibited the metastatic spread of the tumor. More recently, workers observed that either pharmacological inhibition of TG2 or RNAi-induced knockdown of TG2 resulted in decreased EGF-induced stimulation of HeLa cell motility and invasion [48]. Taken together, these divergent findings may indicate that a potential role for TG2 expression as a determinant of tumor invasiveness may be tumor and tissue context-dependent and, moreover, determination of the potential phenotypic effects of TG2 silencing on glioma invasiveness requires further analysis.
Several labs have documented heighted activation of the oncogenic transcription factor NFkB in cells expressing high levels of TG2 [15, 16, 49]. The transcription factor NFkB was originally identified as a transcription factor induced during T cell activation [50] and was subsequently found to be an important modulator of immune response and inflammation [51]. Since this time, NFkB has been found to be a widely-expressed, multi-gene family of proteins that, when the mechanisms that normally restrict NFkB activity are dysregulated, function as a potently oncogenic factors in a variety of tumor types [52]. Acccordingly, it was unsurprising that NFkB was found to be dysregulated in brain tumors [53]. Using immunohistochemistry and Immunoblotting, Wang et al. [54] found a strong correlation between overexpression and activation (as judged by a phospho-specific antibody) of the NFkB subunit p65 (RelA) and tumor grade. Specifically, higher grade tumor types (i.e., gliobastoma multiforme (WHO grade IV), anaplastic oligodendroglioma (WHO grade III), and anaplastic astrocytoma (WHO grade III)) displayed significantly higher levels of p65 expression and activation than lower-grade (WHO grade II) gliomas (i.e., diffuse astrocytoma, oligodendroglioma and oligoastrocytoma). Of note, query of the Rembrandt database suggests that high TG2 expression is also associated with higher tumor grade. Whether a relationship exists between TG2 expression and p65 expression/activation in gliomas is yet to be resolved, these studies suggest that TG2 expression may be used diagnostically and/or prognostically in the clinical management of brain tumors.
Mehta et al. [10] initially observed that heightened levels of TG2 expression in MCF7 breast cancer cells led to resistance to doxorubicin. Specifically, resistance to anthracyclines (e.g., doxorubicin/adriamycin) has been linked to TG2 expression in a wide variety of cell types [11–14]. A role for TG2 in promoting a general drug resistance is not supported by the evidence since Han et al. [13] observed that TG2 expression in lung tumor cells induced resistance to doxorubicin and vincristine, but not cisplatin. In this study, we document that expression of recombinant human TG2 in the glioma line U118MG resulted in strong resistance to doxorubicin. We also observed that TG2 expression induced resistance to the bi-functional alkylating agent CCNU (e.g., Lomustine). This finding is in agreement with the work of Yuan et al. [11] that showed treatment of xenografted glioblastoma tumors with a pharmacological inhibitor of TG2 resulted in increased sensitivity to the related chemotherapeutic BCNU (Carmustine). These findings suggest that TG2 expression could be used as a tumor marker to predict CCNU or BCNU efficacy prior to treating glioma patients with these therapeutics.
Contrary to the observed TG2-associated resistance to doxorubicin and CCNU, we did not observe significant resistance in TG2-expressing U118MG cells treated with the glioma therapeutics vincristine, temozolomide, cisplatin, or cyclophosphamide when compared to cells transfected with empty vector. These findings indicate that TG2 expression does not induce a general drug resistance phenotype in this glioma line. Our observation that TG2 expression does not induce cis-platin resistance in U118MG cells is consistent with published accounts conducted on the human lung cancer line PC-14 [13], but not on other ovarian and lung cancer cell lines [55, 56]. These divergent findings suggest that TG2 exerts cell type-specific effects on cisplatin sensitivity.
We also observed that U118MG cells expressing a transglutaminase-null mutant of TG2 (W241Q) displayed resistance to doxorubicin. This result clearly indicates that transglutaminase activity is not required to induce chemotherapeutic resistance in this cell line. Others have similarly observed that another TGase mutant (i.e., C277 V) was capable of supporting protection from apoptosis induced by a synthetic retinoid analog N-(4-hydroxyphenyl) retinamide [57, 58]. Moreover, Antonyak et al. [57] found that mutants of TG2 that disrupt GTP binding lost their ability to support resistance to this drug, suggesting that molecular mechanisms other than protein cross-linking drive TG2-associated drug resistance. Clearly, more investigation is required to understand the mechanism underlying the resistance phenotype conferred by TG2 overexpression.
In conclusion, we documented that reduced expression of TG2 in cultured human glioma cells stems from aberrant DNA hypermethylation of the proximal promoter region of the TGM2 gene. Analysis of tumor and normal brain tissues and cells indicates that this hypermethylation also occurs in primary brain tumors and we conclude that hypermethylation of the TGM2 gene is a cancer-associated event. Given the role that TG2 plays in ECM integrity, we envision that the reduced TG2 expression resulting from epigenetic silencing may influence the invasive activity of brain tumors. We also document that TG2 is overexpressed in a subset of brain tumors and that is feature is associated with more aggressive forms of the disease and reduced patient survival. Since TG2 overexpression also drives resistance to some, but not all commonly used brain chemotherapeutics, our results suggest that TG2 expression may be a clinically useful biomarker to predict response to drug treatment.
Acknowledgments
The authors are grateful to Mr. Jonathan Green for technical expertise and the Florida Center for Brain Tumor Research (FCBTR) for providing access to clinical samples. Work was supported by a grant (RO1 CA102289) to KDB.
Contributor Information
Lisa M. Dyer, Department of Biochemistry and Molecular Biology, University of Florida College of Medicine, Box 100245, Gainesville, FL 32610-0245, USA. UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA
Kevin P. Schooler, Department of Pediatrics, University of Florida College of Medicine, Gainesville, FL, USA
Lingbao Ai, Department of Biochemistry and Molecular Biology, University of Florida College of Medicine, Box 100245, Gainesville, FL 32610-0245, USA. UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA.
Corinne Klop, UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA.
Jingxin Qiu, Department of Pathology, Immunology and Laboratory Medicine, University of Florida College of Medicine, Gainesville, FL, USA. UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA.
Keith D. Robertson, Department of Biochemistry and Molecular Biology, University of Florida College of Medicine, Box 100245, Gainesville, FL 32610-0245, USA. UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA
Kevin D. Brown, Email: kdbrown1@ufl.edu, Department of Biochemistry and Molecular Biology, University of Florida College of Medicine, Box 100245, Gainesville, FL 32610-0245, USA. UF-Shands Cancer Center Program in Genetics, Epigenetics and Tumor Virology, University of Florida College of Medicine, Gainesville, FL, USA
References
- 1.Weinberg RA. How cancer arises. Sci Am. 1996;275:62–70. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe AP. Chromatin remodeling: why it is important in cancer. Oncogene. 2001;20:2988–2990. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 4.Lorand L, Conrad SM. Transglutaminases. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- 5.Aeschlimann D, Thomazy V. Protein crosslinking in assembly and remodelling of extracellular matrices: the role of transglutaminases. Connect Tissue Res. 2000;41:1–27. doi: 10.3109/03008200009005638. [DOI] [PubMed] [Google Scholar]
- 6.Zemskov EA, Janiak A, Hang J, Waghray A, Belkin AM. The role of tissue transglutaminase in cell-matrix interactions. Front Biosci. 2006;11:1057–1076. doi: 10.2741/1863. [DOI] [PubMed] [Google Scholar]
- 7.Telci D, Griffin M. Tissue transglutaminase (TG2)—a wound response enzyme. Front Biosci. 2006;11:867–882. doi: 10.2741/1843. [DOI] [PubMed] [Google Scholar]
- 8.Haroon ZA, Lai TS, Hettasch JM, Lindberg RA, Dewhirst MW, Greenberg CS. Tissue transglutaminase is expressed as a host response to tumor invasion and inhibits tumor growth. Lab Invest. 1999;79:1679–1686. [PubMed] [Google Scholar]
- 9.Jones RA, Kotsakis P, Johnson TS, et al. Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ. 2006;13:1442–1453. doi: 10.1038/sj.cdd.4401816. [DOI] [PubMed] [Google Scholar]
- 10.Mehta K. High levels of transglutaminase expression in doxorubicin-resistant human breast carcinoma cells. Int J Cancer. 1994;58:400–406. doi: 10.1002/ijc.2910580316. [DOI] [PubMed] [Google Scholar]
- 11.Yuan L, Choi K, Khosla C, et al. Tissue transglutaminase 2 inhibition promotes cell death and chemosensitivity in glioblastomas. Mol Cancer Ther. 2005;4:1293–1302. doi: 10.1158/1535-7163.MCT-04-0328. [DOI] [PubMed] [Google Scholar]
- 12.Chen JS, Agarwal N, Mehta K. Multidrug-resistant MCF-7 breast cancer cells contain deficient intracellular calcium pools. Breast Cancer Res Treat. 2002;71:237–247. doi: 10.1023/a:1014461832403. [DOI] [PubMed] [Google Scholar]
- 13.Han JA, Park SC. Reduction of transglutaminase 2 expression is associated with an induction of drug sensitivity in the PC-14 human lung cancer cell line. J Cancer Res Clin Oncol. 1999;125:89–95. doi: 10.1007/s004320050247. [DOI] [PubMed] [Google Scholar]
- 14.Herman JF, Mangala LS, Mehta K. Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene. 2006;25:3049–3058. doi: 10.1038/sj.onc.1209324. [DOI] [PubMed] [Google Scholar]
- 15.Kim DS, Park SS, Nam BH, Kim IH, Kim SY. Reversal of drug resistance in breast cancer cells by transglutaminase 2 inhibition and nuclear factor-kappaB inactivation. Cancer Res. 2006;66:10936–10943. doi: 10.1158/0008-5472.CAN-06-1521. [DOI] [PubMed] [Google Scholar]
- 16.Mann AP, Verma A, Sethi G, et al. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-{kappa}B in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- 17.Kotsakis P, Griffin M. Tissue transglutaminase in tumour progression: friend or foe? Amino Acids. 2007;33:373–384. doi: 10.1007/s00726-007-0516-1. [DOI] [PubMed] [Google Scholar]
- 18.Chhabra A, Verma A, Mehta K. Tissue transglutaminase promotes or suppresses tumors depending on cell context. Anticancer Res. 2009;29:1909–1919. [PubMed] [Google Scholar]
- 19.Partap S, Fisher PG. Update on new treatments and developments in childhood brain tumors. Curr Opin Pediatr. 2007;19:670–674. doi: 10.1097/MOP.0b013e3282f0eafa. [DOI] [PubMed] [Google Scholar]
- 20.Nieder C, Mehta MP, Jalali R. Combined radio- and chemotherapy of brain tumours in adult patients. Clin Oncol (R Coll Radiol) 2009;21:515–524. doi: 10.1016/j.clon.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Nieder C, Adam M, Molls M, Grosu AL. Therapeutic options for recurrent high-grade glioma in adult patients: recent advances. Crit Rev Oncol Hematol. 2006;60:181–193. doi: 10.1016/j.critrevonc.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Hunter SB, Brat DJ, Olson JJ, Von Deimling A, Zhou W, Van Meir EG. Alterations in molecular pathways of diffusely infiltrating glial neoplasms: application to tumor classification and anti-tumor therapy (Review) Int J Oncol. 2003;23:857–869. [PubMed] [Google Scholar]
- 24.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 25.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 26.Jaworski DM, Kelly GM, Piepmeier JM, Hockfield S. BEHAB (brain enriched hyaluronan binding) is expressed in surgical samples of glioma and in intracranial grafts of invasive glioma cell lines. Cancer Res. 1996;56:2293–2298. [PubMed] [Google Scholar]
- 27.Koochekpour S, Pilkington GJ, Merzak A. Hyaluronic acid/CD44H interaction induces cell detachment and stimulates migration and invasion of human glioma cells in vitro. Int J Cancer. 1995;63:450–454. doi: 10.1002/ijc.2910630325. [DOI] [PubMed] [Google Scholar]
- 28.Koochekpour S, Merzak A, Pilkington GJ. Extracellular matrix proteins inhibit proliferation, upregulate migration and induce morphological changes in human glioma cell lines. Eur J Cancer. 1995;31A:375–380. doi: 10.1016/0959-8049(94)00476-l. [DOI] [PubMed] [Google Scholar]
- 29.Paulus W, Baur I, Dours-Zimmermann MT, Zimmermann DR. Differential expression of versican isoforms in brain tumors. J Neuropathol Exp Neurol. 1996;55:528–533. doi: 10.1097/00005072-199605000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulus W, Tonn JC. Interactions of glioma cells and extracellular matrix. J Neurooncol. 1995;24:87–91. doi: 10.1007/BF01052664. [DOI] [PubMed] [Google Scholar]
- 32.Ai L, Kim WJ, Demircan B, et al. The transglutaminase 2 gene (TGM2), a potential molecular marker for chemotherapeutic drug sensitivity, is epigenetically silenced in breast cancer. Carcinogenesis. 2008;29:510–518. doi: 10.1093/carcin/bgm280. [DOI] [PubMed] [Google Scholar]
- 33.Qiu J, Ai L, Ramachandran C, et al. Invasion suppressor cystatin E/M (CST6): high-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab Invest. 2008;88:910–925. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demircan B, Dyer LM, Gerace M, Lobenhofer EK, Robertson KD, Brown KD. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer. 2009;48:83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalmers-Redman RM, Priestley T, Kemp JA, Fine A. In vitro propagation and inducible differentiation of multipotential progenitor cells from human fetal brain. Neuroscience. 1997;76:1121–1128. doi: 10.1016/s0306-4522(96)00386-7. [DOI] [PubMed] [Google Scholar]
- 36.Yuan L, Siegel M, Choi K, et al. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene. 2006;26:2563–2573. doi: 10.1038/sj.onc.1210048. [DOI] [PubMed] [Google Scholar]
- 37.Verma A, Mehta K. Tissue transglutaminase-mediated chemoresistance in cancer cells. Drug Resist Updat. 2007;10:144–151. doi: 10.1016/j.drup.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Murthy SN, Iismaa S, Begg G, Freymann DM, Graham RM, Lorand L. Conserved tryptophan in the core domain of transglutaminase is essential for catalytic activity. Proc Natl Acad Sci USA. 2002;99:2738–2742. doi: 10.1073/pnas.052715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levicar N, Strojnik T, Kos J, Dewey RA, Pilkington GJ, Lah TT. Lysosomal enzymes, cathepsins in brain tumour invasion. J Neurooncol. 2002;58:21–32. doi: 10.1023/a:1015892911420. [DOI] [PubMed] [Google Scholar]
- 40.Johnson TS, Knight CR, el-Alaoui S, et al. Transfection of tissue transglutaminase into a highly malignant hamster fibrosarcoma leads to a reduced incidence of primary tumour growth. Oncogene. 1994;9:2935–2942. [PubMed] [Google Scholar]
- 41.Jones RA, Nicholas B, Mian S, Davies PJ, Griffin M. Reduced expression of tissue transglutaminase in a human endothelial cell line leads to changes in cell spreading, cell adhesion and reduced polymerisation of fibronectin. J Cell Sci. 1997;110(Pt 19):2461–2472. doi: 10.1242/jcs.110.19.2461. [DOI] [PubMed] [Google Scholar]
- 42.Verderio E, Nicholas B, Gross S, Griffin M. Regulated expression of tissue transglutaminase in Swiss 3T3 fibroblasts: effects on the processing of fibronectin, cell attachment, and cell death. Exp Cell Res. 1998;239:119–138. doi: 10.1006/excr.1997.3874. [DOI] [PubMed] [Google Scholar]
- 43.Balklava Z, Verderio E, Collighan R, Gross S, Adams J, Griffin M. Analysis of tissue transglutaminase function in the migration of Swiss 3T3 fibroblasts: the active-state conformation of the enzyme does not affect cell motility but is important for its secretion. J Biol Chem. 2002;277:16567–16575. doi: 10.1074/jbc.M109836200. [DOI] [PubMed] [Google Scholar]
- 44.Mangala LS, Arun B, Sahin AA, Mehta K. Tissue transglutaminase-induced alterations in extracellular matrix inhibit tumor invasion. Mol Cancer. 2005;4:33. doi: 10.1186/1476-4598-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangala LS, Fok JY, Zorrilla-Calancha IR, Verma A, Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2007;26:2459–2470. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 46.Satpathy M, Cao L, Pincheira R, et al. Enhanced peritoneal ovarian tumor dissemination by tissue transglutaminase. Cancer Res. 2007;67:7194–7202. doi: 10.1158/0008-5472.CAN-07-0307. [DOI] [PubMed] [Google Scholar]
- 47.Verma A, Guha S, Diagaradjane P, et al. Therapeutic significance of elevated tissue transglutaminase expression in pancreatic cancer. Clin Cancer Res. 2008;14:2476–2483. doi: 10.1158/1078-0432.CCR-07-4529. [DOI] [PubMed] [Google Scholar]
- 48.Antonyak MA, Li B, Regan AD, Feng Q, Dusaban SS, Cerione RA. Tissue transglutaminase is an essential participant in the epidermal growth factor-stimulated signaling pathway leading to cancer cell migration and invasion. J Biol Chem. 2009;284:17914–17925. doi: 10.1074/jbc.M109.013037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Kim YS, Choi DH, et al. Transglutaminase 2 induces nuclear factor-kappaB activation via a novel pathway in BV-2 microglia. J Biol Chem. 2004;279:53725–53735. doi: 10.1074/jbc.M407627200. [DOI] [PubMed] [Google Scholar]
- 50.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 51.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 52.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 53.Nagai S, Washiyama K, Kurimoto M, Takaku A, Endo S, Kumanishi T. Aberrant nuclear factor-kappaB activity and its participation in the growth of human malignant astrocytoma. J Neurosurg. 2002;96:909–917. doi: 10.3171/jns.2002.96.5.0909. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–951. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 55.Cao L, Petrusca DN, Satpathy M, Nakshatri H, Petrache I, Matei D. Tissue transglutaminase protects epithelial ovarian cancer cells from cisplatin-induced apoptosis by promoting cell survival signaling. Carcinogenesis. 2008;29:1893–1900. doi: 10.1093/carcin/bgn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park KS, Kim HK, Lee JH, et al. Transglutaminase 2 as a cisplatin resistance marker in non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:493–502. doi: 10.1007/s00432-009-0681-6. [DOI] [PubMed] [Google Scholar]
- 57.Antonyak MA, Singh US, Lee DA, et al. Effects of tissue transglutaminase on retinoic acid-induced cellular differentiation and protection against apoptosis. J Biol Chem. 2001;276:33582–33587. doi: 10.1074/jbc.M105318200. [DOI] [PubMed] [Google Scholar]
- 58.Boehm JE, Singh U, Combs C, Antonyak MA, Cerione RA. Tissue transglutaminase protects against apoptosis by modifying the tumor suppressor protein p110 Rb. J Biol Chem. 2002;277:20127–20130. doi: 10.1074/jbc.C200147200. [DOI] [PubMed] [Google Scholar]