Abstract
The BMP signaling plays a pivotal role in the development of craniofacial organs, including the tooth and palate. BmprIa and BmprIb encode two type I BMP receptors that are primarily responsible for BMP signaling transduction. We investigated mesenchymal tissue-specific requirement of BmprIa and its functional redundancy with BmprIb during the development of mouse tooth and palate. BmprIa and BmprIb exhibit partially overlapping and distinct expression patterns in the developing tooth and palatal shelf. Neural crest specific inactivation of BmprIa leads to formation of an unusual type of anterior clefting of the secondary palate, an arrest of tooth development at the bud/early cap stages, and severe hypoplasia of the mandible. Defective tooth and palate development is accompanied by the down-regulation of BMP responsive genes and reduced cell proliferation levels in the palatal and dental mesenchyme. To determine if BmprIb could substitute for BmprIa during tooth and palate development, we expressed a constitutively active form of BmprIb (caBmprIb) in the neural crest cells in which BmprIa was simultaneously inactivated. We found that substitution of BmprIa by caBmprIb in neural rest cells rescues the development of molars and maxillary incisor, but the rescued teeth exhibit a delayed odontoblast and ameloblast differentiation. In contrast, caBmprIb fails to rescue the palatal and mandibular defects including the lack of lower incisors. Our results demonstrate an essential role for BmprIa in the mesenchymal component and a limited functional redundancy between BmprIa and BmprIb in a tissue specific manner during tooth and palate development.
Keywords: BMP signaling, BmprIA, BmprIB, tooth development, palatogenesis
Introduction
The family of bone morphogenetic proteins (BMPs) comprises over 20 multi-functional cytokines that belong to the TFG-β superfamily. BMPs play many important roles in embryonic development, postnatal growth, and regeneration. BMP signaling is transduced into cell via heteromeric receptor complexes of type I and type II transmembrane serine-threonine kinase receptors. Binding of BMP ligands to a heteromeric receptor complex induces phosphorylation of the type I receptor in the GS domain by the type II receptor. The activated type I receptor further phosphorylates in the cytoplasm the receptor-regulated Smads, primarily Smad-1, −5, and −8, which bind to common Smad (Smad4) and enter the nucleus where the Smad complex interacts with other transcription factors to regulate gene expression (Sieber et al., 2009). Besides this canonical BMP signaling pathway, BMPs can also activate Smad-independent mitogen-activated protein kinase (MAPK) signaling pathways. In addition to the two originally identified type I BMP receptors (BMPR-IA and BMPR-IB), Activin receptor type IA (ActRIa or Alk2) also binds to BMP ligands and transduces BMP signaling (Kawabata et al., 1998; Nohe et al., 2004). While mice deficient for BmprIb are viable with appendicular skeleton defects (Baur et al., 2000; Yi et al., 2000), mutations in either BmprIa or Alk2 lead to embryonic lethality during early gestation stage (Mishina et al., 1995; 1999; Gu et al., 1999), suggesting distinct and potentially redundant roles between these receptors during embryonic development.
The development of mammalian tooth and palate is governed by interactions between pharyngeal ectoderm and cranial neural crest-derived mesenchyme. Among many regulators, BMP signaling plays a pivotal role in mediating the epithelial-mesenchymal interaction during the development of these craniofacial organs (Nie et al., 2006). During palatogenesis, several Bmp genes, including Bmp2, Bmp3, Bmp4, Bmp5, and Bmp7, exhibit dynamic and differential expression patterns along the anterior-posterior (A-P) axis of the developing palatal shelves (Lu et al., 2000; Zhang et al., 2002; Hilliard et al., 2005; Nie, 2005; Levi et al., 2006). In the anterior portion of developing palatal shelves, Bmp4, Msx1, Shh, and Bmp2 form a genetic hierarchy to regulate cell proliferation (Zhang et al., 2002); and BMP signaling is also required for the expression of Shox2 whose inactivation causes formation of a rare type of anterior clefting of the secondary palate in mice (Yu et al., 2005; Gu et al., 2008). In the posterior palate, a balanced BMP activity is essential for the maintenance of palatal epithelial integrity (Xiong et al., 2009; He et al., 2010). Numerous studies have implicated BMP signaling in many aspects of tooth development, from determination of tooth forming sites and tooth types (Neubüser et al., 1997; Tucker et al., 1998), progression from the bud stage to the cap stage and formation of the enamel knot (Chen et al., 1996; Jernvall et al., 1998; Zhang et al., 2000; Zhao et al., 2000), to tooth root formation and tooth eruption (Yamashiro et al., 2003; Hosoya et al., 2008, Huang et al., 2010, Yao et al., 2010). Among those Bmp genes that are expressed in developing tooth, Bmp4 was suggested to play a central role as a morphogen during early tooth morphogenesis (Vainio et al., 1993; Thesleff and Mikkola, 2002).
The crucial role of BMP signaling in tooth and palate development was further revealed by studies that used mice carrying conditionally inactivated type I BMP receptors. In contrast to the lack of any visible craniofacial defect in BmprIb null mice (Baur et al., 2000; Yi et al., 2000), tissue-specific inactivation of BmprIa in the palatal and dental epithelium results in a cleft palate formation and an arrest of molar development at the bud/cap stages, and causes various incisor phenotypes depending on different Cre transgenic mouse lines that were used (Andl et al., 2004; Liu et al., 2005). Furthermore, tissue specific deletion of Alk2 in the neural crest lineage leads to multiple craniofacial defects including cleft palate and hypoplastic mandible; however, a tooth phenotype was not reported in this study (Dudas et al., 2004). Despite that these receptors are highly homologous and can activate both Smad and Smad-independent pathways, and that they may function redundantly to certain extent, each of them mediates specific and non-redundant signaling during embryogenesis (Sieber et al., 2009).
Despite the essential role for BmprIa in the epithelial component for tooth and palate development, the requirement of BmprIa in the mesenchymal component remains unknown. This is likely due to the unavailability of a dental and palatal mesnechyme specific Cre deletor mouse line and the fact that mice bearing deletion of BmprIa in the neural crest cell by Wnt1-Cre die around E12.5 when tooth and palate development just starts (Stottmann et al., 2004). It was recently demonstrated that embryonic lethality in mice lacking BmprIa in the neural crest lineage is due to norepinephrine depletion instead of cardiac defects (Morikawa et al., 2009). Administration of the β-adrenergic receptor agonist isoproterenol prevents embryonic lethality, and Wnt1Cre;BmprIaF/− embryos survive to term, making it possible to examine the role of BmprIa throughout development of neural crest-derived tissues. Taking this advantage, we investigated the role of BmprIa in the mesenchymal tissue and further tested if BMPR-IB mediated signaling is able to substitute for the loss of BmprIa in tooth and palate development.
Materials and methods
Animals and embryo collection
The generation and genotyping of transgenic and gene-targeted animals, including Wnt1Cre, BmprIa+/−, BmprIaF/F, have been described previously (Mishina et al., 1995; Danielian et al., 1998). The pMes-caBmprIb conditional transgenic line contains a constitutively active form (with Gln203 to Asp change) of BMPR-IB (named caBmprIb), which is linked to the 5’ end of the IRES-Egfp sequence and to the 3’ end of the LoxP flanked STOP cassette, under the control of the chick β-actin promoter, as described previously (He et al., 2010). Embryos containing inactivated BmprIa in their neural crest cells (Wnt1Cre;BmprIaF/−) were obtained by crossing Wnt1Cre;BmprIa+/− mice with BmprIaF/F line. To obtain embryos carrying Wnt1Cre;BmprIaF/−alleles and a pMes-caBmprIb transgenic allele, Wnt1Cre;BmprIa+/− mice were crossed with BmprIaF/+;pMes-caBmprIb mice. Mice containing such compounded alleles are referred as Wnt1Cre;BmprIaF/−;caIb.
Embryos with BmprIa deficiency in their neural crest cells (Wnt1Cre;BmprIaF/−) die at E12.5 (Stottmann et al., 2004), due to norepinephrine depletion (Morikawa et al., 2009). Administration of the β-adrenergic receptor agonist isoproterenol prevents embryonic lethality, allowing Wnt1Cre;BmprIaF/− embryos to survive to term (Morikawa et al., 2009). This was done by supplementing the drinking water of dams with 200µg/ml isoproterenol and 2.5 mg/ml ascorbic acid from 7.5 post-coitum (dpc), as described previously (Morikawa and Cserjesi, 2008). To be consistent, all embryos used throughout this study, including the wild type controls, Wnt1Cre;BmprIaF/−, and Wnt1Cre;BmprIaF/−;caIb, were obtained from dams fed with isoproterenol and ascorbic acid.
Embryos were collected from timed-mate pregnant females in ice-cold PBS. Embryonic head samples were dissected and fixed individually in 4% paraformaldehyde (PFA) overnight at 4°C, and processed for paraffin section for histological and in situ hybridization analyses or for frozen section for immunostaining. A tail sample form each embryo was used for PCR-based genotyping (primers information available upon request).
Mouse kidney capsule grafting
For subrenal culture in mice, E13.5 embryos were harvested from mating of Wnt1Cre;BmprIa+/− mice with BmprIaF/+;pMes-caBmprIb mice, and placed in PBS on ice. Embryos carrying Wnt1Cre;BmprIaF/− or Wnt1Cre;BmprIaF/-;caIb compounded alleles exhibited craniofacial abnormalities, and could be easily distinguished from embryos with other genotypes. Wnt1Cre;BmprIaF/-;caIb embryo could be further distinguished from Wnt1Cre;BmprIaF/− mice by the expression of Egfp in craniofacial region. Tail samples from targeted embryos were subjected to genotyping. E13.5 embryos collected from crosses of wild type mice were used as positive control. Mandibular molar germs were isolated from Wnt1Cre;BmprIaF/-;caIb embryos and wild type controls, and were subjected to subrenal culture. Adult CD-1 male mice were used as hosts for subrenal culture. Mice were anesthetized by intraperitoneal injection of Newbutal sodium solution at a dose of 0.01 mg/g of body weight. Kidney capsule grafting was performed following the procedure described in details previously (Zhang et al., 2003). Samples were harvested 2 weeks after subrenal culture.
Histology, in situ hybridization, and immunostaining
For histological and in situ hybridization analyses, paraffin sections were made at 10-µm and subjected for standard Hematoxylin/Eosin staining and non-radioactive in situ hybridization, as described previously (St. Amand et al., 2000). Frozen sections, made at 10-µm, were applied for immunohistochemical staining, as described previously (Xiong et al., 2009). Polyclonal antibodies against p-Smad1/5/8 were purchased from Cell Signaling (cat. #: 9511) and used at the concentration of 1:200. Green fluorescent-conjugated secondary antibodies were obtained from Invitrogen.
Cell proliferation and TUNEL assays
BrdU labeling was performed to determine cell proliferation rate and TUNEL assay was applied to detect apoptotic cells, as described previously (Zhang et al., 2002; Alappat et al., 2005). These were done by using BrdU Labeling and Detection Kit and In Situ Cell Death Detection Kit, both from Roche Diagnostics Corporation. Cell proliferation rates were measured by counting BrdU-positive cells and total cells in defined arbitrary areas in the palatal and dental mesenchyme, respectively. The outcome was presented as percentage of labeled cells among total cells in the defined arbitrary areas. Three continuous sections from each of three individual samples of wild type and mutants were counted, respectively. The sums from both genotypes were subjected to Student’s t-test to determine the significance of difference. Three independent BrdU labeling experiment with 6 samples of each genotype and four independent TUNEL assay with 4 samples of each genotype were performed.
Results
Expression of BmprIa and BmprIb in the developing tooth and palate
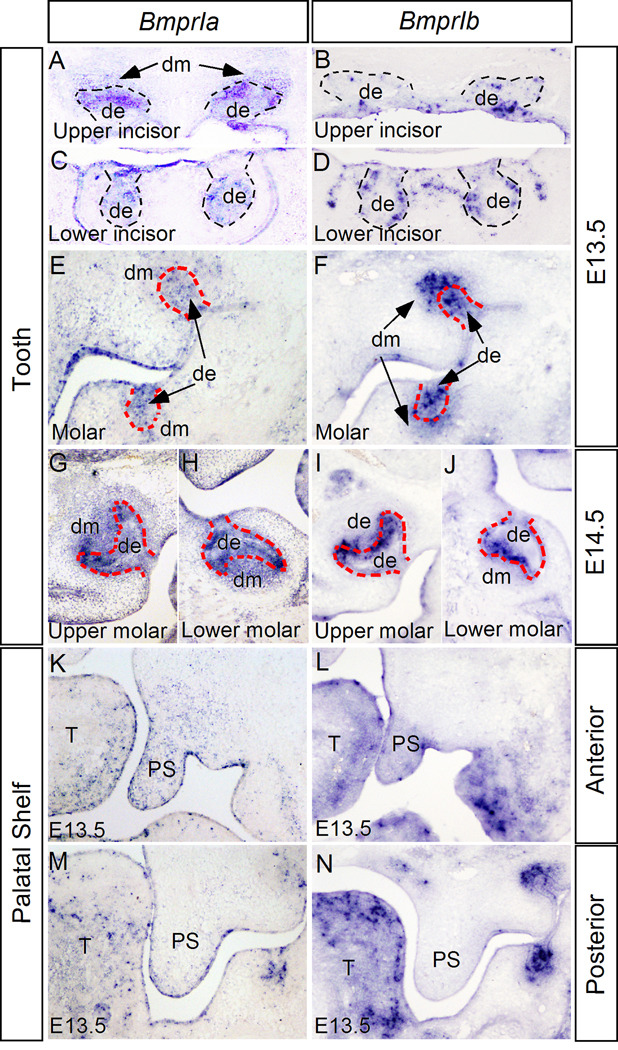
Numerous previous studies have implicated BMP signaling in tooth and palate development. However, detailed expression patterns of BmprIa and BmprIb during tooth and palate development have not been documented. We therefore began with examination of BmprIa expression in the developing tooth at several critical developmental stages, including the bud, the cap, and the bell stages, and in the developing secondary palatal shelves. BmprIb expression was also examined in parallel. In the developing tooth at the E13.5 bud stage, both BmprIa and BmprIb exhibit overlapped but distinct expression patterns (Fig. 1A-F). In the upper (maxillary) incisors, BmprIa is expressed in both dental epithelium and mesenchyme (Fig. 1A), but in the lower (mandibular) incisors, BmprIa expression is restricted in the dental epithelium (Fig. 1C). BmprIb is only expressed in the dental epithelium of both upper and lower incisor germs at this stage (Fig. 1B, D). In the molars, a relatively lower level of BmprIa expression was observed in the dental epithelium and mesenchyme, with a scattered pattern in the mesenchyme (Fig. 1E). BmprIb is also expressed in the epithelium and mesenchyme, with a much stronger level in the upper molar mesenchyme as compared to its expression in the lower molar (Fig. 1F). At the E14.5 cap stage, BmprIa remains its expression in both epithelial and mesenchymal compartments of upper incisors and molars, and in the epithelium of lower incisor (Fig. 1G, H, and Supplemental Figure 1). In contrast, BmprIb is restrictedly expressed in the dental epithelium of all types of tooth germ, with a high level in the future inner enamel organ of molars (Fig. 1I, J, and Supplemental Figure 1). At the E16.5 bell stage, BmprIa expression is mainly restricted in the epithelial component of upper and lower incisors, but an above background expression was also observed in the dental papilla of upper incisors. We also detected BmprIa expression in the inner enamel epithelium as well as odontoblasts in molars (Supplemental Figure 1). At this stage, BmprIb expression becomes weaker and is completely restricted in the dental epithelium of incisors and molars (Supplemental Figure 1).
Figure 1.
Expression of BmprIa and BmprIb in developing tooth and palate. (A, E) At E13.5, BmprIa expression is detected in the epithelium and mesenchyme of upper incisor (A), but is restricted in the epithelium in lower incisor (E). (B, D) BmprIb is expressed in the epithelium of both upper (B) and lower (D) incisor at the same stage. (E, F) In the developing molars at E13.5, BmprIa (E) and BmprIb (F) is expressed in the epithelium and mesenchyme. Note that BmprIb exhibits a higher level of expression in the upper molar mesenchyme as compared to its expression in the lower molar (F). (G-J) At E14.5, BmprIa is continuously expressed in both the epithelial and mesenchymal compartments of upper (G) and lower (H) molars, but BmprIb expression is restricted to molar epithelium, particularly in the inner enamel epithelium (I, J). (K-N) Expression of BmprIa (K, M) and BmprIb (L, N) in E13.5 palatal shelf. In the anterior palate, both BmprIa (K) and BmprIb (L) are expressed in the palatal epithelium and mesenchyme. In the posterior palate, BmprIa expression is observed in the epithelium and mesenchyme at relatively lower levels (M); however, no BmprIb expression is detected (N). Dashed lines demarcate dental epithelium. T, tongue; de, dental epithelium; dm, dental mesenchyme; PS, palatal shelf.
In the developing palatal shelves at E12.5 and E13.5, BmprIa is expressed in the epithelium and mesenchyme of the anterior palate, while in the posterior palate, BmprIa expression is mainly restricted in the palatal epithelium, with an above background level in the mesenchyme (Fig. 1K, M, and Supplemental Figure 1). At the same stages, BmprIb is only expressed in the anterior portion of the palatal shelf in both the epithelium and mesenchyme, but its expression is completely absent in the posterior palatal mesenchyme (Fig. 1L, N, and Supplemental Figure 1).
Neural crest inactivation of BmprIa causes formation of an unusual type of cleft palate
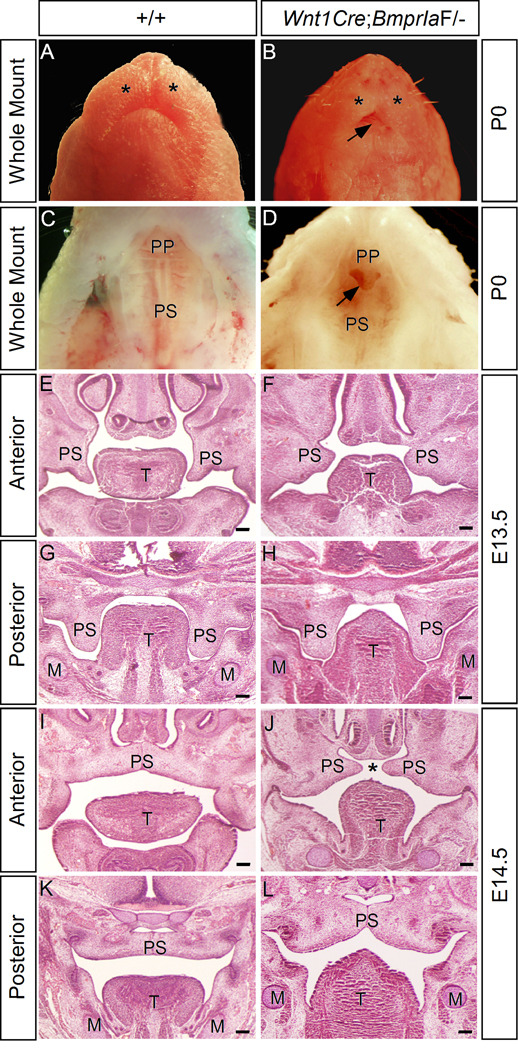
To generate embryos that were deficient for BmprIa in the neural crest cells, we crossed Wnt1Cre;BmprIa+/− mice to BmprIaF/F mice. In order to obtain Wnt1Cre;BmprIaF/− embryos at term, dams were fed with isoproterenol and ascorbic acid in drinking water to prevent embryonic lethality at mid-gestation stage (Morikawa and Cserjesi, 2008; Morikawa et al., 2009). Wild type mice or mice with other genotypes except Wnt1Cre;BmprIaF/− mice were indistinguishable from with type mice from dams fed with normal drinking water. Gross examination of Wnt1Cre;BmprIaF/− mice identified dramatic craniofacial defects, including an extremely shortened mandible, hypoplastic maxillary prominence, and an unusual type of anterior clefting of the secondary palate (Fig. 2A-D). The shortened mandible with hypoplastic distal region exposed the tongue to external view (Fig. 2B). Histological analysis of E13.5 mutant embryo revealed shortened but horizontally positioned palatal shelves in the anterior region (Fig. 1F). However, the posterior palatal shelves in the mutant assumed a vertical position as the wild type control and appeared similar to the control in size and shape (Fig. 2G, H). At E14.5 when the palatal shelves in wild type control have elevated to the position above the tongue and met at the midline, the palatal shelves in the mutant were also positioned above the tongue (Fig. 2I-L). While the posterior palatal shelves in mutants made contact at the midline, the anterior palatal shelves appeared too short to make contact. This unique type of cleft palate defect was also observed in Shox2 mutant mice (Yu et al., 2005; Gu et al., 2008). In addition, a deformed tongue was also seen in mutants. Thus deletion of BmprIa in the neural crest-derived palatal mesenchyme causes a defective growth in the anterior palatal shelves, and ultimately leads to the formation of anterior cleft of the secondary palate, consistent with a restricted expression of BmprIa in the anterior palatal mesenchyme. BmprIa is required not only in the epithelium (Andl et al., 2004; Liu et al., 2005), but also in the mesenchyme for normal palate development.
Figure 2.
Wnt1Cre;BmprIaF/− mice exhibit a unique anterior clefting of the secondary palate. (A-D) Gross examination of wild type (A, C) and Wnt1Cre;BmprIaF/− (B, D) mice at postnatal day 0 (P0) reveals craniofacial defects, including extremely shortened mandible and hypoplastic maxillary prominence (asterisks) (B), and an unusual type of anterior clefting of the secondary palate (arrow) (D). (E-H) coronal sections of E13.5 wild type control (E, G) and Wnt1Cre;BmprIaF/− (F, H) embryos reveal shortened and horizontally positioned palatal shelves in the anterior region of mutant (F). The posterior palatal shelves appear morphologically comparable in wild type control and mutant ( G, H). (I-L) At E14.5 when the palatal shelves meet and fuse at the midline at the anterior (I) and posterior (K) domains in wild type control, the mutant palatal shelves appear too short to meet in the anterior region (J), but do meet and fuse in the posterior region (L). M, Meckel’s cartilage; T, tongue; PP, primary palate; PS, palatal shelf. Asterisks in (A) and (B) mark maxillary prominence. Asterisk in (J) marks palate clefting. Arrow in (B) points to exposed tongue, and in (D) points to the anterior clefting of the secondary palate. Scale bars represent 100 µm.
Deletion of BmprIa in neural crest-derived mesenchyme arrests tooth development
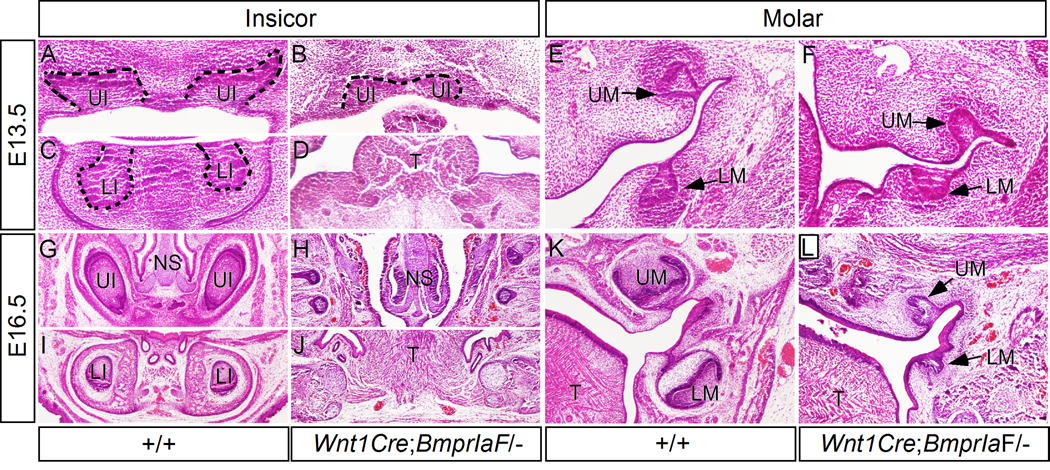
Histological examination failed to identify any definite tooth structure in Wnt1Cre;BmprIa newborn mice (data not shown), suggesting a defective tooth development at early phase. Analysis of mutant embryos at E13.5 revealed formation of molars at the bud stage (Fig. 3F), However, as compared to the wild type controls (Fig. 3E), the molar buds in the mutant appeared a little bit delay in development with less condensed dental mesenchymal cells surrounding the epithelial bud. The incisor phenotype in mutants appeared much dramatic (Fig. 3A-D). The upper incisors formed a single, medially located tooth bud that was arrested at the early bud stage (Fig. 3B), while the lower incisor buds never formed (Fig. 3D), likely due to a substantially shortened mandible (Fig. 2B). At the E16.5 bell stage, in mutants, we could not find any residual structure of upper incisors (Fig. 3H), but observe residual molar germs (Fig. 3L). Among 8 samples examined, all the lower molars were arrested at the bud stage, but 6 residual upper molars appeared to be arrested at the early cap stage (Fig. 3L). Taking together, these observations indicate an absolute requirement of BmprIa in the mesenchyme for tooth development beyond the bud stage or the early cap stage.
Figure 3.
Wnt1Cre;BmprIaF/− mice show defective tooth development. (A, C) coronal sections of an E13.5 wild type embryo show upper incisors (A) and lower incisors (C) at the bud stage. (B, D) Coronal sections of an E13.5 Wnt1Cre;BmprIaF/− embryo show a fused upper incisor at the early bud stage (B) and lack of lower incisors (D). (E, F) Coronal sections of an E13.5 wild type embryo (E) and an E13.5 mutant show molar teeth at the bud stage. Note that the mutant molars appear slightly delay in development and have less condensed dental mesenchyme (F). (G, I) Sections show upper incisors (G) and lower incisors (I) in an E16.5 wild type embryo. (H, J) Sections from an E16.5 mutant embryo show complete lack of upper incisor (H) and lower incisor (J). (K) Coronal section from an E16.5 wild type embryo shows molar teeth at the bell stage. (L) Coronal section from an E16.5 mutant shows residual molar tooth structure. Note that the upper molar is arrested at the early cap stage, while the lower molar is arrested at the bud stage. Dashed lines demarcate dental epithelium. T, tongue; LI, lower incisor; LM, lower molar; NS, nasal septum; UI, upper incisor; UM, upper molar.
Reduced cell proliferation in the palatal and tooth mesenchyme of Wnt1Cre;BmprIaF/− embryos
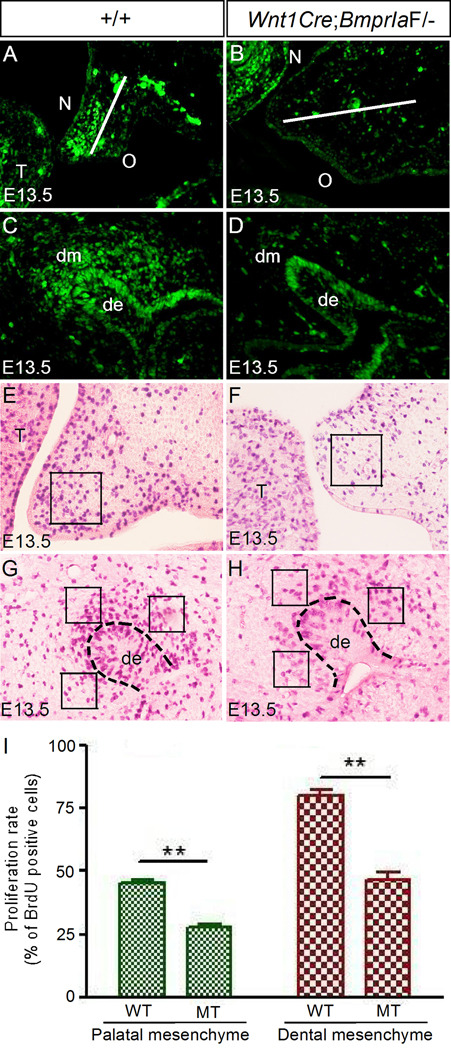
To ensure that deletion of BmprIa in the neural crest cells disrupts BMP signaling in the palatal and dental mesenchyme, we examined the expression of phosphorylated Smad1/5/8 (pSmad1/5/8) by immunohistochemical staining. In wild type controls at E13.5, we detected abundant pSmad1/5/8 positive cells in the anterior palatal mesenchyme primarily on the future nasal side, and in the condensed dental mesenchyme as well as the dental epithelium (Fig. 4A, C). In contrast, as we expected, the number of pSmad1/5/8 positive cells was significantly reduced in the palatal and dental mesenchyme of Wnt1Cre;BmprIaF/− mutants, although the staining in the epithelial compartment remained unchanged (Fig. 4B, D).
Figure 4.
Reduced levels of BMP/Smad signaling activity and cell proliferation in the palatal mesenchyme and dental mesenchyme of Wnt1Cre;BmprIaF/− embryo. (A, C) Immunostaining shows pSmad1/5/8 signals in the palatal shelf (A) and an upper molar (C) of an E13.5 wild type embryo. The white line in (A) bisects the palatal shelf into nasal and oral halves. Note that pSmad1/5/8 signals are mainly detected in the nasal half of the palatal shelf. (B, D) Immunostaining shows significantly reduced pSmad1/5/8 signals in the palatal mesenchyme (B) and dental mesenchyme (D) of an E13.5 mutant. (E-H) BrdU labeling shows reduced cell proliferation rates in the palatal mesenchyme (F) of anterior palate and molar mesenchyme (H) of E13.5 mutant embryos as compared to their wild type counterparts (E, G). Squares indicate the areas where cells were counted. Dashed lines in (G, H) demarcate dental epithelium. (I) Comparison of BrdU-labeled cells in designated areas of palate and molar in controls and mutants. Standard deviation values were shown as error bars, and ** indicate P < 0.001. N designates nasal side of the palatal shelf, and O designates oral side of the palatal shelf. de, dental epithelium; dm, dental mesenchyme; MT, mutant; WT, wild-type.
To investigate cellular defects that may contribute to a cleft palate formation and to the arrest of tooth development in Wnt1Cre;BmprIaF/− mutants, we carried out BrdU labeling and TUNEL assays to examine cell proliferation rate and apoptosis. In the developing palatal shelves of mutants at E13.5, we found a significantly reduced level of cell proliferation in the mesenchyme of the anterior palate, as compared to that in the controls (Fig. 4E, F, I). However, cell proliferation rates in the palatal epithelium and the posterior palatal mesenchyme remained unchanged (data not shown). Similarly, in the developing molar of mutants at this stage, a dramatic reduction in cell proliferation rate was also found in the dental mesenchyme, but not in the dental epithelium (Fig. 4G, H, I; and data not shown). The reduction in cell proliferation rate correlates with a reduced level of pSmad1/5/8 in mutants, indicating a positive role for BMP/Smad signaling in the regulation of cell proliferation. On the other hand, TUNEL assays did not reveal enhanced/ectopic cell apoptosis in the tooth germs and palatal shelves of mutants at this stage (data not shown). Thus this reduced cell proliferation rate in the mesenchymal compartment represents one defective cellular mechanism contributing to a cleft palate formation and an arrest of tooth development in Wnt1Cre;BmprIaF/− mutants.
Altered gene expression in the developing palate and tooth of Wnt1Cre;BmprIaF/− mice
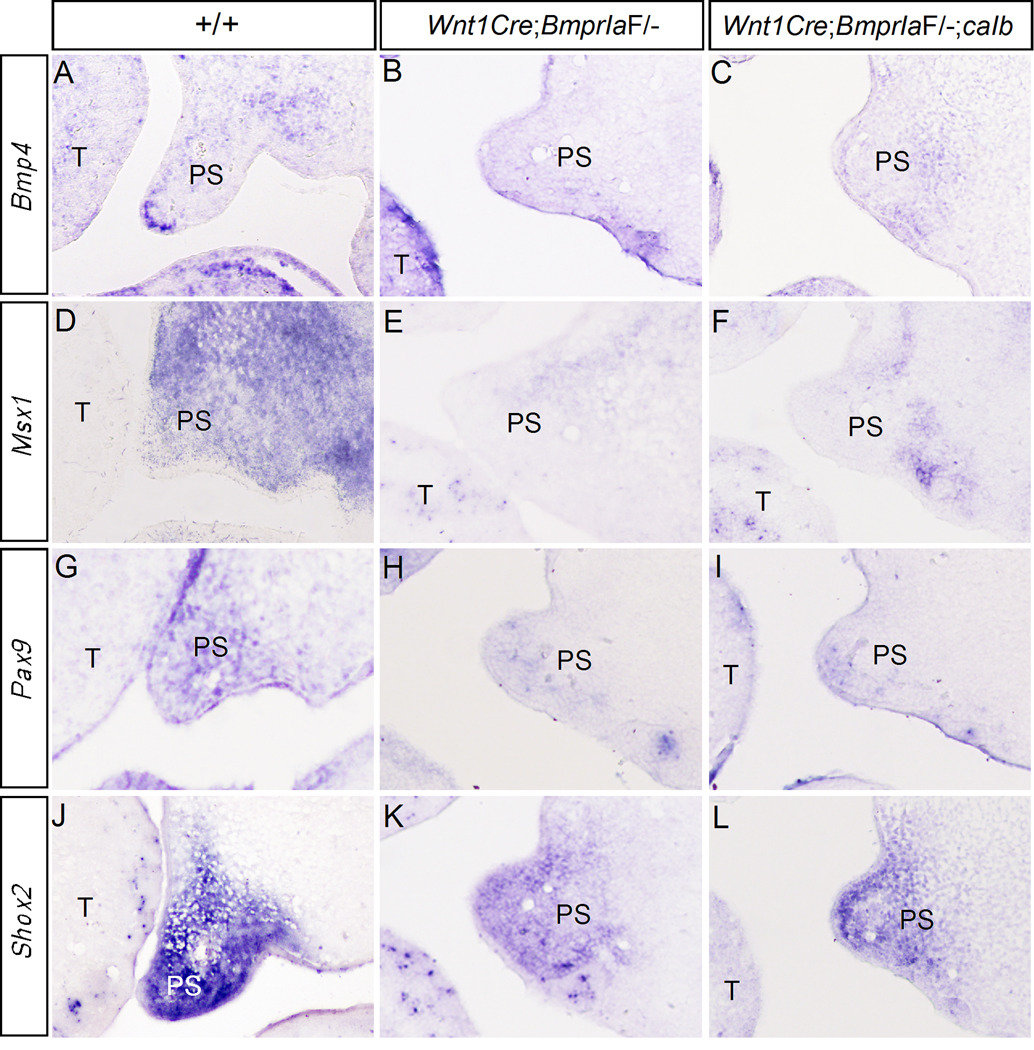
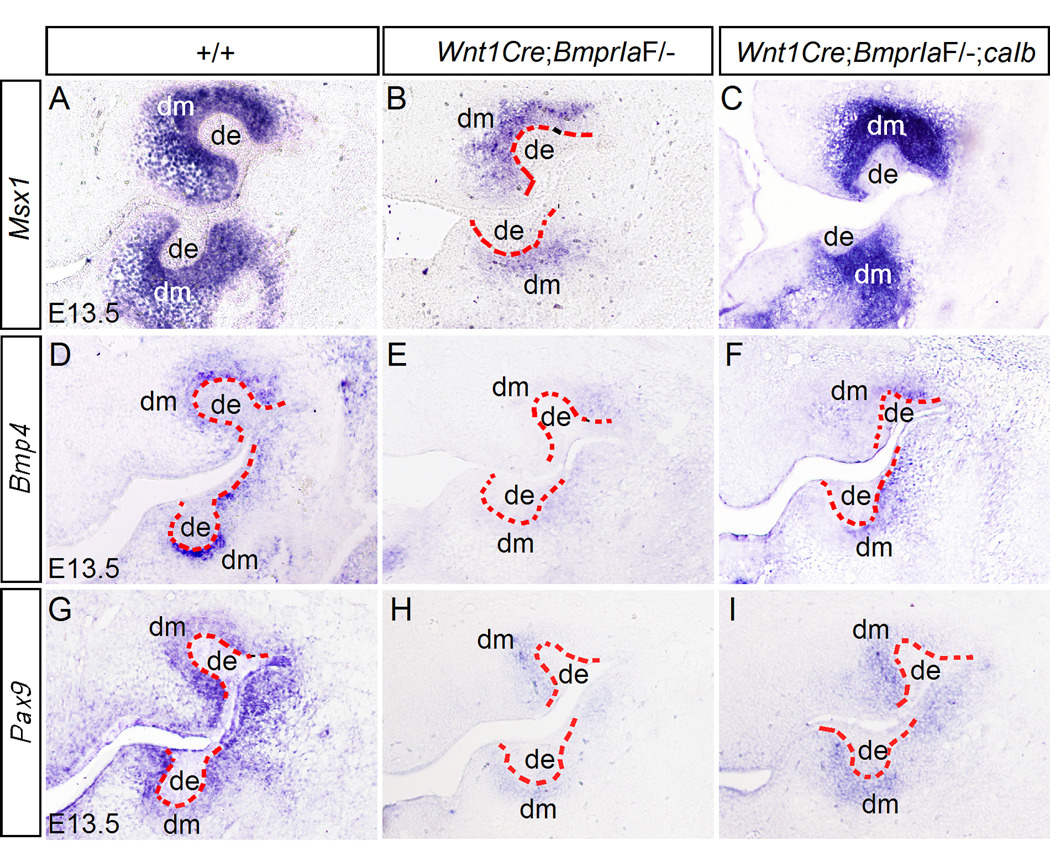
Mutations in several genes that are expressed in the mesenchymal compartment of developing tooth and palate, including Msx1 and Pax9, cause cleft palate defect as well as arrested tooth development at the bud stage (Satokata and Maas, 1994; Peters et al., 1998). Bmp4 is expressed and forms a positive regulatory loop with Msx1 in the anterior palatal mesenchyme and the dental mesenchyme (Chen et al., 1996; Peters et al., 1998; Zhang et al., 2000; Zhang et al., 2002). We therefore set to examine the expression of these genes in the developing palate and tooth of Wnt1Cre;BmprIaF/− mice. Since Shox2 expression pattern overlaps with that of BmprIa in the anterior palatal mesenchyme and inactivation of Shox2 leads to the formation of anterior cleft of the secondary palate in mice (Yu et al., 2005; Gu et al., 2008), we also examined Shox2 expression. Our results revealed a significant down-regulation of Bmp4, Msx1, and Pax9 expression in the anterior palatal mesenchyme of E13.5 Wnt1Cre;BmprIaF/− mice (Fig. 5). Shox2 expression was also reduced in mutants (Fig. 5). Similarly, in the developing tooth germ of mutants, the expression of Bmp4, Msx1, and Pax9 was significantly down-regulated in the mesenchyme at the bud stage (Fig. 6). However, residual expression for all these genes was present in the mutant dental mesenchyme. Interestingly, the upper molars in mutants appeared to retain a relatively higher level of Msx1 expression in the mesenchyme as compared to the lower molars (Fig. 6B). This observation correlates with a stronger BmprIb expression in the upper molar mesenchyme (Fig. 1F) and an arrest of upper molar development at a relatively advanced stage (the early cap stage) in the majority of mutants, suggesting a partially functional redundancy between BmprIa and BmprIb. Nevertheless, our results indicate that BMPR-IA is a major mediator of BMP signaling in the regulation of Msx1 and Pax9 expression, which in turn is required for Bmp4 expression in the palatal and dental mesenchyme (Chen et al., 1996; Peters et al., 1998; Zhang et al., 2000; 2002).
Figure 5.
Gene expression in the developing palatal shelves. (A, D, G, J) in situ hybridization shows expression of Bmp4 (A), Msx1 (D), Pax9 (G) and Shox2 (J) in the anterior palatal shelf of E13.5 wild type embryo. (B, E, H, K) Expression of Bmp4 (B), Msx1 (E), Pax9 (H), and Shox2 (K) is significantly reduced in the anterior palatal shelf of E13.5 Wnt1Cre;BmprIaF/− embryo. (C, F, I, L) Expression of Bmp4 (C), Msx1 (F), Pax9 (I), and Shox2 (L) remains down-regulated in the anterior palatal shelf of E13.5 Wnt1Cre;BmprIaF/−;caIb embryo. T, tongue; PS, palatal shelf.
Figure 6.
Gene expression in developing molars. (A, D, G) In situ hybridization shows expression of Msx1 (A), Bmp4 (D), and Pax9 (G) in the molar mesenchyme of E13.5 wild type embryo. (B, E, H) In situ hybridization shows reduced expression of Msx1 (B), Bmp4 (E), and Pax9 (H) in the molar mesenchyme of E13.5 Wnt1Cre;BmprIaF/− embryo. (C, F, I) In situ hybridization shows a wild type level expression of Msx1 (C), and a partially rescued Bmp4 (F) and Pax9 (I) expression in the molar mesenchyme of E13.5 Wnt1Cre;BmprIaF/−;caIb embryo. Dashed lines demarcate dental epithelium. de, dental epithelium; dm, dental mesenchyme.
caBMPR-IB partially rescues tooth development but not cleft palate defect in Wnt1Cre;BmprIaF/−mice
To determine if BMPR-IB-mediated BMP signaling can substitute for BMPR-IA-mediated signaling in regulating tooth and palate development, we created a conditional transgenic allele that expresses a constitutively active form of BMP receptor-IB (caBmprIb) upon crossing to a Cre mouse line. While activation of the caBmprIb allele in the epidermis by a K14-Cre transgenic allele causes severe ichthyosis skin disease (Yu et al., unpublished), expression of caBmprIb in the neural crest cells activated by Wnt1-Cre does not produce any visible phenotype (data not shown). Wnt1Cre;caBmprIb mice survived normally and were indistinguishable from their wild type littermates. In situ hybridization assay confirmed a wide spread expression of BmprIb in the cranial neural crest-derived mesenchyme, including the palate shelf and tooth germ at E13.5 (Supplemental Fig. 2). We compounded the caBmprIb transgenic allele onto the Wnt1Cre;BmprIaF/− background to generate mice lacking BmprIa but expressing caBmprIb in the neural crest cells (Wnt1Cre;BmprIaF/−;caIb). Grossly, Wnt1Cre;BmprIaF/−;caIb mice exhibited similar craniofacial abnormalities indistinguishable from Wnt1Cre;BmprIaF/− mice, including hypoplastic mandible and anterior clefting of the secondary palate (data not shown). Most Wnt1Cre;BmprIaF/−;caIb embryos died at mid-gestation if the drinking water of dams did not contain isoproterenol and ascorbic acid, except one that was identified at the birth (data not shown). These results indicate that caBmprIb fails to substitute for the loss of BmprIa to regulate craniofacial and peripheral nervous system development.
Histological analyses confirmed the phenotype of anterior palate clefting in Wnt1Cre;BmprIaF/−;caIb mice (data not shown), consistent with failed restoration of Bmp4, Msx1, Pax9 and Shox2 expression in the anterior palatal mesenchyme (Fig. 5). In contrast, although we still could not find any residual structure of lower incisors, we observed tooth structures of upper incisors and both upper and lower molars in Wnt1Cre;BmprIaF/−;caIb newborn mice (Fig. 7A-D). While slight delay in development and smaller in size, the molars in Wnt1Cre;BmprIaF/−;caIb mice indeed formed and appeared comparable to the wild type controls in developmental stage and patterning (Fig. 7B). The rescued tooth development is consistent with an almost completely restored Msx1 expression and partially rescued Bmp4 and Pax9 expression in molar mesenchyme at the bud stage (Fig. 6). However, while the upper incisors developed, they formed adjacently, appeared much smaller in size, and did not extend to the lateral sides of the nasal cavity (Fig. 7D). Closer examination of the upper incisors also revealed a mis-patterned incisor structure. In normal developing incisors, ameloblasts form only in the future labial side (Fig. 7C). However, in the upper incisors of Wnt1Cre;BmprIaF/−;caIb mice, ameloblasts were found around the dental pulp (Fig. 7D’).
Figure 7.
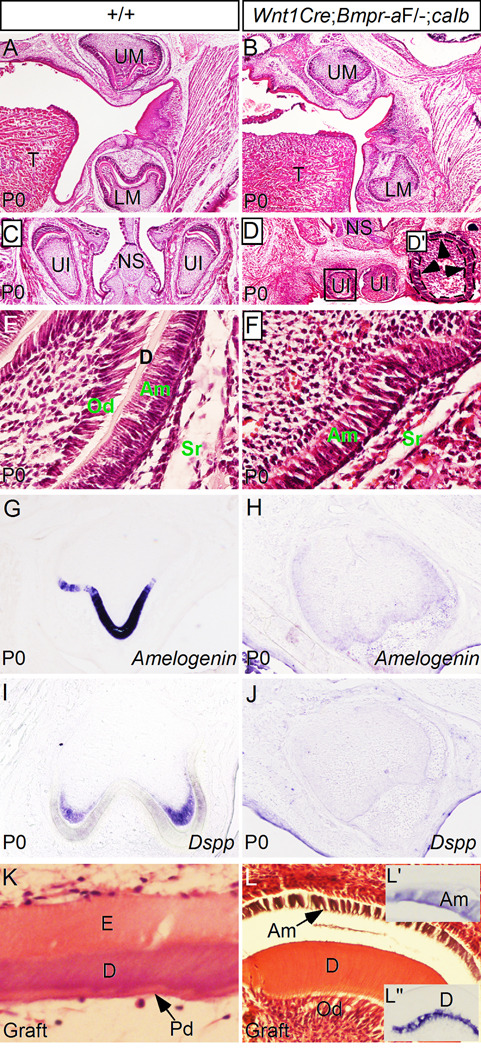
Rescue of tooth development in Wnt1Cre;BmprIaF/−;caIb mice. (A, C) Histological sections show morphology of molars (A) and upper incisors (C) in postnatal day 0 (P0) wild type mice. (B) A histological section from a P0 Wnt1Cre;BmprIaF/−;caIb mouse shows molar structures that are comparable to wild type control in developmental stage and patterning. (D) A P0 Wnt1Cre;BmprIaF/−;caIb mouse shows upper incisor teeth. These two incisors form adjacently and underneath the nasal septum. (D’) Higher magnification of an upper incisor from (D) shows mis-patterned incisor structure, with ameloblasts (arrowheads) forming all around the dental pulp. Black lines demarcate the ameloblast layer. (E) A higher magnification image shows formation of elongated odontoblasts and ameloblasts, and deposit of dentin in a molar of a P0 wild type mouse. (F) A higher magnification image shows formation of elongated ameloblasts, but lack of odontoblasts and dentin deposit in a molar of Wnt1Cre;BmprIaF/−;caIb mouse at P0. (G, I) Expression of Amelogenin in ameloblasts (G) and Dspp in odontoblasts (I) is detected in P0 wild type molars. (H, J) Molars from P0 Wnt1Cre;BmprIaF/−;caIb mouse fail to express Amelogenin (H) and Dspp (J). (K) A molar germ graft from E13.5 wild type embryo shows formation of enamel and dentin after 2-week in subrenal culture. (L) A molar graft from E13.5 Wnt1Cre;BmprIaF/−;caIb embryo shows differentiation of ameloblasts and odontoblasts and dentin deposit. (L’) In situ hybridization shows Amelogenin expression in ameloblasts of molar graft from Wnt1Cre;BmprIaF/−;caIb embryo. (L”) Dspp expression is detected in odontoblasts of molar graft from Wnt1Cre;BmprIaF/−;caIb embryo. D, dentin; E, enamel; Am, ameloblasts; LM, lower molar; NS, nasal septum; Od, odontoblasts; Pd, pre-dentin; Sr, stellate reticulum; UI, upper incisor; UM, upper molar.
Wild type molars at birth (P0) have formed differentiated ameloblasts and odontoblasts, as evidenced by their polarized (elongated) cell structure, expression of Amelogenin and Dspp, and formation of dentin (Fig. 7E, G, I). However, while elongated amebloblasts were observed in the molars of Wnt1Cre;BmprIaF/−;caIb mice at the same age, neither elongated odontoblasts nor dentin were found (Fig. 7F). This was also true for the upper incisors in Wnt1Cre;BmprIaF/−;caIb mice (Fig. 7C, D). The lack of odontoblast differentiation was consistent with the absent expression of the odontoblast differentiation marker Dspp (Fig. 7J). Interestingly, despite their elongated morphology, ameloblasts in the rescued molars and upper incisors of Wnt1Cre;BmprIaF/−;caIb mice expressed an extremely low level, if there is any, of the ameloblast differentiation marker Amelogenin (Fig. 7H, and data not shown).
To determine if the absent Dspp and Amelogenin expression in the rescued teeth of Wnt1Cre;BmprIaF/−;caIb mice at birth represents a delay in tooth development and differentiation, we grafted mandibular molars from E13.5 Wnt1Cre;BmprIaF/−;caIb embryos and wild type controls underneath mouse kidney capsule. After 2 weeks in subrenal culture, the wild type grafts produced well-organized enamel and dentin (Fig. 7K). In Wnt1Cre;BmprIaF/−;caIb grafts, we observed elongated odontoblasts and ameloblasts, and dentin deposit (Fig. 7L). In situ hybridization analyses showed Amelogenin expression in the ameloblasts (Fig. 7L’), and Dspp expression in the odontoblasts (Fig. 7L”). These results confirm a delayed differentiation of odontoblasts and ameloblasts in Wnt1Cre;BmprIaF/−;caIb mice. Thus caBmprIb can substitute for the loss of BmprIa in the cranial neural crest cells to regulate tooth development, but it is unable to fully replace BmprIa function as evidenced by delayed odontoblast and ameloblast differentiation.
Discussion
In this study, we show detailed expression patterns of BmprIa and BmprIb in the developing tooth and secondary palate. While previous studies have uncovered a crucial role for BmprIa in the epithelial component in tooth and secondary palate development (Andl et al., 2004; Liu et al., 2005), our results demonstrate an absolute requirement of BmprIa in the mesenchymal compartment for the development of tooth and secondary palate as well. Cremediated loss of mesenchymal BmprIa leads to an anterior clefting of the secondary palate and an arrest of tooth development at the bud/early cap stage, which is accompanied by reduced expression of BMP signaling downstream genes and defective cell proliferation. Our rescue studies further show a limited functional redundancy between BmprIa and BmprIb in a tissue specific manner in the development of craniofacial organs.
Mesenchymal BmprIa is required for anterior palate development
Increasing evidence has demonstrated heterogeneity, at both the cellular and molecular levels, in the developing secondary palatal shelf along the A-P axis (reviewed in Hilliard et al., 2005; Okana et al., 2006; Gritli-Linde, 2007). At the molecular level, a number of genes exhibit differential expression in the developing palatal shelf along the A-P axis (Zhang et al., 2002; Alappat et al., 2005; Yu et al., 2005; He et al., 2008; Liu et al., 2008; Xiong et al., 2009). At the cellular level, cells from the anterior palate and posterior palate respond differentially to the induction of growth factors in terms of cell proliferation and gene expression (Hilliard et al., 2005). For example, exogenously applied BMP proteins induce cell proliferation and Msx1 expression in the anterior palatal mesenchyme but not in the posterior palatal mesenchyme (Zhang et al., 2002; Hilliard et al., 2005). This could be explained by the restricted expression of both BmprIa and BmprIb in the anterior palatal mesenchyme (Fig. 1). Consistently, tissue-specific inactivation of BmprIa in the palatal mesenchyme results in defective cell proliferation only in the anterior palatal mesenchyme, and consequently leads to an anterior clefting of the secondary palate. These results indicate that BmprIa is a major player in mediating BMP signaling in the regulation of cell proliferation in the palatal mesenchyme. While BmprIb exhibits an expression pattern overlapping with BmprIa in the anterior palate, it apparently cannot compensate for the loss of BmprIa. Therefore, BMPR-IA and BMPR-IB appear to mediate different downstream signaling pathways during palatogenesis. In the anterior portion of palatal shelf, Msx1 and Bmp4 function in an autoregulatory loop to control a genetic cascade to regulate cell proliferation (Zhang et al., 2002). In the absence of BmprIa, expression of Msx1 and Bmp4, in addition to Pax9 which is a key regulatory gene in palate development, is dramatically down-regulated in the palatal mesenchyme. These results indicate an essential role for BMPR-IA mediated BMP4 signaling in the regulation of Msx1 and Pax9 expression. BMPR-IA is also required in the anterior palatal mesenchyme for normal expression of Shox2 whose null mutation leads to the formation of anterior cleft of the secondary palate as well (Yu et al., 2005; Gu et al., 2008). This result is consistent with our previous report that BMP signaling is required for Shox2 expression in the anterior palatal mesenchyme. However, based on the facts that BMP signaling is not sufficient for induction of Shox2 expression and a bioinformatic search fails to identify a potential Smad binding site in the 10-kb upstream region of the mouse Shox2 gene (unpublished data), Shox2 is unlikely a direct target of BMP signaling. Since loss of either Msx1 or Shox2 causes reduced cell proliferation in the anterior palatal mesenchyme (Zhang et al., 2002; Yu et al., 2005), the defective cell proliferation in the anterior palatal mesenchyme of Wnt1Cre;BmprIa mice could be attributed to a compounded effect of reduced expression of Msx1 and Shox2.
It was reported previously that deletion of BmprIa in the palatal epithelium causes a complete clefting of the secondary palate (Andl et al., 2004; Liu et al., 2005). In this study, inactivation of BmprIa in the palatal mesenchyme also results in a cleft palate formation, indicating the requirement of BMPR-IA mediated signaling in both epithelium and mesenchyme for normal palatogenesis. Anterior clefting of the secondary palate is a rare type of cleft palate in humans, and was thought to be caused by a non-genetic mechanism known as post-fusion rupture (Fara, 1971; Mitts et al., 1981; Schupbach, 1983). Together with our previous report that Shox2-deficient mice exhibit anterior clefting of the secondary palate (Yu et al., 2005), our results provide direct evidence for a genetic involvement in the formation of such type of palate clefting.
Mesenchymal BmprIa is required for self-maintenance of Bmp4 expression in early tooth development
Previous studies have established a fundamental role for BMP signaling in many steps of tooth development. Our studies further reveal an essential role for BMPR-IA-mediated BMP signaling in the dental mesenchyme during tooth development. Tissue specific deletion of BmprIa in the cranial neural crest derived dental mesenchyme results in an arrest of molar development at the bud/early cap stage, which is associated with significant reduction in BMP activity and defective cell proliferation. Mesenchymally expressed BMP4 is required for progression of molar development from the bud stage to the cap stage (Chen et al., 1996; Jernvall et al., 1998; Zhang et al., 2000; Zhao et al., 2000). Msx1 and Pax9 act synergistically in dental mesenchyme to regulate Bmp4 expression which in turn maintains Msx1 expression (Chen et al., 1996; Peters et al., 1998; Ogawa et al., 2006; Nakatomi et al., 2010). Our gene expression analyses show dramatically down-regulated expression of Bmp4, Msx1 and Pax9 in Wnt1Cre;BmprIaF/− molar mesenchyme, indicating that BMPR-IA also functions as a major player in the positive regulatory loop involving Bmp4, Msx1 and Pax9. Given the fact that BmprIb expression overlaps with that of BmprIa in molar germs before the cap stage, the residual expression of Bmp4, Msx1 and Pax9 in the Wnt1Cre;BmprIaF/− dental mesenchyme at the bud stage is likely attributed to BmprIb expression. Obviously, the residual Bmp4 expression in the mutant dental mesenchyme is below the threshold that is required for progression of molar development from the bud stage to the cap stage. Interestingly, epithelial deletion of BmprIa also causes an arrest of tooth development at the bud stage (Andl et al., 2004; Liu et al., 2005), suggesting that BMPR-IA most likely also mediates mesenchymally derived BMP4 signaling in the dental epithelium for tooth progression to the cap stage.
Deletion of BmprIa in the mesenchymal compartment leads to different defects in incisors. The mandibular incisor buds never formed in Wnt1Cre;BmprIaF/− embryos. This phenotype is very likely due to the formation of a hypoplastic mandible which had severely defective distal region (Fig. 2B). However, the maxillary incisor buds fuse at the midline to form a single incisor bud which is also arrested at the early bud stage. This phenotype is considered a little bit severer than that observed in molars in the mutants. This could be explained by the complete lack of BmprIb expression in the upper incisor mesenchyme at the bud stage (Fig. 1B), or could be attributed to different requirement for levels of BMP signaling for development of different types of tooth.
BmprIb has limited redundant function with BmprIa in the regulation of craniofacial development
Bmp2 and Bmp4 are expressed in developing craniofacial organs including palate and tooth, and play important roles in the development of these two organs (Nie et al., 2006). Both BMPR-IA and BMPR-IB show high affinity binding to BMP2 and BMP4 (Sieber et al., 2009), and exhibit partially overlapping but distinct expression patterns in the developing tooth and palatal shelf (this study). Despite primary structural differences in their kinase domains, BMPR-IA and BMPR-IB were shown previously to transduce similar intracellular signals in cell cultures (Wozney et al., 1988; ten Dijke et al., 1994; Hoodless et al., 1996; Kretzschmar et al., 1997). Several in vivo studies using gain-of or loss-of-function approaches seemed to be in favor of a functional similarity between BMPR-IA and BMPR-IB in craniofacial development. For examples, mice lacking BmprIb form normal craniofacial structures, suggesting that BmprIa may take over BmprIb’s function during craniofacial development (Baur et al., 2000; Yi et al., 2000). Expression of constitutively active forms of BMPR-IA or BMPR-IB in chicken craniofacial region gave rise to similar phenotypes, indicating that these two receptors may play similar role in regulating bone and cartilage formation during craniofacial development (Ashique et al., 2002). On the other hand, many lines of evidence argue for distinct roles of these two BMP type I receptors in embryogenesis. BmprIa deficient mice die prior to gastrulation, revealing a fundamental role for BmprIa in early embryonic development (Mishina et al., 1995). Expression of mutated forms of BMPR-IA and BMPR-IB in the chicken limb buds produces distinct phenotypes, suggesting that different BMP type I receptors are dedicated to specific functions during organogenesis (Kawakami et al., 1996; Yokouchi et al., 1996; Zou et al., 1997) In this study, we show that, despite their overlapped expression patterns, BmprIb does not share redundant function with BmprIa in the anterior palatal mesenchyme to regulate palate development. BMPR-IA-mediated signaling is irreplaceable for normal palatogenesis, as evidenced by the rescue experiment in which caBmprIb fails to substitute for the loss of BmprIa. In contrast, BmprIb appears to have limited functional redundancy with BmprIa in dental mesenchyme for molar development. This assumption is based on the facts: 1) BmprIb shows a much stronger expression level in the upper molar mesenchyme at the E13.5 bud stage as compared to the lower molar (Fig. 1F); at the cap stage, BmprIb expression is restricted to the dental epithelium of both upper and lower molars (Fig. 1I, J); 2) in Wnt1Cre;BmprIaF/− mice, all lower molars were arrested at the bud stage, but the majority of upper molars developed to the early cap stage (Fig. 3L); 3) Msx1 retains a relatively higher level of expression in the upper molar mesenchyme as compared to its expression in the lower molar mesenchyme in Wnt1Cre;BmprIaF/− embryo at the bud stage (Fig. 6B); 4) caBmprIb was able to partially rescue tooth development (including molars and upper incisor) in Wnt1Cre;BmprIaF/−;caIb mice (Fig. 7). The differential rescue of palate and tooth phenotypes in Wnt1Cre;BmprIaF/−;caIb mice suggests that these two type I receptors mediate different signaling pathways in a tissue specific manner. Taken together, our results reveal an absolute requirement of BmprIa in the mesenchymal component for normal palatogenesis and odontogenesis. BmprIb shares limited redundant function with BmprIa in a tissue specific manner during craniofacial development.
Supplementary Material
Acknowledgments
We thank Dr Yuji Mishina for providing BmprIa mutant mice, and Dr. Yang Chai for sharing in situ probes. This work was supported by NIH grant R01DE14044 to Y.P.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen YP. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev. Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, et al. Epithelial BmprIa regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Ashique A, Fu K, Richman JM. Signaling via type IA and type IB bone morphogenetic protein receptors (BMPR) regulates intramembranous bone formation, chondrogenesis and feather formation in the chicken embryo. Int. J. Dev. Biol. 2002;46:243–253. [PubMed] [Google Scholar]
- Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127:605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- Chen YP, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech. Dev. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Fara M. Congenital defects in the hard palate. Plast. Resconstr. Surg. 1971;48:44–47. [PubMed] [Google Scholar]
- Gu S, Wei N, Yu X, Jiang Y, Fei J, Chen YP. Mice with an anterior cleft of the palate survive neonatal lethality. Dev. Dyn. 2008;237:1509–1516. doi: 10.1002/dvdy.21534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Reynolds EM, Song J, Lei H, Feijen A, Yu L, He W, MacLaughlin DT, van den Eijnden-van Raaij J, Donahoe PK, Li E. The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development. 1999;126:2551–2561. doi: 10.1242/dev.126.11.2551. [DOI] [PubMed] [Google Scholar]
- He F, Xiong W, Wang Y, Matsui M, Yu X, Chai Y, Klingensmith J, Chen YP. Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev. Biol. 2010 doi: 10.1016/j.ydbio.2010.08.014. (In press) http://dx.doi.org/10.1016/j.ydbio.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. MADRa, a MAD-related protein that functions in BMP2 signaling pathway. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- Hosoya A, Kim JY, Cho SW, Jung HS. BMP4 signaling regulates formation of Hertwig’s epithelial root sheath during toot development. Cell Tissue Res. 2008;333:503–509. doi: 10.1007/s00441-008-0655-z. [DOI] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr, Hung YP, Chai Y. Smad-shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J. Bone Miner. Res. 2010;25:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Abert T, Kettunen P, Keranen S, Thesleff I. The life history of embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Ishikawa T, Shimabara M, Tanda N, Enomoto-Iwamoto M, Iwamoto M, Kuwana T, Ueki A, Noji S, Nohno T. BMP signaling during bone pattern determination in the developing limb. Development. 1996;122:3557–3566. doi: 10.1242/dev.122.11.3557. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Levi G, Mantero S, Barbieri O, Cantatore D, Paleari L, Beverdam A, Genova F, Robert B, Merlo GR. Msx1 and Dlx5 act independent in development of craniofacial skeleton, but converge on the regulation of Bmp signaling in palate formation. Mech. Dev. 2006;123:3–16. doi: 10.1016/j.mod.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–3968. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Jin Y, Tipoe GL. Alteration in the expression of bone morphogenetic protein-2,3,4,5 mRNA during pathogenesis of cleft palate in BALB/c mice. Arch. Oral Biol. 2000;45:133–140. doi: 10.1016/s0003-9969(99)00118-1. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Crombie R, Bradley A, Behringer RR. Multiple roles for activin-like kinase-2 signaling during mouse embryogenesis. Dev. Biol. 1999;213:314–326. doi: 10.1006/dbio.1999.9378. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphgenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- Mitts TF, Garrett WS, Hurwits DJ. Cleft of the hard palate with soft palate integrity. Cleft Palate J. 1981;18:204–206. [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ. Res. 2008;103:1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Zehir A, Maska E, Deng C, Schneider M, Mishina Y, Cserjesi P. BMP signaling regulates sympathetic nervous system development through Smad-dependent and –independent pathways. Development. 2009;136:3575–3584. doi: 10.1242/dev.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen YP, Maas R, Peters H. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Neubüser A, Peters H, Ballings R, Martin GR. Antagonistic interactions between FGF and BMP4 signaling pathways: A mechanism for positioning the sites of tooth formation. Cell. 1997;90:147–155. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. BMP signaling in craniofacial development. Int. J. Dev. Biol. 2006;50:511–521. doi: 10.1387/ijdb.052101xn. [DOI] [PubMed] [Google Scholar]
- Nohe A, Keating E, Knaus P, Petersen NO. Signal transduction of bone morphogenetic protein receptors. Cell. Signal. 2004;16:291–299. doi: 10.1016/j.cellsig.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kapadia H, Feng JQ, Raghow R, Peters H, D’Souza RN. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J. Biol. Chem. 2006;281:18363–18369. doi: 10.1074/jbc.M601543200. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Schupbach PM. Experimental induction of an incomplete hard-palate cleft in the rat. Oral Surg. Oral Med. Oral Pathol. 1983;55:2–9. doi: 10.1016/0030-4220(83)90296-7. [DOI] [PubMed] [Google Scholar]
- Sieber C, Kopf J, Hiepen C, Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- St. Amand TR, Zhang Y, Semina EV, Zhao X, Hu YP, Nguyen L, Murray JC, Chen YP. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DI, Ichijo H, Heldin C-H, Miyazono K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J. Biol. Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- Thesleff I, Mikkola M. The role of growth factors in tooth development. Int. Rev. Cytol. 2002;217:93–135. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsok LM, Whitters MJ, Kris RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, Yu X, Zhang Z, Lan Y, Jiang R, Cserjesi P, Chen YP. Hand2 is required in the epithelium for palatogenesis in mice. Dev. Biol. 2009;330:131–141. doi: 10.1016/j.ydbio.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev. Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro T, Tummers M, Thesleff I. Expression of bone morphogenetic proteins and msx genes during root formation. J. Dent. Res. 2003;82:172–176. doi: 10.1177/154405910308200305. [DOI] [PubMed] [Google Scholar]
- Yao S, Prpic V, Pan F, Wise GE. TNF-alpha upregulates expression of BMP-2 and BMP-3 genes in the rat dental follicle—implications for tooth eruption Connect. Tissue Res. 2010;51:59–66. doi: 10.3109/03008200903019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development. 2000;127:621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Sakiyama J, Kameda T, Iba H, Suzuki A, Ueno N, Kuroiwa A. BMP2/-4 mediate programmed cell death in chicken limb buds. Development. 1996;122:3725–3734. doi: 10.1242/dev.122.12.3725. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen YP. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang S, Song Y, Han J, Chai Y, Chen YP. Timing of odontogenic neural crest cell migration and tooth-forming capability in mice. Dev. Dyn. 2003;226:713–718. doi: 10.1002/dvdy.10274. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Z, Zhao X, Yu X, Hu Y, Geronimo B, Fromm SH, Chen YP. A new function of BMP4: Dual role of BMP4 in regulation of Sonic hedgehog expression in the mouse tooth germ. Development. 2000;127:1431–1443. doi: 10.1242/dev.127.7.1431. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen YP. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhang Z, Song Y, Zhang X, Zhang Y, Hu Y, Fromm SH, Chen YP. Transgenically ectopic expression of Bmp4 to the Msx1 mutant dental mesenchyme restores downstream gene expression but represses Shh and Bmp2 in the enamel knot of wild type tooth germ. Mech. Dev. 2000;99:29–38. doi: 10.1016/s0925-4773(00)00467-6. [DOI] [PubMed] [Google Scholar]
- Zou H, Wieser R, Massaque J, Niswander L. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev. 1997;11:2191–2203. doi: 10.1101/gad.11.17.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.