Abstract
Lineage-determination transcription factors coordinate cell differentiation and proliferation by controling the synthesis of lineage-specific gene products as well as cell cycle regulators. GATA-1 is a master regulator of erythropoiesis. Its role in regulating erythroid-specific genes has been extensively studied, whereas its role in controlling genes that regulate cell proliferation is less understood. Ectopic expression of GATA-1 in erythroleukemia cells releases the block to their differentiation and leads to terminal cell division. An early event in reprogramming the erythroleukemia cells is induction of the cyclin-dependent kinase inhibitor p21. Remarkably, ectopic expression of p21 also induces the erythroleukemia cells to differentiate. We now report that GATA-1 directly regulates transcription of the p21 gene in both erythroleukemia cells and normal erythroid progenitors. Using reporter, electrophoretic mobility shift, and chromatin immunoprecipitation assays, we show that GATA-1 stimulates p21 gene transcription by binding to consensus binding sites in the upstream region of the p21 gene promoter. This activity is also dependent on a binding site for Sp1/KLF-like factors near the transcription start site. Our findings indicate that p21 is a crucial downstream gene target and effector of GATA-1 during red blood cell terminal differentiation.
Keywords: p21, GATA-1, transcription, differentiation, erythroid, Sp1, KLF1
Introduction
Differentiation of precursor cells into more mature cells involves both the expression of tissue-specific functions and progressive restriction of proliferative capacity. Ultimately, both processes are controlled by specific master regulatory transcription factors. The regulation of tissue-specific gene expression programs by such factors has been studied extensively. On the other hand, much less is known about their role in regulating genes involved in cell proliferation.
GATA-1 is a Zn-finger DNA binding protein that is required for development of erythrocytes and megakaryocytes.1–3 Many tissue-specific genes that are directly regulated by GATA-1 have been described, including globins and components of the heme biosynthetic pathway in red cells, as well as platelet factor 4 and GPIbβ in megakaryocytes (reviewed in refs. 1 and 2). GATA-1 is also likely to be involved, either directly or indirectly, in controlling proliferation in cells undergoing terminal differentiation. For example, female mice heterozygous for a hypomorphic mutation in the X-linked GATA-1 gene accumulate immature cells in hematopoietic organs and exhibit a disorder similar to myelodysplastic syndrome which progresses to acute leukemia.4 GATA-1 mutations in humans with Trisomy 21 are associated with transient myeloproliferative disorder and acute megakaryoblastic leukemia,5–8 as well as other disorders of the megakaryocytic and erythrocytic lineages (reviewed in ref. 9).
Several cell culture systems in which GATA-1 levels can be modulated directly are available and have been used to study its effects on erythroid differentiation and cell proliferation. For example, G1E cells are an immortalized GATA-1 null erythroid line that proliferates indefinitely as immature erythroblasts until GATA-1 activity is restored, whereupon the cells undergo differentiation and terminal arrest.9 Murine erythroleukemia (MEL) cells are transformed erythroblasts that are blocked from differentiating due to spleen focus forming virus (SFFV) insertional activation of the PU.1 transcription factor.10–13 PU.1 binds to and inhibits GATA-1’s ability to promote transcription and erythroid differentiation.14–16 Remarkably, simply expressing an activated form of ectopic GATA-1 (GATA-1/ER) in these highly malignant cells reverses the block to differentiation and leads to terminal cell division and loss of tumorigenicity.17
The ability of GATA-1 to induce terminal growth arrest, as well as to activate expression of phenotypic markers of mature erythroid cells, suggests that it may exert control over regulators of cell proliferation. Indeed, gene expression profiling of G1E cells undergoing erythroid differentiation in response to GATA-1 showed that it induces changes in expression of numerous genes involved in cell cycle regulation, including core cell cycle components such as cyclin-dependent kinases (CDKs) and CDK inhibitors (CDKIs), c-Myc, and other genes associated with changes in the rate of cell proliferation.18 Likewise, the levels of many of the same core cell cycle components were observed to change during reprogramming of MEL cells by GATA-1/ER.17 Nevertheless, it is not known whether these changes in cell cycle regulators are due to direct effects of GATA-1 on the corresponding genes or due to other processes, for example indirect effects mediated by downstream gene targets of GATA-1.
A clue to which cell cycle regulators may be directly controlled by GATA-1 was provided by our previous report in which we tested the ability of several CDKIs to affect the differentiation program of MEL cells. Among four CDKIs tested (p15, p16, p21 and p27), only p21 was sufficient to reprogram the leukemia cells to terminal differentiation.19 The differentiation program induced by p21 in MEL cells is similar to that induced by GATA-1/ER in these cells, including synthesis of hemoglobin, morphological differentiation and terminal arrest. Thus, p21 mimics the actions of GATA-1 in promoting differentiation of MEL cells.
The observation that ectopic expression of either of two such disparate molecules as p21 and GATA-1 alone leads to resumption of terminal differentiation in MEL cells prompted us to ask whether GATA-1 controls p21 gene expression. We report here that GATA-1 regulates transcription of the p21 gene. These results illustrate how a master transcriptional regulator restricts cell proliferation while also promoting a tissue-specific gene expression program during terminal differentiation.
Results
GATA-1 transactivates the p21 promoter
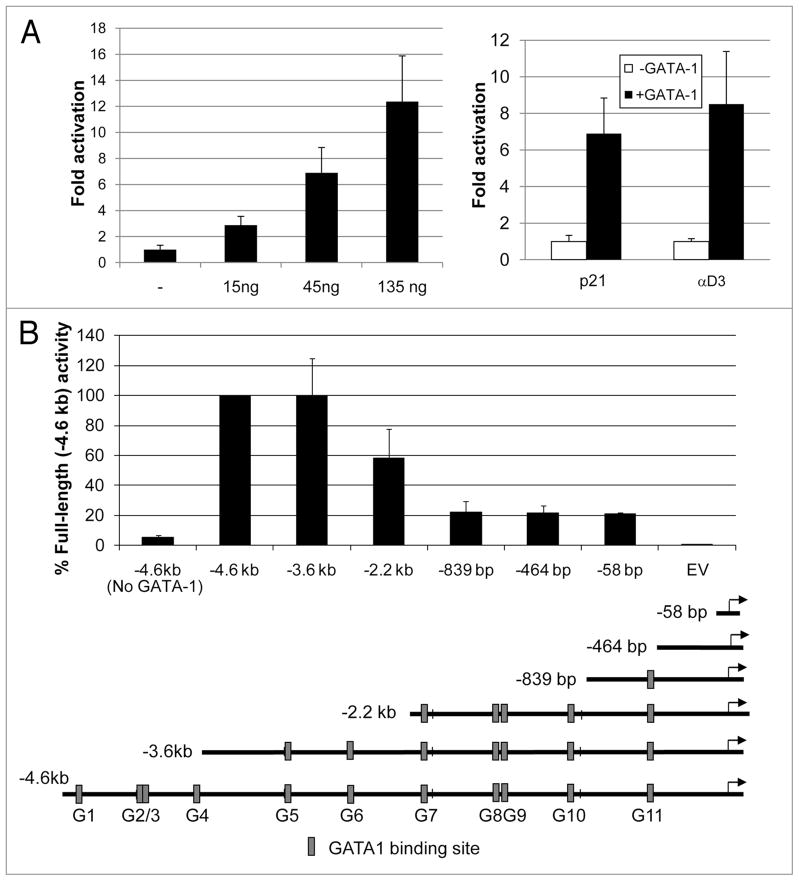
To investigate a potential role for GATA-1 in regulating p21 transcription, we examined the murine p21 promoter sequence for consensus GATA-1 binding sites (WGATAR).20,21 Within the 4.5 kb DNA sequence upstream of the p21 transcription start site there are eleven consensus GATA-1 binding sites (Fig. 1B). To determine whether GATA-1 is able to regulate the transcriptional promoter activity of this segment, we carried out transfection experiments to study the effect of GATA-1 on reporter activity of a plasmid consisting of this segment, which includes the transcription start site and 90 bp of the p21 5′UTR (+117 downstream of the TATA box). The p21 promoter reporter plasmid was co-transfected along with a GATA-1 expression plasmid into HeLa cells that do not express endogenous GATA-1. As shown in Figure 1A, GATA-1 stimulated production of luciferase activity in a dose-dependent manner and to a similar extent as its stimulation of the chicken alpha-globin promoter (αD3), a well-characterized GATA-1 target gene.22 GATA-1 stimulation of p21 promoter activity is independent of the tumor suppressor p53 because similar results were obtained in reporter assays with a p21 promoter construct harboring mutations in each of the two p53 binding sites at −2,809 bp and −1,915 bp (data not shown). To identify the GATA-1 responsive region within the −4.6 kb p21 promoter fragment, reporter plasmids containing progressively smaller portions of the −4.6 kb fragment were constructed and assayed. Removal of nearly 1 kb from the 5′ end of the fragment, eliminating four potential GATA-1 binding sites, had no effect on reporter activity. However, removing sequences between −3.6 kb and −2.2 kb and between −2.2 kb and −839 bp (relative to the TATA box) substantially reduced reporter activity (Fig. 1B). These deletions remove two and four potential GATA-1 binding sites, respectively. The −839 bp promoter segment contains a single consensus GATA-1 site. However, removing it by deleting the p21 promoter fragment to either −464 bp or −58 bp did not further affect reporter activity. These results indicate that sequences lying between −3.6 kb and −839 bp (relative to the TATA box) are primarily responsible for GATA-1-mediated stimulation of p21 promoter activity.
Figure 1.
GATA-1 stimulates the p21 promoter. (A) Left: luciferase reporter assays were carried out in HeLa cells transfected with 7.5 ng of the −4.6 kb p21 promoter-luciferase reporter with and without (−) the indicated amounts of pXM-GATA-1 expressing murine GATA-1. 48 hours after transfection cell extracts were prepared and analyzed for luciferase activity as described in Materials and Methods. Luciferase activity was normalized with respect to the protein content of the cell extracts. GATA-1-stimulated reporter activity is expressed relative to that of the −4.6 kb reporter construct in the absence of pXM-GATA-1. Right: luciferase reporter assays using 7.5 ng of the −4.6 kb p21 or −65 bp chicken α-globin (αD3) promoter-reporter constructs with or without (− GATA-1) 45ng pXM-GATA-1. (B) A series of luciferase reporter plasmids containing the indicated regions of the murine p21 promoter were constructed in pGL3-Basic as described in Materials and Methods and were assayed as in (A) with or without (no GATA-1) 45 ng pXM-GATA-1. Luciferase activity was expressed relative to that of the −4.6 kb reporter construct in the presence of pXM-GATA-1. EV: empty vector (pGL3 Basic vector without p21 promoter). The diagram below the figure indicates the positions of potential GATA-1 binding sites (G1–G11) in the p21 promoter. Error bars indicate the standard deviations of triplicate assays. Similar results were obtained in at least three experiments.
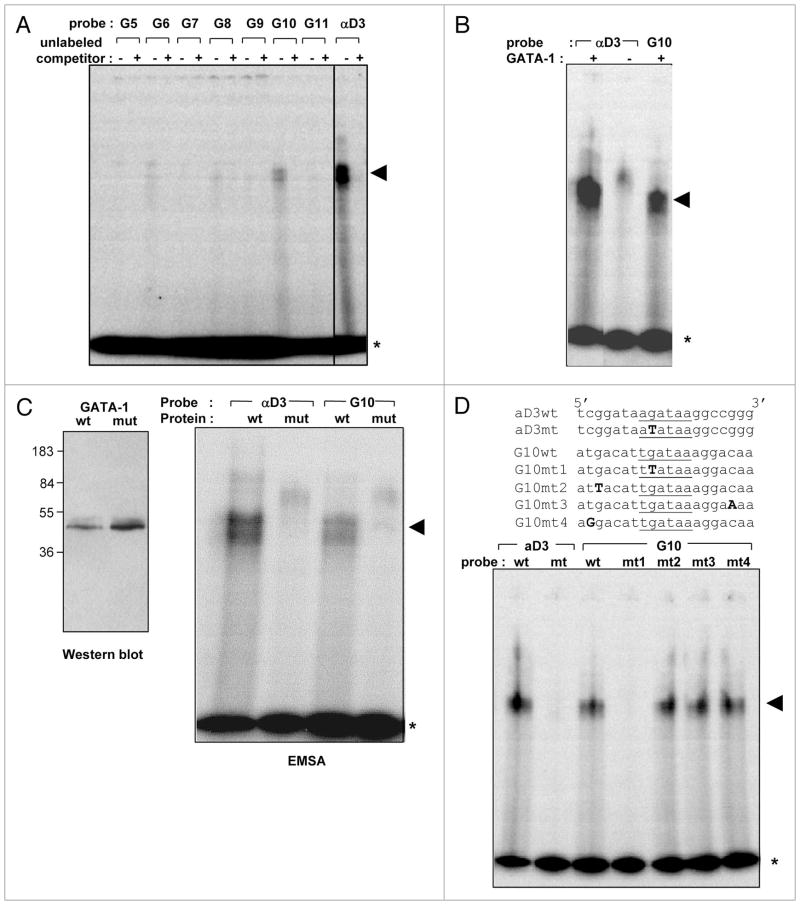
To determine whether the six potential consensus GATA-1 binding sites lying between −3.6 kb and −839 bp in the p21 promoter are actually able to bind GATA-1, we performed electrophoretic mobility shift assays with 20 bp oligonucleotides containing sequences encompassing each of the sites (Suppl. Table 1). An oligonucleotide encompassing the GATA-1 binding site in the chicken alpha globin promoter (αD3) served as a positive control. The source of GATA-1 was an extract of 293T cells transfected with the GATA-1 expression plasmid. GATA-1 bound oligonucleotides corresponding to several sites, including G6, G8 and G10 at −2,678 to −2,673 bp, −1,650 to −1,645 bp, and −1,153 to −1,148 bp, respectively, relative to the TATA box (Fig. 2A). However, the extent of binding of GATA-1 to these sequences was substantially less than to the GATA-1 binding sequence in the chicken alpha globin promoter. The sequence at site G10 at −1,153 to −1,148 bp appeared to bind most strongly (Fig. 2A). Similar results were obtained in EMSAs with a GST-GATA-1 fusion protein expressed in bacteria (data not shown). The binding was specific as demonstrated by competition with the corresponding unlabeled competitor oligonucleotide (Fig. 2A) and the requirement for GATA-1 expression in the 293T cell lysate (Fig. 2B). Specificity for GATA-1 was further demonstrated by showing that extracts of 293T cells transfected with a mutant GATA-1 defective in DNA binding23 are unable to bind the probes (Fig. 2C). Binding of GATA-1 to the G10 probe is due to the GATA-1 consensus sequence in the probe, and not to interactions with other residues of the probe, as shown by the failure of GATA-1 to bind to a mutant G10 oligonucleotide in which the core G residue is mutated to T and the inability of mutations outside of the core GATA-1 motif to disrupt this interaction (Fig. 2D).
Figure 2.
GATA-1 binds WGATAR sequences in the p21 promoter. (A) EMSA assays were performed as described in Material and Methods with 0.5 ng of 20 bp 32P end-labeled DNA probes corresponding to the indicated sequences in the p21 promoter (G5–G11, see the legend to Fig. 1 and Suppl. Table 1) or the GATA-1 binding sequence in the chicken α-globin promoter (αD3). Each probe was incubated with 15 μg of a protein extract from 293T cells transfected with pXM-GATA-1. Where indicated, a 125-fold molar excess of unlabeled competitor oligonucleotide was included in the reaction mixture. (B) EMSAs were performed as in (A) with αD3 and G10 probes and untransfected (− GATA-1) or pXM-GATA-1-transfected (+ GATA-1) 293T lysates. (C) EMSAs were performed with αD3 and G10 probes as in (A) except that the protein extracts consisted of 293T cells transfected with either pXM-GATA-1 (wt) or a version of pXM-GATA-1 encoding a mutant GATA-1 (mt) in which the first two cysteines of the C-terminal Zn finger domain were mutated to glycine.23 Left panel shows a western blot of 293T cell lysates transfected with pXM-GATA-1 (wt—left lane) or pXM-GATA-1 encoding the mutated GATA-1 (mt—right lane) using rat anti-GATA-1 (N6, Santa Cruz). (D) EMSA assays were performed as in (A) except that G10 probes with both wild-type (wt) and mutated (mt 1–4) sequences (shown on top) were used. Asterisk indicates free probe and arrowhead indicates shifted complex.
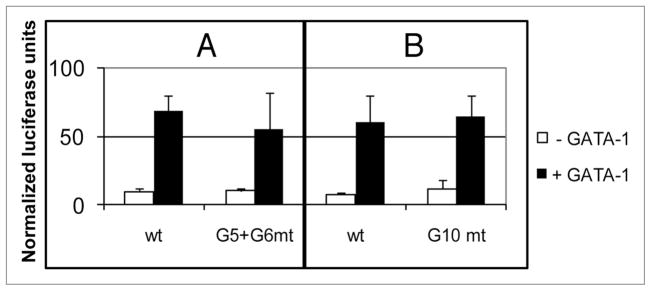
The presence of six GATA-1 binding sequences within the two GATA-1 responsive promoter segments between −3.6 kb and −839 bp, and the observation that GATA-1 can bind several of these sites, makes it unlikely that a single GATA-1 consensus sequence is responsible for the GATA-1 stimulation of p21 promoter activity. Since site G10 appeared to have the highest binding affinity for GATA-1, we constructed a 4.6 kb promoter-reporter plasmid in which site G10 was mutated so that it cannot bind GATA-1 (Fig. 3B). GATA-1 stimulated activity of this reporter as well as the unmutated reporter plasmid. Similar results were obtained with a G5, G6 doubly mutated plasmid (Fig. 3A). These results suggest that GATA-1 can use more than one of the six consensus sequences between −3.6 kb and −839 bp to stimulate p21 transcription.
Figure 3.
The G5 + G6 or G10 GATA-1 binding sites alone are not required for GATA-1-stimulated transcription of the p21 promoter. Luciferase reporter assays were performed in HeLa cells as in Figure 1 with −4.6 kb p21 promoter-reporter plasmids containing either the wild-type (wt) sequence or one in which the G5 + G6 sites (A) or the G10 site (B) was mutated (WGATAR → WTATAR). Error bars indicate the standard deviations of triplicate assays. Similar results were obtained in three experiments.
GATA-1 occupies the p21 promoter in normal erythroid progenitors and erythroleukemia cells
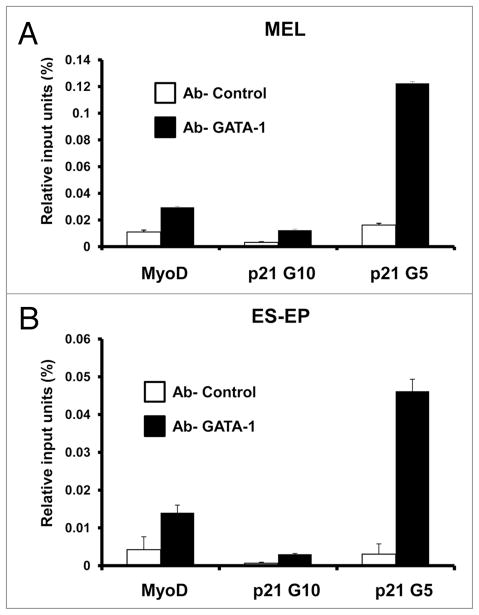
The data presented in the previous section strongly suggest that GATA-1 directly regulates transcription of the p21 gene. If this conclusion is correct, then GATA-1 should be detectable at the p21 promoter in erythroid cells in which GATA-1 promotes terminal differentiation. To determine whether GATA-1 occupies the p21 upstream regulatory region in such cells, we carried out quantitative chromatin immunoprecipitation (qChIP) experiments with a GATA-1 antiserum. Because we and others have reported that p21 gene expression is induced during erythroid differentiation,17–19,24 we studied GATA-1 occupancy of the p21 promoter in differentiating erythroid cells. qChIP was performed with chromatin from both normal embryonic stem cell-derived erythroid progenitors (ES-EP) undergoing erythroid differentiation25,26 and differentiating MEL cells. GATA-1 was found to occupy the region near the G5 GATA-1 consensus binding site (see Fig. 1) in differentiating ES-EP and MEL cells (Fig. 4). It was not observed near the G10 site that exhibited the strongest in vitro binding. These results indicate that GATA-1 binds the p21 promoter in vivo and regulates the p21 gene in differentiating erythroid cells.
Figure 4.
GATA-1 occupies the p21 promoter in erythroid cells. qChIP was performed as described in Materials and Methods on cross-linked chromatin from (A) MEL cells expressing a GATA-1-estrogen receptor (ER) fusion protein treated with 17β-estradiol for 48 hours and (B) ES-EP, with anti-GATA-1 antibody or anti-HA antibody as a control. ES-EP were induced to differentiate for 24 hours as described in Materials and Methods. The amounts of the indicated specific DNA fragments present in immunoprecipates were quantitated by real-time PCR. The bars indicate the percentages of input DNA fragments present in specific immunoprecipitates. Error bars indicate the standard deviations of triplicate PCRs. Similar results were obtained in three experiments. For other details see Materials and Methods.
GATA-1 transactivation of the p21 promoter depends on a binding site for Sp1/KLF-like factors near the transcription start site
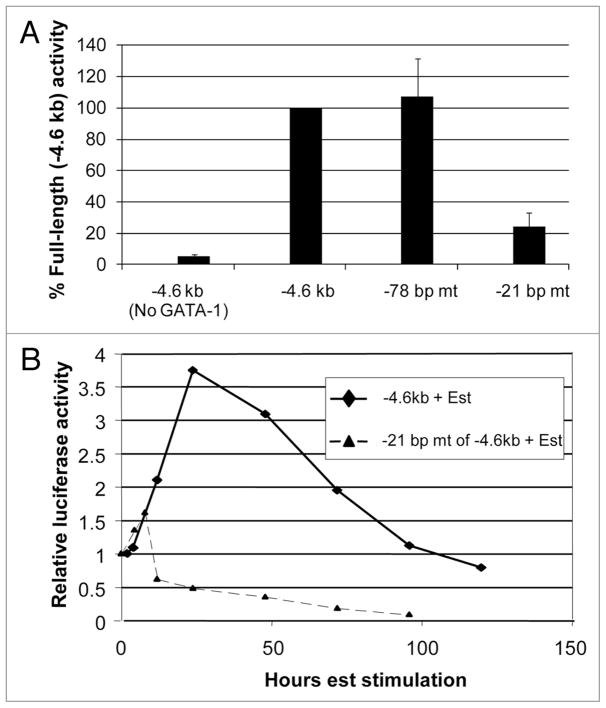
Despite the relatively weak in vitro binding of GATA-1 to its consensus sequences between −3.6 kb and −839 bp (Fig. 2), the ability of GATA-1 to stimulate the p21 promoter-reporter plasmid is quite robust in vivo. These observations suggest that GATA-1-mediated transactivation of the p21 promoter may depend upon other factors. GATA-1 is known to functionally interact with other zinc-finger transcription factors, including the widely expressed Sp1 and the related Kruppel-like, erythroid-specific factor KLF1.23,27,28 There are several reports of the involvement of Sp1/KLF family members in p21 gene transcription, including its cooperation with other transcription factors through interactions in the proximal region of the p21 promoter.39–42 Sp1 is expressed in HeLa cells used in the reporter assays.29 Sp1 and KLF1 are also known to bind directly to GATA-1 and to tether it to promoters, increasing GATA-1-stimulated transcription.23 Interestingly, reporter constructs containing −464 bp and −58 bp of the p21 promoter region that lack GATA-1 consensus binding sequences exhibit considerable GATA-1-stimulated transcription (Fig. 1B). There are consensus binding sites for Sp1/KLF-like factors in the p21 promoter at −78 to −71 bp and at −21 to −13 bp (relative to the TATA box). To determine whether these sites contribute to GATA-1-stimulated expression of the p21 promoter in HeLa cells, we mutated each of the sites separately in the −4.6 kb promoter-reporter construct. Remarkably, mutating the site between −21 to −13 bp markedly diminished reporter activity, whereas mutating the site between −78 to −71 bp had no effect (Fig. 5A).
Figure 5.
GATA-1 stimulation of the p21 promoter depends upon a binding site for Sp1/KLF-like factors near the transcription start site. (A) Luciferase reporter assays were performed in HeLa cells as in Figure 1 with −4.6 kb p21 promoter-reporter plasmids containing either the wild-type sequence (−4.6 kb) or sequences in which the consensus binding site for Sp1/KLF-like factors at −78 to −71 bp or −21 to −13 bp is mutated from GGGCGG to GTTTTG. (B) The wild-type p21 promoter-reporter plasmid (−4.6 kb) and the reporter plasmid in which the −21 to −13 bp sequence is mutated were stably transfected into MEL cells expressing a GATA-1-estrogen receptor (ER) fusion protein. Pools of transfected cells were isolated as described in Materials and Methods. GATA-1 was activated by treatment of the cells with 17β-estradiol (+ Est) and at the indicated times cell extracts were prepared and luciferase activity was assayed as described in Materials and Methods. Luciferase activity is expressed relative to the activity present in untreated cells (0 hours).
To determine whether the binding site at −21 to −13 bp also contributed to GATA-1-stimulated transcription in erythroid cells, we generated stable transfectants of MEL cells containing the wild-type −4.6 kb reporter plasmid and a version in which the site at −21 to −13 bp is mutated. Induction of GATA-1 activity caused a stimulation of the wild-type construct, but it did not stimulate the mutant plasmid (Fig. 5B). These results indicate that a factor(s) present in HeLa cells and erythroid cells that is capable of recognizing the site at −21 to −13 bp plays an important role in GATA-1-stimulated transcription of the p21 promoter. Such a factor(s) might stabilize binding of GATA-1 to one or more of the relatively weak GATA-1 consensus sites in the upstream regulatory region.
Discussion
GATA-1 is a Zn-finger DNA binding transcription factor that is required for development of erythrocytes and megakaryocytes. Numerous genes that are expressed specifically in these lineages have been shown to be regulated by GATA-1.18,30–33 In addition to promoting differentiation of these cell types, expression of GATA-1 also leads ultimately to cessation of cell proliferation. Microarray transcriptome analysis of G1E cells undergoing erythroid differentiation in response to GATA-1 indicates that it controls an extensive gene expression program in these cells that includes many genes involved in cell cycle regulation.18 However, for the vast majority of these genes, it is not known whether or not they are directly regulated by GATA-1. One gene, c-Myc, that was found to be negatively controlled by GATA-1 was also shown by ChIP to have GATA-1 bound to its promoter, suggesting direct repression of this important pro-proliferation gene by GATA-1.18
One of the most striking examples of the ability of GATA-1 to induce terminal arrest in erythroid cells is the demonstration that ectopic expression of GATA-1/ER in MEL tumor cells is sufficient to induce erythroid differentiation and terminal cell division, leading to loss of tumorigenicity.17 A very early event in this process is the induction of the CDK inhibitor p21.17,19 Even more remarkably, we found that ectopic expression of p21 is also sufficient to restore erythroid differentiation and terminal cell division in the erythroleukemia cells. This effect is specific for p21, since p15 and p27 were not active; p16 also exhibited activity but only in combination with the chemical CDK inhibitor roscovitine.19 Thus, GATA-1 and p21 share a unique ability to reprogram erythroleukemia cells from their transformed state towards normal erythroid differentiation and terminal growth arrest.
Having found that two such disparate molecules as GATA-1, a transcription factor, and p21, a CDK inhibitor, both can reprogram MEL tumor cells into terminal differentiation, we were prompted to ask whether GATA-1 controls p21 gene expression. Several lines of evidence presented here demonstrate that GATA-1 directly controls transcription of the p21 gene. The evidence includes: (1) reporter assays showing that the p21 promoter is highly stimulated by ectopic expression of GATA-1 in heterologous cells (Fig. 1). This reporter is also induced by activation of ectopic GATA-1/ER in differentiating MEL cells (Fig. 5); (2) EMSA analysis showing that GATA-1 can bind to several different sequences found in the upstream region of the p21 promoter (Fig. 2). These sequences conform to the WGATAR consensus binding sequence for GATA-1. Although in vitro binding of GATA-1 to these sequences is weaker than to a well-characterized GATA-1 site in the chicken α-globin promoter, binding is strictly dependent upon the core G nucleotide in the sequence. The stronger binding of GATA-1 to the chicken α-globin probe may be due to the presence of both a consensus WGATAR motif as well as an overlapping direct repeat minor site (GGATAA) that has been shown to increase the affinity of GATA-1 for DNA by interacting with the GATA-1 N-terminal Zn finger.34 This arrangement is not present in the p21 promoter probes (see Suppl. Table 1); (3) qChIP assays showing that GATA-1 occupies the p21 promoter in both differentiating MEL cells and normal erythroid progenitors undergoing differentiation in response to erythropoietin (Fig. 4). The fact that GATA-1 occupies the p21 promoter in normal erythroid cells suggests that p21 plays an important role not only in MEL cells, but also during the later stages of normal erythroid differentiation. Indeed, p21 mRNA levels increase during GATA-1-stimulated differentiation of GIE cells.18 Induction of p21 was also seen during differentiation of ES-EP, as well as normal fetal liver erythroid cells (Ujhelly O and Skoultchi AI, unpublished observations).
The p21 gene can be activated by both p53-dependent and p53-independent mechanisms.24 Our data indicate that GATA-1 activation of the p21 promoter is independent of p53. Mutation of the two consensus p53 binding sites in the p21 promoter does not attenuate GATA-1 stimulation of the promoter in reporter assays performed in HeLa cells (data not shown). The observed occupancy of GATA-1 at the p21 promoter in MEL cells by ChIP (Fig. 4) is also very likely to be independent of p53 because most Friend virus-induced murine erythroleukemia cell lines contain inactivated forms of p53.35–37 These results suggest that the occupancy of the p21 promoter by GATA-1 in normal erythroid progenitors (Fig. 4), which presumably contain wild-type p53, as well as GATA-1-stimulated p21 promoter activation in these cells, is similarly independent of p53.
Our results suggest that GATA-1 is able to bind weakly to several consensus binding sites in the p21 promoter and that it does not depend on the presence of a particular site to stimulate promoter activity. Progressive removal of segments containing multiple consensus GATA-1 binding sites decreased GATA-1-stimulated promoter activity (Fig. 1), indicating that this activity depends upon the overall number of available GATA-1 binding sites, especially those between −3.6 and −839 bp. Mutation of the G5 + G6 or G10 binding sites did not affect GATA-1-stimulation of the full-length, −4.6 kb p21 promoter (Fig. 3), suggesting that although GATA-1 can bind these sequences, in their absence it can bind other sites and activate the promoter. However, in vivo there may be a preference for the G5 binding site since qChIP assays demonstrated GATA-1 binding in this region (Fig. 4).
Our results also suggest that GATA-1-stimulated transcription of the p21 gene depends on other transcription factors. We found that full activation of the p21 promoter by GATA-1 is dependent on the presence of an intact Sp1/KLF-like factor binding site at −21 to −13 bp (relative to the TATA box). Sp1 and the erythroid-specific factor KLF1 can physically interact with GATA-1, and they have been shown to synergistically activate promoters by two mechanisms: (1) at low concentrations, Sp1/KLF1 and GATA-1 cooperatively bind their respective DNA binding sites, even when separated by hundreds of base pairs, and (2) at high concentrations, Sp1/KLF1 tether GATA-1 to a promoter devoid of consensus GATA-1 binding sites, and vice versa.23 There are now many reports of interactions between Sp1 family members and other transcription factors, including GATA-1,28 that lead to synergistic regulation of transcription of target genes.38 Given the relatively weak in vitro binding of GATA-1 to the consensus GATA-1 binding sites in the p21 promoter, compared with its binding to the chicken α-globin promoter, it is tempting to speculate that Sp1/KLF family members, in particular KLF1, physically interact with GATA-1 on the p21 promoter and stabilize its binding to consensus GATA-1 binding sites. A looping mechanism suggested by Merika et al.23 to account for synergy between Sp1/KLF1 and GATA-1 could explain the dependence we observed for GATA-1-stimulated transcription of the p21 promoter on the −21 to −13 bp GGGCGG site in both Hela cells and erythroid cells.
The results reported here confirm and extend the observations of Rylski et al.18 indicating that GATA-1 plays a major role in coordinating the proliferation and differentiation programs in erythroid cells. The involvement of GATA-1 in regulating erythroid-specific expression of genes such as globins and heme biosynthetic enzymes is well established. As mentioned, Rylski et al.18 found that expression of GATA-1 in G1E cells also causes changes in expression of numerous genes involved in cell cycle control. At least one of those genes, the pro-proliferation c-Myc gene, was suggested to be directly repressed by GATA-1. The studies reported here demonstrate that GATA-1 directly stimulates transcription of the anti-proliferation p21 gene. Importantly, both c-Myc and p21 have been shown to have profound effects on the ability of MEL cells to resume erythroid differentiation. Ectopic expression of c-Myc in MEL cells blocks differentiation,43–47 whereas ectopic expression of p21, like GATA-1, is sufficient to drive MEL cells into terminal erythroid differentiation.19 These results suggest that GATA-1 coordinates the proliferation and differentiation programs in erythroid cells by regulating a network of genes, at least some of which, like c-Myc and p21, can themselves control the decision of the cells to proliferate versus differentiate. This principle of network control initiated by the erythroid master regulator GATA-1 is likely to be a general property of lineage-determination transcription factors. Recently, we reported that the transcription factor PU.1, which promotes myeloid differentiation but inhibits erythroid differentiation, directly stimulates transcription of the CDK6 gene.48 Interestingly, CDK6, like PU.1, can block erythroid differentiation.49 In the future, it will be very interesting to determine what proportion of the direct gene targets, like c-Myc, p21 and CDK6, of such master regulatory transcription factors actually participate in the cross-talk between the proliferation and differentiation programs in lineage-committed cells. An even greater challenge will be to define the network and the control mechanisms that link the two programs regulated by these crucial, lineage-determination transcription factors.
Materials and Methods
Cell culture, transfection and differentiation
HeLa cells and MEL cells (clone DS19) were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C, in a humidified chamber containing either 5% CO2 (HeLa) or 10% CO2 (MEL). MEL cells stably expressing a GATA-1-estrogen receptor (ER) fusion protein in which GATA-1 is fused to the ER ligand binding domain (GATA-1/ER MEL cells) were generated, cultured and differentiated with 17β-estradiol as described previously.17 Embryonic stem cell-derived erythroid progenitors (ES-EP) were isolated, cultured and differentiated as described previously.26,48 To generate stable transfectants of GATA-1/ER MEL cells, 2.5 × 106 cells were transfected as described previously50 with a 1:10 molar ratio of pGK-Neo51 and either the −4.6 kb p21 promoter-luciferase reporter plasmid or a plasmid in which the potential Sp1/KLF1 binding site at −21 bp to −13 bp (relative to the 5′ nucleotide of TATA box) of the p21 promoter was mutated. Pools of cells representing approximately 25 stably transfected clones were selected for 10–14 days in 5 μg/ml puromycin and 1 mg/ml G418 and propagated in the same medium. Stable integration of the luciferase reporter gene into cellular DNA was verified by PCR for the luciferase coding sequence using purified genomic DNA as a template. For EMSAs, 293T cells were transfected with 5 μg wild-type or mutated GATA-1 in 100 mm dishes using Lipofectamine Plus (Invitrogen) as described by the manufacturer.
Reporter assays
HeLa cells, plated the night before at 30,000 cells per well in 24-well plates, were transfected with 7.5 ng of the indicated p21 promoter-reporter plasmid and, where indicated, 45 ng of GATA-1/pXM52 using Lipofectamine Plus (Invitrogen). The total DNA content per well was maintained at 225 ng with addition of plasmid pEBB.53 48 hours after transfection, cell lysates were prepared as described by the manufacturer and assayed for luciferase activity using Luciferase Assay Substrate (Promega) and a TD-20/20 (Turner Designs) or LMax II 384 (Molecular Devices) luminometer. Luciferase activity was normalized to the protein content of each extract. Luciferase activity present in cell extracts of stable MEL cell transfectants was assayed by the same procedure.
Construction of p21 promoter luciferase reporter plasmids
The −4.6 kb p21 promoter-luciferase reporter plasmid (p21/pGL3) contains −4,542 bp to +117 bp (relative to the 5′ nucleotide of TATA box) of the murine p21 promoter subcloned into pGL3 Basic (Promega). The wild-type construct and one deleted of both p53 binding sites were originally constructed by Xiao et al.54 and were gifts of Dr. Jill Pelling (Northwestern University Feinberg School of Medicine, Chicago, IL). Promoter-luciferase reporter plasmids containing progressively smaller portions of the wild-type −4.6 kb fragment were constructed by digesting p21/pGL3 with either ApaL1, Tth111I, HincII, BsmI or SmaI, creating blunt ends with Klenow fragment of DNA polymerase I (New England Biolabs), digesting with HindIII to generate fragments consisting of −3628/+117, −2230/+117, −839/+117, −464/+117 bp and −58/+117 of mouse p21 promoter (relative to TATA box), and ligating gel-purified fragments into pGL3 Basic that had been digested with HindIII and SmaI. Point mutations were introduced by standard techniques55 or with the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and were confirmed by nucleotide sequencing.
Electrophoretic mobility shift assay (EMSA)
Single-stranded oligonucleotides, corresponding to wild-type or mutated murine p21 promoter DNA sequences (sites G5–G11) or the GATA-1 binding site of the chicken α-globin promoter (αD322) flanked by seven nucleotides on each end, were annealed and end-labeled using γ-32P ATP and polynucleotide kinase (Invitrogen). 10 μl binding reaction mixtures, consisting of 2 μl (0.5 ng) double-stranded end-labeled oligonucleotide, 3 μl (15 μg) wild-type or disrupted C-terminal Zn finger23 GATA-1-transfected 293T cell lysate, 1 μl (1 μg) poly dI:dC, and 2 μl 5× buffer (50 mM HEPES pH 7.9, 250 mM KCl, 25 mM MgCl2, 5 mM EDTA, 5 mM DTT, and 25% glycerol), were incubated for 20 minutes at room temperature in the presence or absence of a 125-fold molar excess of unlabeled oligonucleotide. The reaction mixtures were electrophoresed in 6% nondenaturing polyacrylamide gels, and the dried gels were processed for autoradiography using a Storm 860 Molecular Imager (Molecular Devices) and ImageQuant software (Molecular Devices).
Quantitative chromatin immunoprecipitation (qChIP)
ChIP was carried out as described previously48,56 with antisera against GATA-1 (a gift of Emery Bresnick57) or HA (Santa Cruz Y11). qPCR was performed with the following primers: MyoD Fwd: TAA CCT TCC ACT CCC CTC ACA GA, Rev: TGT TCT GTG TCG CTT AGG GAT GC; p21 amplicon near GATA-1 site G5 Fwd (−2,931 bp relative to TATA box): TGC AAG GCT GCA TCA GTC CT, Rev (−2,826 bp relative to TATA box): TAG TCC CCA CCC AGG ACT GAA; p21 amplicon that includes GATA-1 site G10 Fwd (−1,206 bp relative to TATA box): GTC TTA CTG CTA TGT CTG TC, Rev (−1,119 bp relative to TATA box): AAG ATC CAG ACA GTC CAC TA using SYBR green master mix (ABI) and the ABI Prism 7900HT real time-PCR machine.
Supplementary Material
Acknowledgments
We thank Harmut Beug for providing ES-EP cells, Emery Bresnick for providing GATA-1 antiserum and Elly Antone for technical assitance. This work was supported by NIH grant HL078381 to A.I.S. M.P. was supported by NIH grant 1F32 HL077242-0102 and S.N.W. was supported by NIH/MSTP Grant 5T32GM07288. T.S. was supported by IGA and MSMT Grants from the Czech Republic (10310-3 & 2B06077). A.I.S. also receives support from National Cancer Institute Cancer Center Grant 2P30CA13330.
M.P., S.N.W. and T.S. designed experiments, performed research, analyzed data, and edited the manuscript. A.I.S. designed experiments, analyzed data and wrote the manuscript.
Footnotes
References
- 1.Migliaccio AR, Rana RA, Vannucchi AM, Manzoli FA. Role of GATA-1 in normal and neoplastic hemopoiesis. Ann NY Acad Sci. 2005;1044:142–58. doi: 10.1196/annals.1349.019. [DOI] [PubMed] [Google Scholar]
- 2.Ohneda K, Yamamoto M. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 2002;108:237–45. doi: 10.1159/000065660. [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Transcription factor GATA-1 in megakaryocyte development. Stem Cells. 1998;16:79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S, et al. Leukemogenesis caused by incapacitated GATA-1 function. Mol Cell Biol. 2004;24:10814–25. doi: 10.1128/MCB.24.24.10814-10825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–70. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rainis L, Bercovich D, Strehl S, Teigler-Schlegel A, Stark B, Trka J, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–6. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 7.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–52. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 8.Xu G, Nagano M, Kanezaki R, Toki T, Hayashi Y, Taketani T, et al. Frequent mutations in the GATA-1 gene in the transient myeloproliferative disorder of Down syndrome. Blood. 2003;102:2960–8. doi: 10.1182/blood-2003-02-0390. [DOI] [PubMed] [Google Scholar]
- 9.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–51. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goebl MK. The PU.1 transcription factor is the product of the putative oncogene Spi-1. Cell. 1990;61:1165–6. doi: 10.1016/0092-8674(90)90676-6. [DOI] [PubMed] [Google Scholar]
- 11.Marks PA, Sheffery M, Rifkind RA. Induction of transformed cells to terminal differentiation and the modulation of gene expression. Cancer Res. 1987;47:659–66. [PubMed] [Google Scholar]
- 12.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–80. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 13.Paul R, Schuetze S, Kozak SL, Kabat D. A common site for immortalizing proviral integrations in Friend erythroleukemia: molecular cloning and characterization. J Virol. 1989;63:4958–61. doi: 10.1128/jvi.63.11.4958-4961.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–51. [PubMed] [Google Scholar]
- 15.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–10. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe KS, Radparvar F, Matushansky I, Rekhtman N, Han X, Skoultchi AI. Reversal of tumorigenicity and the block to differentiation in erythroleukemia cells by GATA-1. Cancer Res. 2003;63:6363–9. [PubMed] [Google Scholar]
- 18.Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, et al. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–42. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matushansky I, Radparvar F, Skoultchi AI. Reprogramming leukemic cells to terminal differentiation by inhibiting specific cyclin-dependent kinases in G1. Proc Natl Acad Sci USA. 2000;97:14317–22. doi: 10.1073/pnas.250488697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, King DC, Dore LC, Zhang X, Zhou Y, Zhang Y, et al. Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 2008;18:1896–905. doi: 10.1101/gr.083089.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci. 1988;85:5976–80. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans T, Felsenfeld G. Trans-activation of a globin promoter in nonerythroid cells. Mol Cell Biol. 1991;11:843–53. doi: 10.1128/mcb.11.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–47. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, et al. p53-dependent and independent expression of p21 during cell growth, differentiation and DNA damage. Genes Dev. 1995;9:935–44. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 25.Carotta S, Pilat S, Mairhofer A, Schmidt U, Dolznig H, Steinlein P, Beug H. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104:1873–80. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- 26.Dolznig H, Kolbus A, Leberbauer C, Schmidt U, Deiner EM, Mullner EW, Beug H. Expansion and differentiation of immature mouse and human hematopoietic progenitors. Methods Mol Med. 2005;105:323–44. doi: 10.1385/1-59259-826-9:323. [DOI] [PubMed] [Google Scholar]
- 27.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–76. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 28.Fischer KD, Haese A, Nowock J. Cooperation of GATA-1 and Sp1 can result in synergistic transcriptional activation or interference. J Biol Chem. 1993;268:23915–23. [PubMed] [Google Scholar]
- 29.Grinstein E, Jundt F, Weinert I, Wernet P, Royer HD. cell cycle phase specific transcription factor Sp1 as G1 in epithelial cells. Oncogene. 2002;21:1485–92. doi: 10.1038/sj.onc.1205211. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–81. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szalai G, LaRue AC, Watson DK. Molecular mechanisms of megakaryopoiesis. Cell Mol Life Sci. 2006;63:2460–76. doi: 10.1007/s00018-006-6190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–47. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 33.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–95. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16:2238–47. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben David Y, Prideaux VR, Chow V, Benchimol S, Bernstein A. Inactivation of the p53 oncogene by internal deletion or retroviral integration in erythroleukemic cell lines induced by Friend leukemia virus. Oncogene. 1988;3:179–85. [PubMed] [Google Scholar]
- 36.Mowat M, Cheng A, Kimura N, Bernstein A, Benchimol S. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature. 1985;314:633–6. doi: 10.1038/314633a0. [DOI] [PubMed] [Google Scholar]
- 37.Munroe DG, Peacock JW, Benchimol S. Inactivation of the cellular p53 gene is a common feature of Friend virus-induced erythroleukemia: relationship of inactivation to dominant transforming alleles. Mol Cell Biol. 1990;10:3307–13. doi: 10.1128/mcb.10.7.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wierstra I. Sp1: emerging roles—beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophy Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 39.Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–84. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–8. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem. 2001;276:29116–25. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]
- 42.Sowa Y, Orita T, Hiranabe-Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites. Ann NY Acad Sci. 1999;886:195–9. doi: 10.1111/j.1749-6632.1999.tb09415.x. [DOI] [PubMed] [Google Scholar]
- 43.Bar-Ner M, Messing LT, Cultraro CM, Birrer MJ, Segal S. Regions within the c-Myc protein that are necessary for transformation are also required for inhibition of differentiation of murine erythroleukemia cells. Cell Growth Differ. 1992;3:183–90. [PubMed] [Google Scholar]
- 44.Coppola JA, Cole MD. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–3. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 45.Dmitrovsky E, Kuehl WM, Hollis GF, Kirsch IR, Bender TP, Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986;322:748–50. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- 46.Lachman HM, Cheng GH, Skoultchi AI. Transfection of mouse erythroleukemia cells with myc sequences changes the rate of induced commitment to differentiate. Proc Natl Acad Sci USA. 1986;83:6480–4. doi: 10.1073/pnas.83.17.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochownik EV, Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature. 1986;322:848–50. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- 48.Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates CDK6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. J Biol Chem. 2010;285:3044–52. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene. 2003;22:4143–9. doi: 10.1038/sj.onc.1206484. [DOI] [PubMed] [Google Scholar]
- 50.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–31. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 51.McBurney MW, Sutherland LC, Adra CN, Leclair B, Rudnicki MA, Jardine K. The mouse Pgk-1 gene promoter contains an upstream activator sequence. Nucleic Acids Res. 1991;19:5755–61. doi: 10.1093/nar/19.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–98. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka M, Gupta R, Mayer BJ. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–37. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao H, Hasegawa T, Miyaishi O, Ohkusu K, Isobe K. Sodium butyrate induces NIH3T3 cells to senescence-like state and enhances promoter activity of p21WAF/CIP1 in p53-independent manner. Biochem Biophys Res Commun. 1997;237:457–60. doi: 10.1006/bbrc.1997.7158. [DOI] [PubMed] [Google Scholar]
- 55.Cormack B. Directed mutagenesis using the polymerase chain reaction. Curr Protoc Mol Biol. 2001;8:5–8. doi: 10.1002/0471142727.mb0805s37. [DOI] [PubMed] [Google Scholar]
- 56.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–23. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–70. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.