Summary
We report a general method to examine the recognition of post-translational modifications (PTMs) by antibodies and proteins. We use this method to evaluate the binding of modification-specific antibodies and chromatin-associating factors to an array of high-purified, biotinylated peptides (derived from human histone sequences) harboring multiple PTMs printed onto streptavidin-coated glass slides. We find that modification-specific antibodies are both more promiscuous in their PTM recognition than expected and highly influenced by neighboring PTMs. Binding of chromatin-associating factors is also influenced by combinatorial PTMs, giving further support for the “Histone Code” hypothesis. Thus we report the first thorough characterization of PTM influence on antibody recognition and describe a tool for the rapid and inexpensive assessment of chromatin-associating factor binding specificity.
Results and Discussion
Post-translational modifications (PTMs) of proteins such as phosphorylation, methylation, acetylation, and ubiquitination regulate many processes such as protein degradation, protein trafficking, and mediation of protein-protein interactions[1]. Perhaps the best-studied PTMs are those found associated with histone proteins. More than one hundred histone PTMs have been described and they largely function by recruiting protein factors to chromatin, which in turn, drive processes such as transcription, replication, and DNA repair[2]. Likewise, dozens of chromatin-associating factors have been identified that bind to particular histone PTMs and hundreds of modification-specific histone antibodies have been developed to understand the in vivo function of these modifications[3].
The enormous number of potential combinations of histone PTMs represents a major obstacle toward our understanding of how PTMs regulate chromatin-templated processes, as well as our ability to develop high-quality diagnostic tools for chromatin and epigenetic studies. The same obstacle applies to other proteins regulated by combinatorial PTMs – for example, p53, RNA polymerase, or nuclear receptors[4-6]. To that end, we developed a peptide array-based platform to begin to address how both proteins and antibodies recognize combinations of PTMs. We focused primarily on the recognition of PTMs associated with the N-terminal tail of histone H3, but this approach is useful for the study of other histone modifications and combinatorial PTMs found on other proteins.
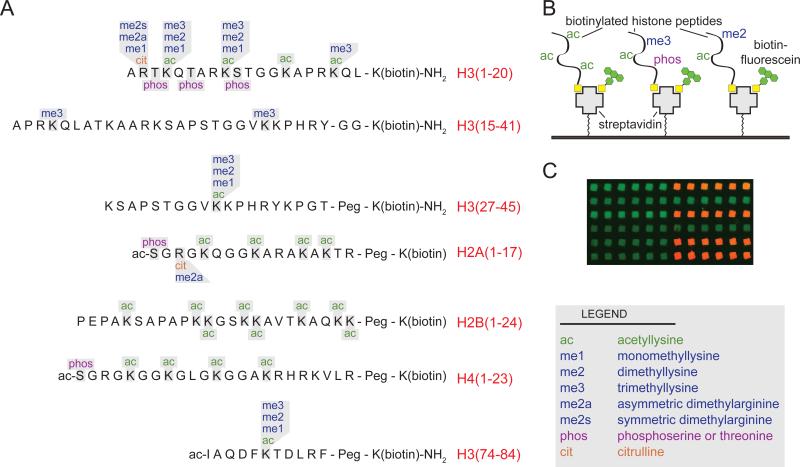
We generated a library of 110 synthetic histone peptides bearing either single or combinatorial PTMs and a biotin moiety for immobilization (Figure 1 and Table S2). Prior to printing, all peptides were subjected to rigorous quality control to verify their accuracy (see http://www.med.unc.edu/~bstrahl/Arrays/index.htm for complete details). This is significant, as extensive peptide purification and mass spectrometric analysis is not possible with other recently described array technologies used to study combinatorial histone PTMs[7]. Another significant advancement in our method was the introduction of a biotinylated fluorescent tracer molecule, which served as a positive control for the quality of our printing in all experiments. Lastly, peptides were printed as a series of 6 spots, two times per slide by two different pins, yielding 24 independent measurements of every binding interaction per slide. These measures were adopted to minimize binding artifacts due to pin variation or inconsistencies on slide surface. Thus, these arrays and the technical approaches described herein are the first to offer a large number of extensively characterized histone peptide substrates suitable for the assessment of protein or antibody binding.
Figure 1.
Composition of histone peptide arrays. (A) Peptides synthesized for this study with possible sidechain modifications (in single or combinatorial fashion) are indicated for each amino acid. (B) Depiction of array surface. Streptavidin-coated glass slides were spotted with a library of histone peptides containing different combinations of post-translational modifications (PTMs) (see also Table S2 for complete peptide list). Biotin-fluorescein was mixed with the peptides and used as an internal control for spotting efficiency. (C) Fluorescent image from a sample array. Positive binding interactions are shown as red spots where only the printing control (green) is visible for negative interactions.
We initially used our arrays to ask two fundamental questions regarding the recognition of histone PTMs: 1) How well do modification-directed antibodies recognize their intended epitope? and 2) what impact, if any, do combinatorial PTMs have on antibody recognition? We tested more than 20 commercially available antibodies raised against individual modifications on histone tails (see Table S4 and http://www.med.unc.edu/~bstrahl/Arrays/index.htm for experimental conditions and complete datasets). Generally, we found that antibodies were reasonably proficient at recognizing their target modification (Figure S3) however we found several exceptions – notably the discrimination between different methyllysine states by methyl-specific antibodies and the recognition of histone H3 lysine 14 acetylation (H3K14ac).
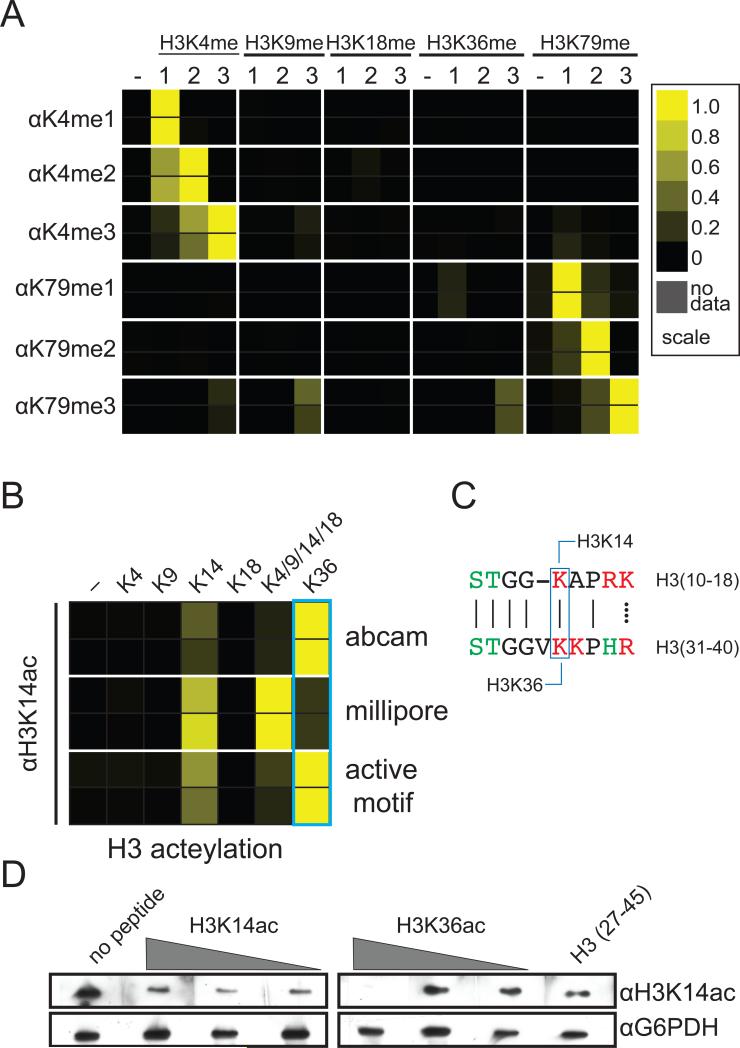
To explore methyllysine recognition, we tested the specificity of commercial antibodies raised against the three different methylated forms (mono-, di-, and trimethyl) of H3 at lysine 4 and 79 (H3K4me and H3K79me) (Figure 2). These antibodies were generally specific for their target lysine residue - however, both the trimethyl- and dimethyl-directed antibodies show measurable cross-reactivity with dimethyllysine and monomethyllysine, respectively (Figure 2A and Figure S1). This finding has particular biological importance, as each methylation state of a given histone lysine residue is thought to mediate different biological outcomes through the recruitment of distinct chromatin-associated factors[8]. For example, H3K4me3 is well correlated with transcriptional activation through the recruitment of histone acetyltransferases and the preinitiation complex of transcription[9-11]. Conversely, H3K4me2 was reported to recruit the Set3 histone deacetylase complex[8]. The ability to distinguish between these methyl states is therefore necessary to dissect how H3K4 methylation controls the balance of histone acetylation/deacetylation at transcribed genes.
Figure 2.
Antibody binding to histone peptide microarrays. Results of two independent arrays consisting of 24 independent spots for each peptide are depicted as heatmaps of the normalized mean intensity and plotted on a scale from 0 to 1 with 1 (yellow) being the most significant (see Methods). (A) Interactions of H3K4- and H3K79-specific antibodies with methylated peptides derived from the N-terminus of histone H3 (antibodies used are given in Table S1 and further information in Figure S1 and S3). (B) Recognition of histone H3 acetyllysine peptides by H3K14ac antibodies. (C) Alignment of sequence surrounding H3K14 and H3K16. (D) Western blot of yeast whole cell extract probed with H3K14ac antibody preincubated with various concentrations of histone H3 peptides.
We also tested a number of antibodies raised against acetyllysine found at position 14 of histone H3 (H3K14ac). Unlike lysine methylation, our arrays detected that several of these antibodies had difficulty in recognizing their target sequence, preferring acetylation at lysine 36 (H3K36ac) instead (Figure 2B). Additionally, peptide competition assays verified the interaction between the H3K14 antibodies and the H3K36ac peptide (Figure 2D). This result is likely explained by the fact that H3K14 and H3K36 are found in very similar sequence contexts and are acetylated by the same enzyme in vivo (Figure 2C). Acetylation of both H3K14 and H3K36 is catalyzed by the histone acetyltransferase Gcn5[12], however, H3K14ac is reported to be recognized by the RSC complex in yeast whereas H3K36ac has been reported to be recognized by the bromodomain of PCAF in human cells[13, 14]. Thus misdetection of H3K36ac using H3K14ac-directed antibodies by either western blot or chromatin immunoprecipitation may have implication for the studies of chromatin-templated processes regulated by H3K14 acetylation.
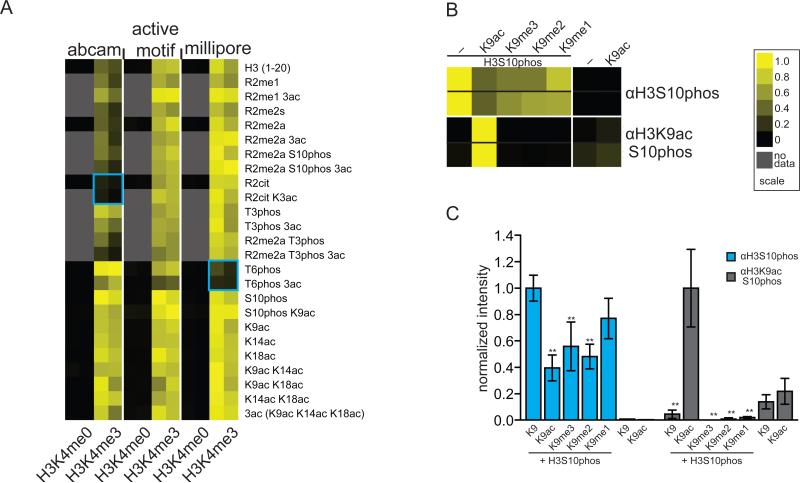
The large number of synthetic peptides containing combinatorial PTMs allowed us to also ascertain how PTM recognition is influenced by neighboring modifications. We therefore did further analysis of the H3K4me3 antibodies to determine how adjacent modifications affect substrate recognition. We observed that a monoclonal antibody widely used against H3K4me3 (Abcam; cat # ab1012) is perturbed mainly by modification at Histone H3 arginine 2 (H3R2) (Figure 3A). Contrastingly, a widely used polyclonal antibody from Millipore (#07-473) was negatively influenced by H3T6 phopshorylation and a similar antibody from Active Motif (#39160) was not particularly sensitive to any neighboring modification (Figure 3A).
Figure 3.
Effect of neighboring modifications on histone antibody recognition. Results of two independent arrays consisting of 24 independent spots for each peptide are depicted as heatmaps of the normalized mean intensity and plotted on a scale from 0 to 1 with 1 (yellow) being the most significant (see Methods). (A) Heatmap of neighboring modification effect on H3K4me3-specific antibody recognition. (B) Recognition of H3S10 phosphorylation by mono- and dual-specific PTM antibodies. (C) bar graph of data in (B). Differences in intensities were compared using two-way ANOVA analyses and confidence intervals (* 95% and ** 99%) are indicated for individual comparisons. Further information is available in Figure S3.
We also examined the well-characterized PTM “switch” region on histone H3, where H3K9 is modified by either acetylation or methylation and the neighboring serine 10 (H3S10) is a target for phosphorylation[15]. A polyclonal antibody (Active Motif; #39253) raised against H3S10 phosphorylation showed a statistically significant reduction in binding to peptides also modified at H3K9 (Figure 3B and 3C). In contrast, an antibody raised against both H3S10phos and H3K9ac (Cell Signaling; #9711) showed nearly absolute specificity for the peptide containing both modifications (Figure 3B and 3C). These data can be interpreted to suggest that biological changes in acetylation and methylation at H3K9 would influence the ability of antibodies derived against H3S10 phosphorylation to appropriately detect this mark. Such findings are significant, as H3S10 phopshorylation levels have already been correlated to change during the cell cycle, and in response to histone deacetylase inhibitors [16-18].
Collectively, our analysis of histone PTM-specific antibodies enabled us to uncover recognition of related but “off” target sequences, in addition to adjacent PTM affects. To our knowledge this is the first major assessment of how neighboring PTMs influence the recognition of histone PTM-specific antibodies. Given other proteins are predicted to have PTM “codes” within their sequences that regulate activity, our work has potential implications in other areas of biological regulation as well [19].
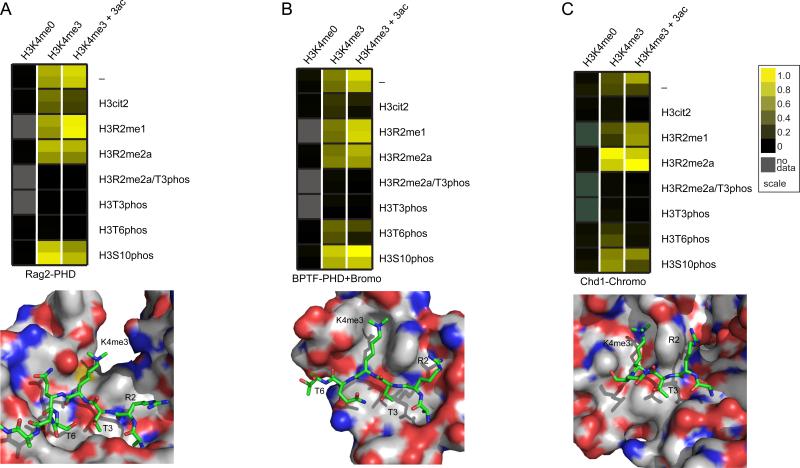
In addition to being a powerful diagnostic tool for the characterization of PTM-derived antibodies, we used our peptide array technology to measure how PTM codes affect the interaction of chromatin-associated proteins. Accordingly, we measured the binding of several domains known to interact with H3K4me3. We found that the PHD domain from the VDJ recombination factor Rag2 was specific for H3K4me3, and was blocked by phosphorylation at either H3T3 or H3T6 (Figure 4A). From the structure of the Rag2 PHD domain bound to H3K4me3 peptide[20], it can clearly be seen how H3T3 phosphorylation may disrupt binding. Timmers and coworkers very recently published that H3T3 phosphorylation acts as a switch to control the binding of TAF3 PHD domain[21]. Thus, this may be a general mechanism for controlling gene expression during mitosis (when H3T3 is phosphorylated). Similarly, Denu and coworkers found that H3T6 phosphorylation may disrupt this binding[22].
Figure 4.
Chromatin-associating domain binding to histone peptide arrays. (A) (top) Molecular representation of the Rag2 PHD domain binding to an H3K4me3-containing peptide (PDB accession 2V83). (bottom) Heatmap of Rag2 PHD domain binding to histone H3 peptides. (B) (top) Molecular representation of the BPTF PHD domain binding to an H3K4me3-containing peptide (PDB accession 2F6J). (bottom) Heatmap of Rag2 PHD-Bromo domain binding to histone H3 peptides. (C) (top) Molecular representation of the CHD1 chromodomain binding to an H3K4me3-containing peptide (PDB accession 2B2W). (bottom) Heatmap of CHD1 chromodomain binding to histone H3 peptides. All models were constructed using PyMol software. Additional information is also contained in Figures S2 and S4.
We next examined the tandem Bromo–PHD domains of BPTF (subunit of the NURF ATP-dependent remodeling complex[23]). Our studies showed that the tandem domain was specific for H3K4me3 and also showed reduced binding in the presence of either H3T3 or H3T6 phosphorylation (Figure 4B). However, both Rag2 and BPTF are blocked by citrulline, but not methylation at position 2, suggesting a role for the positive charge of H3R2 in PHD domain binding. Notably, converting H3R2 to citrulline results in a loss of cationic charge and likely loss of ionic and hydrogen bonding interactions within the pockets of the two PHD domains (Figure 4A and 4B). Interestingly, our ability to synthesize and print longer peptides allowed us to observe greater interactions of BPTF (PHD-Bromo) with H3K4me3 peptides also harboring acetylation. We found multiple acteylations on H3 enhanced the binding of BPTF to H3K4me3 (Figure 4B and Figure S2), suggesting coordination between the methyl-binding PHD domain and the acetyl-binding bromodomain to recognize multiple modifications on the Histone H3 tail.
The chromodomain of human CHD1 is also known to recognize H3K4me3, but has a structurally distinct binding pocket from the PHD domains. We found that CHD1, like Rag2 and BPTF, preferentially binds H3K4me3 and is also negatively influenced by phosphorylation at H3T3 and H3T6 (Figure 4C). Interestingly, we also found that methylation of H3R2 appears to slightly enhance binding of CHD1 whereas citrullination blocks this binding. While the finding that H3R2 methylation reduces binding affinity of human CHD1 to H3K4me3 is in opposition to a previous report [24], this discrepancy may be due to the fact that Khorasanizadeh and coworkers used peptides labeled at the N-terminus with fluorescein in their binding studies, which may have contributed to the binding. Consistent with our CHD1 findings, we and others have found that H3R2 methylation does not decrease CHD1 binding to H3K4me3 by either isothermal titration calorimetry (data not shown) or by fluorescence polarization using C-terminally labeled peptides (Marcey Waters personal communication). H3R2 methylation and H3K4me3 have been found to be mutually exclusive in yeast and humans[25, 26]. Thus, H3R2 methylation and H3K4me3 may function to prevent the binding of effector proteins that promote gene transcription while facilitating the recruitment of CHD1 (and possibly other factors) to genes in order to promote gene silencing.
The complex patterns of histone PTMs are critical determinants of chromatin structure and function, but also represent a significant challenge for future study. While many protein domains that bind selectively to particular PTMs have been identified, little is known regarding how neighboring modifications inhibit or contribute to these interactions. Of equal importance is our understanding of how patterns of PTMs influence antibody recognition. In this case, detection of biologically important events could be blocked or misrepresented if neighboring modifications interfere with epitope recognition. Thus, our work underscores a need for more rigorous testing and characterization of histone-specific antibodies. The datasets for the antibodies and proteins described here plus numerous additional antibodies are available in the supplemental data and from our website (http://www.med.unc.edu/~bstrahl/Arrays/index.htm). In addition, we will continue to characterize histone antibody specificities and post the data to our website as an ongoing resource for chromatin community.
Finally, while several other peptide array approaches have been used to measure binding to histone PTMs[7, 27-29], our arrays are a significant improvement over these other technologies. Specifically, our array displays a larger number of peptides carrying multiple PTMs allowing us to measure the influence of neighboring PTMs on binding. Additionally, we report full characterization of all synthesized peptides. Lastly, the high density of spotting allows us to perform statistical analysis of binding interactions. Liu et al. recently reported a similarly semi-quantitative approach, however their arrays were largely limited to peptides containing single PTMs and the peptides were labeled via their N-terminus, which could potentially occlude proteins and antibodies from recognizing modifications such as H3K4 methylation[28]. In addition, A very elegant bead-based approach has been used to generate even larger peptide libraries and successfully characterized the binding of several protein factors to combinatorial histone PTMs[22], however, our approach offers advantages in that we obtain binding data for each individual peptide and do not require sophisticated mass spectrometry for analysis. Importantly, post-translationally modified peptides can be synthesized relatively inexpensively and can also be purchased from commercial sources. Furthermore slides for peptide immobilization are relatively inexpensive making this approach feasible for the study of other systems regulated by PTMs as well.
EXPERIMENTAL METHODS
Antibodies
All primary antibodies tested are commercially available and are listed in Table S1. Secondary antibodies were Alexafluor 647-conjugated goat anti-rabbit IgG (Cat A21244) and Alexfluor 647-conjugated rabbit anti-mouse IgG (Cat. A21239) antibodies from Invitrogen.
Peptide Synthesis
All reagents were obtained from commercial suppliers (AnaSpec, EMD, and Apptec). The peptides, biotinylated at their C-termini, were synthesized on either NovaPEG Rink amide resin (histone H3 peptides) or Biotin-PEG NovaTag resin (histone H2A, H2B, and H4 peptides) using Fmoc (Fluorenylmethyloxycarbonyl) chemistry on a PS-3 automated peptide synthesizer (PTI) (See Table S2 for the complete list of peptides). All standard amino acids were coupled using HATU and N-methylmorpholine in DMF. Fmoc deprotection was performed using 20% piperidine in DMF. Modified amino acid residues were coupled using HATU, HOAt, and N,N,-diisopropyletylamine in NMP and the coupling of these residues was monitored using ninhydrin test and repeated when needed. Peptides were cleaved from the resins using a 2.5% TIS, 2.5% water in trifluoroacetic acid (TFA). After TFA evaporation and washing with diethyl ether, the peptides were lyophilized from and acetonitrile/water solution and purified via preparative HPLC using water-acetonitrile gradient (0.1%TFA in both solvents) on a Waters SymmetryShield RP-18 5μm 19×150mm column. All peptides were analyzed using MALDI-TOF MS and analytical HPLC. The average purity of peptides was over 90% (analytical HPLC). Analytical data for all peptides mentioned in this paper is available on our website.
Array Fabrication
Biotinylated peptides (25 μM final concentration) in printing buffer (10 mg/ml bovine serum albumin (Amresco), 0.3% Tween-20, and 10 μM biotin-conjugated fluorescein added to 1X ArrayIt protein printing buffer) were arrayed onto SuperStreptavidin-coated slides (ArrayIt) using SMP6 stealth pins (~200 μm spot diameter) and a OmniGrid100 arrayer (Digilab/Genomic Solutions) at ambient temperature and humidity (50-60%) using the following printing parameters. To minimize effects from individual pins or localized imperfections in the substrate arrays, samples were arrayed as a series of six spots, two times on each slide at a spacing of 375 μm as indicated in Table S3 and each peptide was printed by two different pins on each slide. After printing, slides were incubated overnight at 4°C in a humidified environment to facilitate interaction between the biotinylated peptide and the streptavidin surface. Slides were then blocked for 1 h at 4° C with Biotin-blocking buffer (ArrayIt), washed three times with phosphate-buffered saline, dried with air, and stored at 4°C and used within 60 days.
Antibody Binding
Antibody dilutions were made in PBS containing 1% BSA (~10 mg/ml) and 0.3% Tween-20 and exact concentration for each array is summarized in Table S4. Antibodies were incubated with printed slides for 90-180 minutes at 4°C (with the exception of the H3K4me3 monoclonal antibody from Abcam which was incubated overnight) and washed three times with cold PBS. Arrays were then probed with the appropriate Alexafluor 647–conjugated secondary antibody (Invitrogen) for 30–60 minutes at 4°C, washed three times with cold PBS and dried. Arrays were then scanned using a Typhoon TRIO+ imager (GE Healthcare) at 10μm resolution using the 526nm and 670nm filter sets for the biotin-fluorescein and secondary antibody respectively. Interactions were quantified using ImageQuant array software (GE Healthcare).
Protein Binding
Prior to binding, arrays were blocked in PBS containing 5% BSA (~ 50 mg/mL) and 0.3% Tween-20 for one hour at 4°C to reduce non-specific binding. GST-tagged protein (~25 μM) in the same buffer was overlaid on each array (200 μl total volume) and incubated in a hybridization chamber at 4°C overnight. Slides were washed three times with cold PBS. Anti-GST primary antibody was incubated with slides for 90-180 minutes at 4°C and washed three times with cold PBS. Arrays were then probed with the Alexafluor 647–conjugated anti-rabbit secondary antibody (Invitrogen) for 30–60 minutes at 4°C, washed three times with cold PBS and dried. Arrays were then scanned using a Typhoon TRIO+ imager (GE Healthcare) at 10μm resolution using the 526nm and 670nm filter sets for the biotin-fluorescein and secondary antibody respectively. Interactions were quantified using ImageQuant array software (GE Healthcare).
Statistical Analysis
Complete data sets are available for all arrays tested through our online database (http://www.med.unc.edu/~bstrahl/Arrays/index.htm). Briefly, printing of individual spots was evaluated based on the intensity of the fluorescein-biotin cospotted with each peptide. Spots with control intensities of less than 5% of the average intensity for all peptides were labeled as “not spotted” and omitted from subsequent analysis. Data were treated as four individual subarrays to account for small changes in intensity across the slide, each subarray containing all 110 peptides spotted six times. Alexafluor 647 intensities (corresponding to a positive interaction) were normalized for all spots by dividing the intensity by the sum of all intensities within a subarray. The six spots for each peptide were averaged (outliers were removed using a Grubbs test) and treated as a single value for a given subarray. The normalized intensities for the four subarrays were used to calculate the mean, and the error is reported as the standard error of the mean. For data displayed as heat maps, mean values were normalized to either the highest calculated value across all peptides or against the peptide for which a given antibody was supposed to interact. Heat maps were created using Java Treeview and all data plotted on a scale from 0 to 1 (Figure S3). Statistical analyses were performed using Graph Pad Prism software. ANOVA analyses were used to compare interactions and confidence intervals are reported as 95% (*), 99% (**), or 99.9% (***).
Supplementary Material
Supplementary Figure S1 Comparison of H3K4 methyllysine-specific antibodies for different methylation states (H3K4me1 – Millipore 07-436, H3K4me2 – Active Motif 39142, H3K4me3 – Active Motif 39160). Data are plotted as the mean with SEM for the indicated peptide from a single array. The results of two independent arrays are shown. Differences in intensities were compared using two-way ANOVA analyses and confidence intervals (* 95% and *** 99.9%) are indicated for individual comparisons.
Supplementary Figure S2. Effect of neighboring acetylation on BPTF binding. Data are plotted as the mean with SEM for the indicated peptide from a single array. Differences in intensities were compared using two-way ANOVA analyses and confidence intervals (* 95%, ** 99% and *** 99.9%) are indicated for comparisons to the H3K4me3 peptide with no other modifications (left).
Supplementary Figure S3. Heat map of all experimental antibody data. Data was normalized to the strongest interaction plotted on a scale from 0 to 1 with 1 (yellow) being the most significant.
Supplementary Figure S4. Scatter plots comparing two arrays for Rag2, BPTF, and CHD1.
ACKNOWLEDGEMENTS
This work was supported by a National Institutes of Health (NIH) EUREKA award to B.D.S. and a NIH NRSA Postdoctoral Fellowship (GM80896) to S.M.F. The authors would like to thank Gary Johnson for temporary use of the mass spectrometer, David Klapper for assistance with peptide synthesis, and Andrew Nobel for suggestions with statistical analysis, and Michael Keogh for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Assessing recognition of post-translational modifications using peptide arrays
- Neighboring PTMs modulate antibody recognition of histone modifications
- Peptide arrays identified positive and negative influences on chromatin factor recognition
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochim Biophys Acta. 2009;1789:26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 16.Davies GF, Ross AR, Arnason TG, Juurlink BH, Harkness TA. Troglitazone inhibits histone deacetylase activity in breast cancer cells. Cancer Lett. 2010;288:236–250. doi: 10.1016/j.canlet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi-Takanaka Y, Yamagata K, Nozaki N, Kimura H. Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. J Cell Biol. 2009;187:781–790. doi: 10.1083/jcb.200904137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang Z, Chen G, Zhao M, Wang D, Du Z, Xu Y, Yu X. FK228 induces mitotic catastrophe in A549 cells by mistargeting chromosomal passenger complex localization through changing centromeric H3K9 hypoacetylation. Acta Biochim Biophys Sin (Shanghai) 2010;42:677–687. doi: 10.1093/abbs/gmq077. [DOI] [PubMed] [Google Scholar]
- 19.Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 20.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HM. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J. 2010 doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat Chem Biol. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 25.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 26.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, Gozani O. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Galka M, Iberg A, Wang Z, Li L, Voss C, Jiang X, Lajoie G, Huang Z, Bedford MT, Li SS. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J Proteome Res. 2010;9:5827–5836. doi: 10.1021/pr100597b. [DOI] [PubMed] [Google Scholar]
- 29.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Comparison of H3K4 methyllysine-specific antibodies for different methylation states (H3K4me1 – Millipore 07-436, H3K4me2 – Active Motif 39142, H3K4me3 – Active Motif 39160). Data are plotted as the mean with SEM for the indicated peptide from a single array. The results of two independent arrays are shown. Differences in intensities were compared using two-way ANOVA analyses and confidence intervals (* 95% and *** 99.9%) are indicated for individual comparisons.
Supplementary Figure S2. Effect of neighboring acetylation on BPTF binding. Data are plotted as the mean with SEM for the indicated peptide from a single array. Differences in intensities were compared using two-way ANOVA analyses and confidence intervals (* 95%, ** 99% and *** 99.9%) are indicated for comparisons to the H3K4me3 peptide with no other modifications (left).
Supplementary Figure S3. Heat map of all experimental antibody data. Data was normalized to the strongest interaction plotted on a scale from 0 to 1 with 1 (yellow) being the most significant.
Supplementary Figure S4. Scatter plots comparing two arrays for Rag2, BPTF, and CHD1.