Abstract
Growing evidence supports the hypothesis that narcolepsy with cataplexy is an autoimmune disease. Using genome-wide association (GWA) in narcolepsy patients versus controls, with replication and fine mapping across three ethnic groups (3406 individuals of European ancestry, 2414 Asians, and 302 African Americans), we found a novel association between SNP rs2305795 in the 3′UTR of the purinergic receptor subtype 2Y11 (P2RY11) gene and narcolepsy (p(Mantel Haenszel)=6.1×10-10; odds ratio 1.28; n=5689). The disease-associated allele is correlated with a 3-fold lower expression of P2RY11 in CD8+ T lymphocytes (p=0.003) and natural killer (NK) cells (p=0.031) but not in other peripheral blood mononuclear cell (PBMC) types. The low expression variant is also associated with decreased P2RY11 mediated resistance to adenosine triphosphate (ATP) induced cell death in T lymphocytes (p=0.0007) and NK cells (p=0.001). These results identify P2RY11 as an important regulator of immune cell survival, with possible implications in narcolepsy and other autoimmune diseases.
Narcolepsy-cataplexy affects 1 in 2,000 individuals and is primarily caused by the loss of around 70,000 hypocretin (hcrt, also known as orexin) producing neurons in the hypothalamus1,2. The disease is associated with HLA-DQB1*06023, and the T cell receptor alpha locus (TRA@)4. Further, autoantibodies against Tribbles homolog 2 (Trib2), a protein expressed in hcrt cells, have recently been detected in the sera of recent onset narcoleptic patients5,6,7. These findings strongly suggest narcolepsy is caused by an autoimmune attack on the hypocretin neurons. This disease may thus offer a unique model to study immune surveillance of neurons, a topic of growing importance.
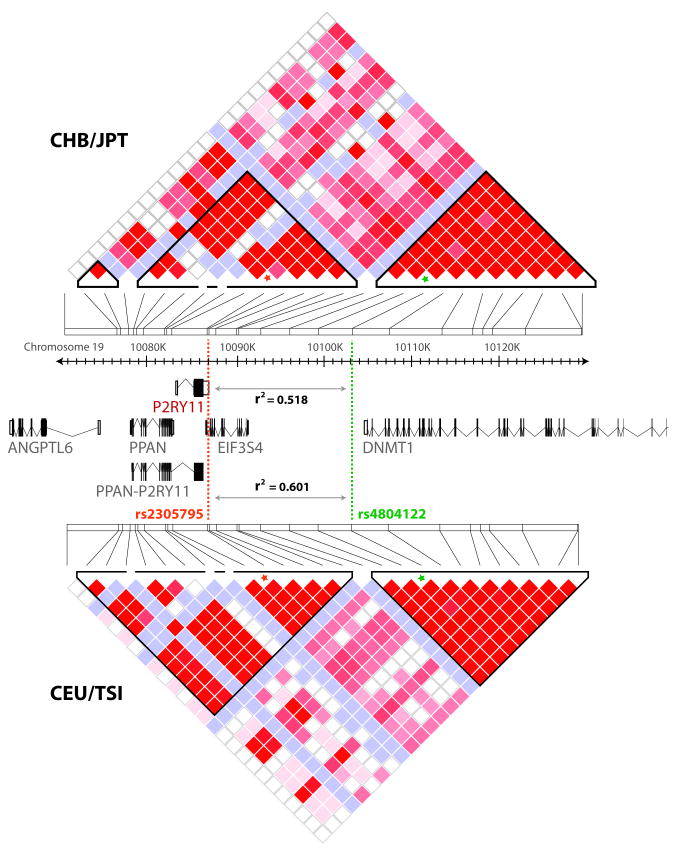
Following on our recently published GWA study of 807 narcolepsy-cataplexy patients versus 1,074 HLA DQB1*0602 positive controls, we conducted replication of 10 loci in 1,525 individuals of European ancestry (594 cases and 931 controls). Single Nucleotide Polymorphisms (SNPs) for replication were selected as having P-values less than p= 5×10-6. A total of 18 SNPs, representing 10 genomic regions met this criterion (Supplementary Table 1). Of these only one, rs4804122, replicated strongly, being still significant after Bonferroni correction for 18 markers (p<0.01, see Table 1 for nominal p value). This SNP is located downstream of P2RY11 on chromosome 19 in a region of high linkage disequilibrium (LD) spanning several genes (PPAN, P2RY11, EIF3G, and DNMT1, Figure 1).
Table 1. Association of rs4804122 with narcolepsy.
| Cohorts (n) | MAF cases (n) | MAF controls (n) | OR (95% CI) | Nominal P | P (BD) |
|---|---|---|---|---|---|
| European ancestry | |||||
| - original sample (1881) | 0.383 (614) | 0.468 (911) | 0.71 (0.61-0.82) | 3.69×10-6 | 0.285 |
| - replication sample (1525) | 0.391 (603) | 0.453 (907) | 0.77 (0.66-0.89) | 5.42×10-4 | 0.252 |
| - all (3406*) | 0.395 (1391) | 0.459 (1968) | 0.76 (0.69-0.84) | 1.16×10-7 | 0.173 |
| Asians | |||||
| - Chinese (1269) | 0.264 (582) | 0.279 (681) | 0.93 (0.78-1.10) | 0.389 | 0.617 |
| - Japanese (869) | 0.221 (424) | 0.245 (432) | 0.87 (0.70-1.09) | 0.224 | N/A |
| - Koreans (276) | 0.238 (105) | 0.240 (169) | 0.99 (0.66-1.49) | 0.967 | N/A |
| - all (2414) | 0.245 (1111) | 0.262 (1282) | 0.91 (0.80-1.04) | 0.172 | 0.894 |
| African Americans | |||||
| - all (302) | 0.314 (113) | 0.307 (189) | 1.04 (0.72-1.48) | 0.851 | N/A |
| All samples (6122) | 0.328 (2615) | 0.377 (3439) | 0.83 (0.76-0.89) | 1.03×10-6 | 0.1568 |
MAF: minor allele frequency. BD: Breslow Day test (performed in combination with Mantel Haenszel test of association).
Including genotypes for 174 cases, 150 controls from original sample that were typed on Affymetrix 500K array set.
Figure 1.
Risk locus on 19q13.2, showing gene organization and linkage disequilibrium in the region of interest (10,071,000-10,130,000). Top: D′/LOD-based LD plot using data from combined Chinese and Japanese populations (CHB-JPT). Bottom: D′/LOD-based LD plot for individuals of European ancestry only (CEU-TSI). D′ values are calculated from HapMap v3R2 CHB, JPT, CEU, and TSI populations. In addition, r2 values between the original marker, rs4804122 (green) and the best transethnic marker rs2305795 (orange) derived from our own data are indicated. rs2305795 falls in the 3′UTR of P2RY11.
The last of these 18 markers is rs9275523 a SNP located between HLA-DQB1 and HLA-DQA2 (p= 5.1×10-6; OR 0.59; MAF 0.071 in 799 cases, 0.116 in 1068 controls). Association with this marker is consistent with prior data indicating that the genetic influences of HLA on narcolepsy predisposition are not mediated solely through DQB1*0602 heterozygosity3. DQB1*0602 homozygotes or DQB1*0602/0301 heterozygotes are, for example, at higher risk for narcolepsy compared to DQB1*0602 heterozygotes in general, whereas DQB1*0602/0601, DQB1*0602/0501 and DQB1*0602/0603 are at decreased risk3,8. Heterodimerisation of DQA1*0102 and DQB1*0602 with other DQA1 and DQB1 alleles of the DQ1 group may explain these protective effects, by reducing abundance of the disease susceptibility DQA1*0102/DQB1*0602 heterodimer9. The finding of a secondary association in the HLA-DQ locus was thus expected and further indicates a complex influence of HLA-DQ or other loci in high LD with HLA-DQ on narcolepsy. A recently published study also reported an association of a SNP located in the HLA-DQA2 region, rs2858884, with narcolepsy independent ofo DQB1*06028. In our initial GWA study sample of 1,881 individuals of European ancestry, rs2858884 had a nominal p-value of 0.013, an effect well below rs9275523 (Supplementary Table 1).
Our next step was to attempt replication of the new chromosome 19 association in other ethnic groups. Surprisingly, rs4804122 had no effect in 2,414 Asians and a small African American sample (Table 1). This led us to explore differential LD patterns for this marker across ethnic groups. Based on data from the International HapMap Project, we selected 5 SNPs in high LD with rs4804122 in individuals of European ancestry but lower LD in Asians. One additional marker, rs3745601, a functional SNP located in P2RY11 previously reported to be associated with myocardial infarction and elevated levels of C-reactive protein10 was also genotyped, even though it is in low LD with rs4804122. These 6 SNPs were genotyped in 3,406 individuals of European ancestry (1,401 patients and 2,005 controls), 2,414 Asians (1,130 patients and 1,284 controls), and 302 African Americans (113 patients and 189 controls). A SNP located in the 3′ untranslated region (3′UTR) of the P2RY11 gene, rs2305795, showed the highest association with narcolepsy across all ethnic groups (Table 2 and Figure 1). The rs2305795 association was significant in individuals of European ancestry (p=3.1×10-7), Asians (p=0.025), and overall (p=3.7×10-9) after Bonferroni correction for the 6 fine typing markers in the replication sample. These findings identify this locus as a novel narcolepsy susceptibility factor (rs2305795 nominal p value = 6.1×10-10; odds ratio 1.28). The replication across ethnic groups also illustrates the value of transethnic mapping in narcolepsy, as previously found for HLA and TCR@ studies3,4,11. No significant interaction was found between rs2305795 and previously identified loci (data not shown).
Table 2. Fine-typing of SNP markers in chromosome 19 region.
| Cohort (n) | SNP | Position (bp) | Associated Allele | Freq Cases (n) | Freq Controls (n) | OR (95% CI) | Χ2 (MH*) | P (MH*) | P (BD) |
|---|---|---|---|---|---|---|---|---|---|
| European ancestry (3406) | rs11666402 | 10080076 | G | 0.476 (1313) | 0.437 (1816) | 1.18 (1.07-1.31) | 10.34 | 0.00130 | 0.933 |
| rs12460842 | 10083195 | G | 0.603 (1283) | 0.546 (1593) | 1.25 (1.12-1.39) | 16.27 | 5.50×10-5 | 0.338 | |
| rs3745601 | 10085548 | A | 0.134 (1216) | 0.109 (1587) | 1.27 (1.08-1.50) | 7.99 | 0.00470 | 0.061 | |
| rs2305795 | 10087052 | A | 0.608 (1311) | 0.542 (1802) | 1.33 (1.20-1.47) | 29.64 | 5.19×10-8 | 0.482 | |
| rs4804122a | 10102944 | T | 0.605 (1391) | 0.541 (1968) | 1.32 (1.19-1.45) | 28.09 | 1.16×10-7 | 0.173 | |
| rs11880388 | 10114573 | A | 0.536 (1312) | 0.496 (1828) | 1.16 (1.05-1.30) | 9.068 | 0.00260 | 0.832 | |
| rs2228611 | 10128077 | A | 0.538 (1316) | 0.497 (1839) | 1.18 (1.06-1.30) | 9.345 | 0.00224 | 0.850 | |
| Asians (2414)b | rs11666402 | 10080076 | G | 0.421 (558) | 0.406 (644) | 1.03 (0.87-1.21) | 0.093 | 0.7610 | 0.487 |
| rs12460842 | 10083195 | G | 0.758 (501) | 0.721 (631) | 1.20 (0.99-1.45) | 3.549 | 0.0596 | 0.884 | |
| rs3745601 | 10085548 | A | 0.885 (104) | 0.712 (288) | 1.38 (0.99-1.93) | 3.646 | 0.0562 | 0.358 | |
| rs2305795 | 10087052 | A | 0.728 (1105) | 0.689 (1249) | 1.20 (1.06-1.37) | 8.209 | 0.00417 | 0.770 | |
| rs4804122a | 10102944 | T | 0.755 (1111) | 0.738 (1282) | 1.10 (0.96-1.25) | 1.862 | 0.1724 | 0.894 | |
| rs11880388 | 10114573 | G | 0.590 (563) | 0.579 (638) | 1.09 (0.92-1.28) | 0.897 | 0.3437 | 0.310 | |
| rs2228611 | 10128077 | G | 0.594 (561) | 0.577 (644) | 1.11 (0.94-1.32) | 1.637 | 0.2007 | 0.199 | |
| African Americans (302) | rs11666402 | 10080076 | G | 0.273 (108) | 0.256 (117) | 1.09 (0.72-1.66) | 0.162 | 0.6875 | NA |
| rs12460842 | 10083195 | G | 0.574 (95) | 0.474 (116) | 1.49 (1.01-2.17) | 4.146 | 0.0417 | NA | |
| rs3745601 | 10085548 | A | 0.122(74) | 0.121 (112) | 1.01 (0.53-1.91) | 0.00098 | 0.9749 | NA | |
| rs2305795 | 10087052 | A | 0.684 (106) | 0.621 (116) | 1.32 (0.89-1.96) | 1.952 | 0.1624 | NA | |
| rs4804122a | 10102944 | C | 0.314 (113) | 0.307 (189) | 1.04 (0.72-1.48) | 0.035 | 0.8514 | NA | |
| rs11880388 | 10114573 | G | 0.491 (106) | 0.430 (114) | 1.28 (0.88-1.86) | 1.632 | 0.2014 | NA | |
| rs2228611 | 10128077 | G | 0.472 (107) | 0.430 (115) | 1.18 (0.81-1.72) | 0.772 | 0.3795 | NA | |
| All samples (6122) | rs11666402 | 10080076 | G | 0.449 (1979) | 0.421 (2577) | 1.14 (1.04-1.24) | 8.482 | 0.00359 | 0.820 |
| rs12460842 | 10083195 | G | 0.643 (1892) | 0.600 (2439) | 1.25 (1.14-1.37) | 22.31 | 2.32×10-6 | 0.662 | |
| rs3745601 | 10085548 | A | 0.313 (1394) | 0.290 (1987) | 1.28 (1.10-1.48) | 10.87 | 0.00098 | 0.159 | |
| rs2305795 | 10087052 | A | 0.664 (2522) | 0.603 (3167) | 1.28 (1.19-1.39) | 38.28 | 6.14×10-10 | 0.727 | |
| rs4804122a | 10102944 | T | 0.672 (2615) | 0.623 (3439) | 1.22 (1.12-1.32) | 23.87 | 1.03×10-6 | 0.157 | |
| rs11880388 | 10114573 | A | 0.499 (1981) | 0.481 (2580) | 1.08 (0.99-1.17) | 2.962 | 0.08524 | 0.133 | |
| rs2228611 | 10128077 | A | 0.500 (1984) | 0.482 (2598) | 1.08 (0.99-1.17) | 2.817 | 0.09327 | 0.096 | |
Mantel Haenszel (MH) test was performed on all cohorts except for African Americans in order to account for diverse subgroups. Breslow Day (BD) test of heterogeneity results are presented in context of MH testing. For ease of comparison of Odds Ratio (OR), results for the associated allele (rather than minor allele) are presented for each cohort. Most significant SNP is marked with bold.
Original GWA SNP.
Differences in Asian genotype counts reflect samples genotyped at Stanford vs Samples at Stanford combined with those genotyped in China.
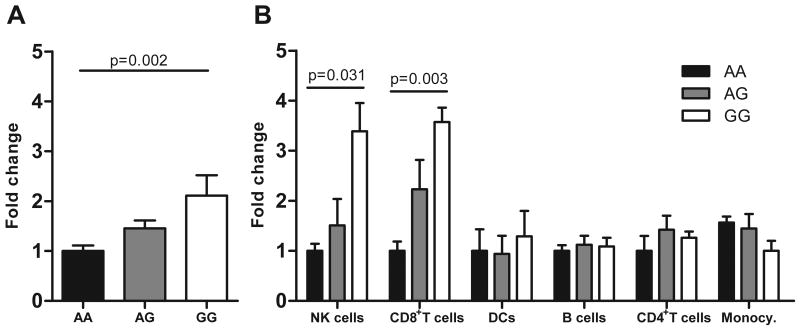
To determine whether the presence of the disease associated SNP was correlated with a significant change in expression of any of the genes in the linkage region (Figure 1), we quantified mRNA expression in peripheral blood mononuclear cells (PBMCs) of PPAN, P2RY11, PPAN-P2RY11, EIF3G and DNMT1 in a Caucasian sample of 60 narcoleptic and 56 control subjects. Expression of P2RY11 mRNA correlated strongly with rs2305795 genotype (2-fold lower expression with the disease associated allele, p=0.002, Figure 2A), sex (lower in females, p=0.039), but not disease status, HLA-DQB1*0602 genotype, age, or body mass index (BMI). The lack of effect of disease status is not surprising considering the current narcolepsy-cataplexy model suggesting rapid hypocretin cell destruction with minimal residual immune response once the destruction is complete (a “hit and run” hypothesis). A weaker correlation was also found with DNMT1 mRNA expression (lower with disease associated allele, p=0.029). As expression of DNMT1 positively correlates with P2RY11 independently of rs2305795 (R2=0.48, p<0.0001), this effect is likely secondary. Expression of the readthrough PPAN-P2RY11 transcript12 was found to be very low (15.5 fold lower than P2RY11 expression) and to vary with sample storage conditions, thus was not further analyzed. Gene expression levels of PPAN and EIF3G did not correlate significantly with rs2305795 or disease status (Supplementary Table 2). These results indicate that rs2305795A, the disease-associated allele, decreases P2RY11 mRNA expression in PBMCs.
Figure 2.
P2RY11 mRNA expression in peripheral blood mononuclear cells (PBMCs). A) Expression in PBMCs from 116 subjects with various rs2305795 genotypes (Mean + SEM, 60 patients and 56 controls; AA n=49, AG n=51, GG n=16). As no direct effect of disease status on P2RY11 expression was observed, subjects are grouped by genotype. The P2RY11 rs2305975AA genotype results in a 50% reduction in P2RY11 expression compared to the rs2305795GG genotype and is associated with increased risk of narcolepsy. B) P2RY11 expression by rs2305795 genotype in various immune cell subsets (Mean + SEM, n=7-8 normal controls per genotype category). NK cells= CD56+ natural killer cells; B Cells = CD19+ B cells; Monocy.=CD14+ monocytes; DCs=myeloid/plasmacytoid dendritic cells. Shown are Bonferroni corrected one-way ANOVA p-values.
P2RY11 is a member of a large family of more than 20 purinergic receptors. Purinergic signaling plays a fundamental role in immune regulation, modulating proliferation, apoptosis, and chemotaxis in lymphocytes, monocytes, and polymorphonuclear granulocytes13,14. Unlike most other purinergic receptors, P2YR11 is a low affinity receptor and detects high concentrations of extracellular ATP. P2RY11 is unique among purinergic receptors, as it is coupled to both Gq and Gs, with activation leading to increases in both cAMP and IP315. In healthy tissue, ATP is mostly localized intracellularly (mM range) and not extracellularly (nM range). During inflammation, however, ATP levels rise in the extracellular space16 and produce a cascade of concentration-dependent effects on the immune system. At lower concentrations, ATP induces immune cell chemotaxis through the stimulation of P2Y2 and P2Y6 receptors17,18,19. High levels of ATP are typically cytotoxic, an effect mediated by the P2X7 receptor20,21,22,23. Because P2RY11 is a pseudogene in rodents, it has been difficult to study its function. P2RY11 expression is widespread, pronounced in both human brain and white blood cells24. In neutrophils, stimulation of P2RY11 delays apoptosis25, and P2RY11 has also been shown to inhibit the migratory capacity of monocyte-derived dendritic cells (MoDCs) and Cd1a+ dermal DCs26. In addition to the lack of P2RY11 in rodents, functional studies have also been hampered by a lack of specific ligands. Most studies have used NF157, a partially selective antagonist27. Only recently have a more specific antagonist (NF340) and an agonist (NF546) been developed28.
To further explore how P2RY11 might regulate the immune system, we next quantified receptor expression in CD4+ T cells, CD8+ T cells, CD56+ natural killer (NK) cells, CD19+ B cells, CD14+ monocytes, and myeloid/plasmacytoid dendritic cell (DC) subsets (a combination of CD1c+, CD141+, and CD304+ cells). P2RY11 expression has previously been shown to be higher in DCs compared to monocytes and CD4+ T cells29, but in that study expression in CD8+ and CD19+ cells was not measured. We found that P2RY11 expression is widespread in immune cells but notably higher in CD8+ cells compared to DCs (2.7-fold lower), CD19+ B cells (2.9-fold lower), CD4+ T cells (4-fold lower), and CD14+ monocytes (6.6-fold lower). Further, the effect of the disease associated allele, rs2305795A, on gene expression was apparent in both CD8+ T cells and NK cells (3-fold reduction across genotypes, Figure 2B), but not in other PBMC subtypes. The smaller genotype effect on expression in PBMCs (Figure 2A) is consistent with a primary effect in CD8+ T cells and NK cells, which represent roughly 25% of total PBMCs.
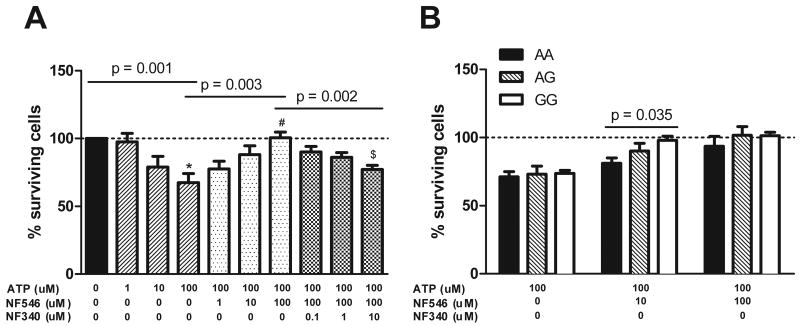
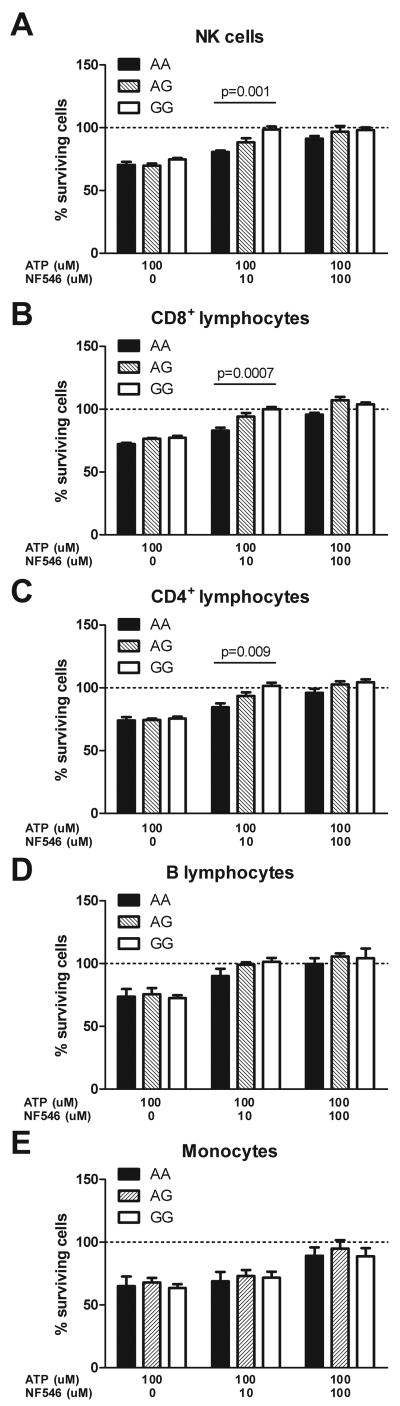
As changes in gene expression do not necessarily translate into differential functional effects, we next studied whether ATP and P2RY11 has genotype-dependent effects on immune cells. As previously reported12, we found that increasing concentrations of ATP induce PBMC cell death (Figure 3A), an effect likely mediated by P2X7 receptor stimulation. Using the recently developed P2RY11 agonist NF546 and antagonist NF340, we further discovered that P2RY11 stimulation mitigates this effect, suggesting that immune cell death in the presence of high ATP is controlled by a balance of activation of multiple purinergic receptors that includes P2RY11. A similar survival effect of P2RY11 stimulation by ATP has been reported in neutrophils24, and endothelial cells following NK cell mediated killing30. Comparing cells with various rs2305795 genotypes, we found that the protecting effect of P2RY11 stimulation was less pronounced in subjects carrying the narcolepsy-associated, low expression rs2305795A genotype, as demonstrated by the lower P2RY11 induced survival in PBMCs of this genotype (Figure 3B). To determine whether these effects vary by immune cell subsets, we used fluorescence activated cell sorting (FACS) and found significant genotype effects in NK (p=0.001), CD8+ T (p=0.0007) and CD4+ T cells (p=0.009), but not in monocytes or B cells (Figure 4). This result is in line with expression data reported in Figure 2B, although we also found genotype-dependent effects in CD4+ T cells, a population without P2RY11 expression differences, a finding possibly reflecting differential P2RY11 responses in various CD4+ T cell subsets.
Figure 3.
PBMC cell death induced by ATP is inhibited by the stimulation of P2RY11 and varies by rs2305795 genotype. A) Effect of ATP on cell viability and dose-response of co-incubation with the P2RY11 specific agonist NF546 and antagonist NF340 (Mean + SEM, n= 7-8, rs2305795AG control subjects). Overall one-way ANOVA p-values are shown, with Tukey's post test: * significantly different from control with no ATP, p<0.01; # significantly different from treatment with 100 μM ATP but no NF546, p<0.01; $ significantly different from treatment with 100 μM ATP and 100 μM NF546, p<0.01. B) Effect of the rs2305795 genotype on the percent of cells rescued from ATP induced cell death by P2RY11 stimulation. 10 μM NF546 has a less potent effect on cell survival after ATP-induced cell death with the rs2305795AA genotype compared to the rs2305795GG genotype. Heterozygote subjects fall in between. Mean + SEM, n=9 subjects in each group.
Figure 4.
Effect of ATP and P2RY11 co-stimulation on different immune cell subtypes. PBMCs were co-incubated with 100 uM ATP and the P2RY11 specific agonist NF546 in different doses and the effect on different cell fractions was determined by FACS. An effect by rs2305795 genotype was seen in T lymphocytes and NK cells but not in B lymphocytes and monocytes. Shown are mean + SEM, n=8/column, p-values are from one-way ANOVAs.
How could reduced P2RY11 function, associated with rs2305795A, be involved in narcolepsy susceptibility? Our results demonstrate clear effects of the polymorphism on immune cell viability. A possible pathway may thus be modulation by P2RY11 of immune response to an important infectious trigger, such as Streptococcus Pyogenes31, or a modulatory effect of the autoimmune process leading to hypocretin cell destruction. Although our results suggest a novel function for P2RY11 in T cells and NK cells, relevant effects on other cells not measured here are possible, if not likely. For example, P2RY11 induces thrombospondin-1 secretion and inhibits lipopolysaccharide-stimulated interleukin-12 (IL-12) release in monocyte-derived DCs32, an effect that could have a cascade of indirect effects on the immune system. Further, activation of P2RY11 on DCs induces maturation and stimulates IL-8 release33. As IL-8 is an important mediator of neutrophil chemotaxis, modulation of the innate immune system could also be involved. Indeed, it has recently been shown that P2RY11 stimulation modulates NK cell chemotaxis in response to CX(3)CL1 and CXCL1230. Finally, direct effects of P2RY11 on hypocretin cell apoptosis or microglial activation are also possible as virtually nothing is known regarding localized expression and function of P2RY11 in the human brain.
In summary, we report on a novel association of P2RY11 and rs2305795A in narcolepsy. This receptor is highly expressed in CD8+ T cells and NK cells, and modulates immune cell viability. Additional studies of this receptor in narcolepsy and other autoimmune diseases are warranted.
Methods
Subjects
Narcolepsy-cataplexy cases were selected as previously described4. The initial GWA sample was comprised of 807 cases and 1,074 Caucasian DQB1*0602 positive controls: 415 cases and 753 controls were recruited from the United States and Canada; 392 cases and 321 controls were recruited from European centers. Analysis of the GWA data (549,596 SNPs) was performed as previously described4. The Caucasian replication sample contained 1526 individuals, of whom 1195 were recruited from the United States and Canada (404 cases, 791 controls) and 331 from Europe (211 Cases, 120 controls). The Asian sample included 1269 Chinese (588 cases, 681 controls), 869 Japanese (437 cases, 432 controls), and 276 Koreans (105 cases, 171 controls). Finally, we studied 302 African Americans (113 cases, 189 controls). Interaction studies were conducted in the initial set and in replication sets (cases and controls) using a test for epistatic effects implemented in the PLINK software package (v1.06 26/April/2009), which performs a logistic regression including main genotype effects plus an interaction term.
Fine mapping of the chromosome 19 locus, and replication of published SNPs
For fine mapping of the associated locus, we used the same case and control samples as described above. Based on LD and r2 data from the International HapMap Project (Hapmap.org), we chose 5 markers in high LD with our initial hit in individuals of European ancestry but lower linkage in other ethnic groups. The chosen SNPs were rs11666402, rs12460842, rs2305795, rs11880388, and rs2228611. We also typed the potential functional SNP rs3745601 in P2RY11. Previously reported SNP rs2858884 was also genotyped for replication in our sample. For the genotyping we used predesigned Taqman SNP genotyping assays (Applied Biosystems, Carlsbad, CA, USA). Standard D′/LOD plots were generated using Haploview 4.2. Genotyping was performed at Stanford University except for Chinese samples, which were genotyped at Beijing University by one of us (LL).
Selection of cells for mRNA expression analysis
To compare mRNA expression in controls and narcoleptic subjects, 116 subjects (60 patients and 56 controls) were randomly selected. Twenty-four controls were added on the basis of their rs2305795 genotype (age, sex and gender matched within genotypic groups). PBMCs were purified using Ficoll-Hypaque gradient centrifugation, total RNA isolated from ∼4×106 cells (RNeasy Mini Kit #74104, QIAGEN), and RNA concentration/quality determined by absorbance at 260 and 280 nm (SpectraMax M2e, Molecular Devices). In the 24 subjects selected by genotype, ∼ 1×107 PBMCs were sequentially sorted based on CD4, CD8, CD14, and CD19 positivity using Dynabeads (Invitrogen Dynal AS, Oslo, Norway). Two independent replications were performed. In the first, CD19 was separated first, followed by CD14, CD4, and CD8. In the second, CD14 was separated first, followed by CD8, CD4, and CD19. CD8+ T cells and NK cells were separated independently from new sets of ∼1×107 PBMCs. In this experiment, NK cells were purified using MACS CD56+ MicroBeads (NK cell Isolation Kit #130-092-657, Miltenyi Biotec, Bergisch Gladbach, Germany) while CD8+ T-cells were purified from the CD56- fraction (CD8+ T Cell Isolation Kit #130-091-154, Miltenyi Biotec, Bergisch Gladbach, Germany). Finally, for purification of dendritic cells, we performed negative selection using Dynabeads Human DC Enrichment Kit (Invitrogen Dynal AS, Oslo, Norway) followed by positive selection using Blood Dendritic Cell Isolation Kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) on ∼1×108 PBMCs from 21 healthy control blood samples of the 3 genotypes. Purity of the cell fractions was addressed by quantitative RT-PCR of the different cell surface marker mRNAs (see below) and also a smaller sample was sorted using flow compatible Dynabeads (#113.61D, #113.62D, #113.67D, and #125-06D, Invitrogen Dynal AS, Oslo, Norway) and checked by flow cytometry as described below. The MACS Microbead sorted fractions was checked directly with flow cytometry. Purities of the fractions were (mean ± SD): CD4: 97.6% ± 0.4; CD8: 98% ± 1.2; CD14: 92.3% ± 5.9; CD19: 76.1% ± 11.2; CD56: 89.6% ± 4.9.
Gene expression using Real-Time (RT) PCR
cDNA was synthesized from 200 ng total RNA (cell subsets) or 400 ng total RNA (PBMCs) using High Capacity cDNA Reverse Transcription Kit (#4374966, Applied Biosystems). Gene expression was determined by RT-PCR (ABI 7000, Applied Biosystems) using TaqMan gene expression assays (Applied Biosystems). Probe numbers were P2RY11 (Hs01038858_m1), PPAN (Hs00220301_m1), PPAN-P2RY11 (Hs01568729_m1), EIF3G (Hs00959170_m1), DNMT1 (Hs00945899_m1), CD4 (Hs00181217_m1), CD8 (Hs00233520_m1), CD14 (Hs00169122_g1), CD19 (Hs00174333_m1), CD56 (Hs00941830_m1), CD1c (Hs00233509_m1), BDCA-2/CD303 (Hs00369958_m1), NRP1/CD304 (Hs00826128_m1), B2M (#4333766F), UBC (Hs00824723_m1), GAPDH (#4333764F), and ACTB (#4333762F), the latter four serving as endogenous control genes. RT-PCR of CD4, CD8, CD14, CD19, CD56, CD1c, CD303, and CD304 mRNAs were used to verify purity of each sorted cell fraction and samples. Cell fractions had a 30-69000 fold difference in expression between wanted and unwanted markers, except CD19 cells where differences were 12-24 fold and dendritic cells where differences where 5.5-27 fold. Relative quantities of target mRNAs were calculated using the comparative threshold method (Ct-method), with the geometric mean of UBC, GAPDH, and ACTB expression as endogenous controls. Standard deviations (SD) on fold changes were calculated as SD=2ΔΔCt·ln2·SD(ΔCt) with SD(ΔCt) being the SD of ΔCt of all samples in the group.
ATP induced cell death
PBMCs (1×106 cells/ml) from twelve controls selected on the basis of their rs2305795 genotype (age, sex and gender matched between genotypic groups) were incubated 1 or 2 hours in the presence of ATP in different concentrations (0.1, 1, 10, 100 μM), combined with NF546 (0.1, 1, 10, 100 μM) and/or NF340 (0.1, 1, 10 μM) (both compounds were synthesized as described previously27,28). Both compounds were also tested alone (NF546 0.1, 1, 10, 100, and 500 μM; NF340 0.1, 1, 10, and 100 μM), and no effect was seen on cell viability except a tendency to a decrease with 1 μM NF546. All cell work was performed using Ultra Low Attachment plates (24W: #3473, 96W: #7007, Costar, Corning Inc., Corning, NY), and care was taken to flush loosely attached cells of the plates for analysis. The cells were counted in a hemocytometer using Trypan Blue exclusion of dead and dying cells. All measurements were performed in duplicates. In a second setup all surviving cells were subsequently analysed by FACS to determine their immune phenotypes.
FACS analysis of cell phenotypes
Purity of the different cell fractions were checked on a FACscan using the following antibody combinations (antibodies are from BD Biosciences, San Jose, CA). 1) αCD14-FITC (#555397) and αCD4-PerCP-Cy5.5 (#560650) 2) αCD8-FITC (#555366), αCD56-PE (#555516), and αCD3-PerCP, 3) αCD19-PE (#555413) and αCD3-PerCP. For analyzing the phenotypes of the PBMCs surviving the ATP treatment we used a 7-marker panel consisting of: αCD14-FITC (#555397), αCD4-PerCP-Cy5.5 (#560650), αCD3-Pacific Blue (#558117), αCD19-APC (#555415), αCD56-PE (#555516), and αCD8-PE-Cy7 (#557746) also including Aqua Amine Live/Dead cell stain (L34957, Invitrogen). This analysis was performed on a BD LSRII (BD Biosciences) in duplicates. Data was combined with cells counts as described above.
Statistical analysis
Genotype data was maintained in our database (Progeny Lab 7). Allelic tests of association were performed using the PLINK software package (v1.06 26/April/2009). Genome wide association analysis of the original Affymetrix sample has been described previously4. When studying multiple ethnic groups or subgroups (Taiwanese, Chinese, Japanese, Koreans) the Mantel Haenszel test was used together with the Breslow Day test of homogeneity of the Odds Ratio. For statistical analysis of expression data, we used student T-test, one- and two-way ANOVAs in GraphPad Prism Version 5.00 where appropriate, and general linear regression models in Systat 12 Version 12.00.08, with control of relevant covariates (age, sex, BMI), if significant.
Supplementary Material
Acknowledgments
We are indebted to all the participants of the study, most notably narcoleptic patients. Without their contributions this study would not have been possible. This study was supported by National Institutes of neurological Disease and Stroke P50 NS2372. Additional funding included Danish Medical Council 09-066348/FSS to B.R. Kornum, Stanford training grant: Molecular and Cellular Immunobiology, 5 T32 AI07290 to K. Weiner, National Institutes of Mental Health R01 MH080957 to E. Mignot, 5U01 MH079470 to D. Levinson, and National Institutes of Health NS-044199 to M.M. Ohayon. We are also grateful to GAIN (the Genetic Association Information Network, NIH) and KORA (Kooperative Gesundheitsforschung in der Region Augsburg, Germany). The authors extend their thanks to Patricia Chang, Anna Voros, and Jiang Zhang for technical assistance, and Carl Grumet for brainstorming and constant support.
Footnotes
Accession numbers: P2RY11: NM_002566.4; PPAN: NM_020230.4; EIF3G: NM_003755.3; DNMT1: NM_001130823.1.
Author Contributions: EM, BRK, JH, and NR designed the study with valuable input from RCA, HK, LS, KT, and PYK. BRK, MKa, LL, SH, RCA, HK, KW, JLE, and TMi generated molecular data. AH and MUK provided P2RY11 agonist and antagonist. BRK, JF, JH, EM, and NR participated in the data analysis. BRK and EM wrote the manuscript. JF, SW, MKv, DFL, NR and JH read and substantially commented the manuscript. EM, FH, SK, JL, XD, GP, SN, SCH, YH, MH, BH, JM, PB, DK, YSH, ME, AD, ER, PEH, FPo, FPi, BF, JHJ, SPL, KPS, WTL, MMO, and PJ contributed narcolepsy samples. TR, JW, TGNT, MD, GTN, HEW, GAR, CG, TMe, PP, and TY provided samples and/or genotypes. EM provided financial support.
References
- 1.Thannickal TC, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–74. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peyron C, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–7. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 3.Mignot E, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallmayer J, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–11. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cvetkovic-Lopes V, et al. Elevated Tribbles homolog 2–specific antibody levels in narcolepsy patients. J Clin Invest. 2010;120:713–9. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawashima M, et al. Anti-Tribbles homolog 2 (TRIB2) autoantibodies in narcolepsy are associated with recent onset of cataplexy. Sleep. 2010;33:869–74. doi: 10.1093/sleep/33.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyoda H, et al. Anti-Tribbles homolog 2 autoantibodies in Japanese patients with narcolepsy. Sleep. 2010;33:875–8. doi: 10.1093/sleep/33.7.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hor H, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet. 2010;42:786–9. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- 9.Hong SC, et al. DQB1*0301 and DQB1*0601 modulate narcolepsy susceptibility in Koreans. Hum Immunol. 2006;68:59–68. doi: 10.1016/j.humimm.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Amisten S, Melander O, Wihlborg A, Berglund G, Erlinge D, et al. Increased risk of acute myocardial infarction and elevated levels of C-reactive protein in carriers of the Thr-87 variant of the ATP receptor P2Y11. Eur Heart J. 2007;28:13–8. doi: 10.1093/eurheartj/ehl410. [DOI] [PubMed] [Google Scholar]
- 11.Matsuki K, et al. DQ (rather than DR) gene marks susceptibility to narcolepsy. Lancet. 1992;339:1052. doi: 10.1016/0140-6736(92)90571-j. [DOI] [PubMed] [Google Scholar]
- 12.Communi D, Suarez-Huerta N, Dussossoy D, Savi P, Boeynaems JM. Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. J Biol Chem. 2001;276:16561–6. doi: 10.1074/jbc.M009609200. [DOI] [PubMed] [Google Scholar]
- 13.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Therap. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Di Virgilio F, Boeynaems J, Robson SC. Extracellular nucleotides as negative modulators of immunity. Cur Opin Pharmacol. 2009;9:507–13. doi: 10.1016/j.coph.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Communi D, Govaerts C, Parmentier M, Boeynaems JM. Cloning of a human purinergic P2Y receptor coupled to phospholipase C and adenylyl cyclase. J Biol Chem. 1997;272:31969–73. doi: 10.1074/jbc.272.51.31969. [DOI] [PubMed] [Google Scholar]
- 16.Bodin P, Burnstock G. Increased release of ATP from endothelial cells during acute inflammation. Inflam Res. 1998;47:351–4. doi: 10.1007/s000110050341. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, et al. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 2006;314:1792–5. doi: 10.1126/science.1132559. [DOI] [PubMed] [Google Scholar]
- 18.Elliott MR, et al. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature. 2009;461:282–6. doi: 10.1038/nature08296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben Yebdri F, Kukulski F, Tremblay A, Sévigny J. Concomitant activation of P2Y(2) and P2Y(6) receptors on monocytes is required for TLR1/2-induced neutrophil migration by regulating IL-8 secretion. Eur J Immunol. 2009;39:2885–94. doi: 10.1002/eji.200939347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L, Bradley CJ, Wiley JS. P2Z purinoceptor, a special receptor for apoptosis induced by ATP in human leukemic lymphocytes. Chin Med J. 1999;112:356–62. [PubMed] [Google Scholar]
- 21.Aswad F, Kawamura H, Dennert G. High sensitivity of CD4+CD25+ regulatory T cells to extracellular metabolites nicotinamide adenine dinucleotide and ATP: a role for P2X7 receptors. J Immunol. 2005;175:3075–83. doi: 10.4049/jimmunol.175.5.3075. [DOI] [PubMed] [Google Scholar]
- 22.Aswad F, Dennert G. P2X7 receptor expression levels determine lethal effects of a purine based danger signal in T lymphocytes. Cell Immunol. 2006;243:58–65. doi: 10.1016/j.cellimm.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor SRJ, et al. Sequential shrinkage and swelling underlie P2X7-stimulated lymphocyte phosphatidylserine exposure and death. J Immunol. 2008;180:300–8. doi: 10.4049/jimmunol.180.1.300. [DOI] [PubMed] [Google Scholar]
- 24.Moore DJ, et al. Expression pattern of human P2Y receptor subtypes: a quantitative reverse transcription-polymerase chain reaction study. Biochim Biophys Acta. 2001;1521:107–19. doi: 10.1016/s0167-4781(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 25.Vaughan KR, et al. Inhibition of neutrophil apoptosis by ATP is mediated by the P2Y11 receptor. J Immunol. 2007;179:8544–53. doi: 10.4049/jimmunol.179.12.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnurr M, et al. ATP gradients inhibit the migratory capacity of specific human dendritic cell types: implications for P2Y11 receptor signaling. Blood. 2003;102:613–620. doi: 10.1182/blood-2002-12-3745. [DOI] [PubMed] [Google Scholar]
- 27.Ullmann H, et al. Synthesis and structure-activity relationships of suramin-derived P2Y11 receptor antagonists with nanomolar potency. J Medic Chem. 2005;48:7040–8. doi: 10.1021/jm050301p. [DOI] [PubMed] [Google Scholar]
- 28.Meis S, et al. NF546 [4,4′-(carbonylbis(imino-3,1-phenylene-carbonylimino-3,1 (4-methyl-phenylene)-carbonylimino))-bis(1,3-xylene-alpha,alpha'-diphosphonic acid) tetrasodium salt] is a non-nucleotide P2Y11 agonist and stimulates release of IL-8 from human monocyte-derived dendritic cells. J Pharmacol Exp Therapeu. 2010;332:238–47. doi: 10.1124/jpet.109.157750. [DOI] [PubMed] [Google Scholar]
- 29.Duhant X, et al. Extracellular adenine nucleotides inhibit the activation of human CD4+ T lymphocytes. J Immunol. 2002;169:15–21. doi: 10.4049/jimmunol.169.1.15. [DOI] [PubMed] [Google Scholar]
- 30.Gorini S, et al. ATP secreted by endothelial cells blocks CX3CL1-elicited natural killer cell chemotaxis and cytotoxicity via P2Y11 receptor activation. Blood. 2010 doi: 10.1182/blood-2009-12-260828. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 31.Aran A, et al. Elevated anti-streptococcal antibodies in patients with recent narcolepsy onset. Sleep. 2009;32:979–83. doi: 10.1093/sleep/32.8.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marteau F, et al. Involvement of multiple P2Y receptors and signaling pathways in the action of adenine nucleotides diphosphates on human monocyte-derived dendritic cells. J Leukoc Biol. 2004;76:796–803. doi: 10.1189/jlb.0104032. [DOI] [PubMed] [Google Scholar]
- 33.Wilkin F, et al. The P2Y11 receptor mediates the ATP-induced maturation of human monocyte-derived dendritic cells. J Immunol. 2001;166:7172–7. doi: 10.4049/jimmunol.166.12.7172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.