Abstract
The extracellular domain of human CD38 is a multifunctional enzyme involved in the metabolism of two Ca2+ messengers, cyclic ADP-ribose (cADPR) and nicotinic acid adenine dinucleotide phosphate (NAADP). When NAD is used as a substrate, CD38 predominantly hydrolyzes it to ADP-ribose with a trace mount of cADPR produced through cyclization of the substrate. However, a mutation of a key residue at the active site, E146A, inhibits the hydrolysis activity of CD38 but greatly increases its cyclization activity. To understand the role of the residue, E146, in the catalytic process, we determined the crystal structure of the E146A mutant protein with a substrate analogue, ara-2’F-NAD. The structure captured the enzymatic reaction intermediates in six different conformations in a crystallographic asymmetric unit. The structural results indicate a folding-back process for the adenine ring of the substrate and provides the first multiple snap shots of the process. Our approach of utilizing multiple molecules in the crystallographic asymmetric unit should be generally applicable for capturing the dynamic nature of enzymatic catalysis.
Keywords: CD38, cyclization, hydrolysis, cADPR, NAD analogue
Human CD38 was first identified as a T lymphocyte differentiation antigen 1 that is involved in many cellular functions including differentiation, proliferation and apoptosis 2; 3. Its expression is associated with some human diseases such as B cell chronic lymphocyte leukemia 4; 5, AIDS 6 and diabetes 7. The extracellular domain of CD38 is also a multi-functional enzyme 8; 9 that catalyzes not only the hydrolysis of NAD and cyclic ADP-ribose (cADPR) to ADP-ribose, but also the cyclization of NAD to cADPR. Furthermore, at acidic conditions, it can also catalyze the base-exchange reaction using NADP and nicotinic acid as substrates to produce NAADP 10. Both cADPR and NAADP are Ca2+ messengers regulating mobilization of intracellular Ca2+ stores in different types of cells ranging from protozoan to plant to human 11; 12.
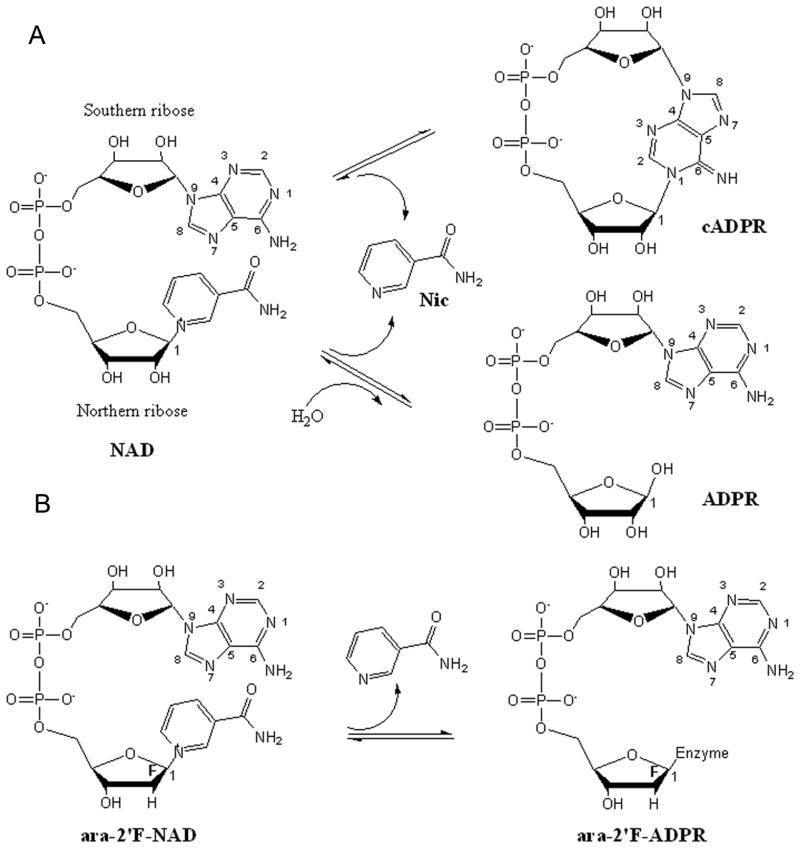
Human CD38 is a type II transmembrane glycoprotein with a short cytoplasmic tail, a single transmembrane segment and a large extracellular domain. A previous crystallographic study has shown that the extracellular domain of human CD38 can be subdivided into two domains, one -helix-rich N domain and one β-strand-rich C domain 13. Between these two domains is located the cavity where catalytically important residues have been identified by site-directed mutation assays and crystallographic studies 14; 15; 16; 17. Of these residues, E226 was shown to be the catalytic residue, while W189 and W125 were responsible for substrate binding. During catalysis, the substrate, NAD, enters the active site cavity with its nicotinamide group stacking with W189 through hydrophobic interaction and forming a hydrogen bond with E146. The 2′-OH and 3′-OH groups of the nicotinamide ribose then form hydrogen bonds with E226 while the diphosphate forms hydrogen bonds with the main chain amine groups of T221 and F222. The first step of the catalytic reaction is the cleavage of the nicotinamide group, leaving a nucleophilic C-1′ atom at the ribose. Either a linear or cyclic product is produced subsequently depending on the nucleophilic group accessible. If it is a water molecule, NAD is hydrolyzed to a linear product, ADP-ribose (ADPR). On the other hand, if the nucleophilic attack comes from the adenine ring that is folded back, NAD is cyclized into cADPR (Fig. 1A).
Figure 1.
Schematic diagram of the CD38-catalyzed reactions of NAD and ara-2′F-NAD. (A) Hydrolysis and cyclization of NAD catalyzed by human CD38. Human CD38 catalyzes the cleavage of the nicotinamide group from the northern ribose of NAD. In the hydrolysis reaction, a water molecule attacks the newly formed C-1′ atom producing linear product ADPR. In the cyclization reaction, the N1 atom of the adenine ring attacks the C-1′ atom producing cyclic product cADPR. (B) Hydrolysis of ara-2′F-NAD. The nicotinamide group is cleaved from the northern ribose and the newly generated C-1′ atom forms a covalent bond with the enzyme.
For wild type CD38, the main product from NAD is the linear ADPR (>99.9%) and the minor circular product cADPR is less than 0.1% 18. A single amino acid mutation at E146 to alanine dramatically changes the reaction balance, producing 3 times more cADPR than the linear ADPR 19. To further understand the role of E146 in the catalysis process, we crystallized this mutant protein with a substrate analog, ara-2′F-NAD (Fig. 1B). The crystal structure captured the CD38-ligand complex in six different conformations in the asymmetric unit. The results provide the first multiple snapshots of the cyclization process.
RESULTS
Overall structure of E146A mutant
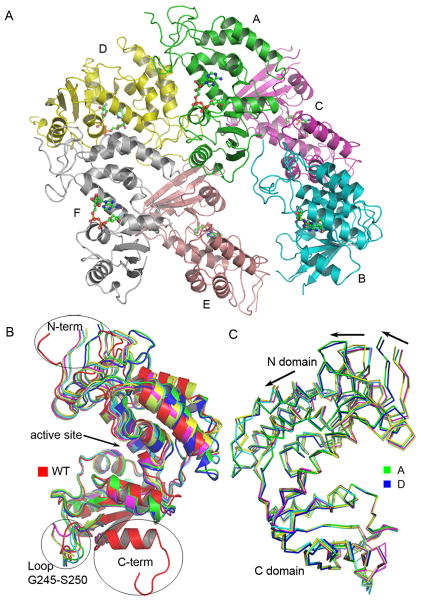
The space group of the CD38 E146A mutant crystals is P212121 and there are six CD38 molecules in the asymmetric unit as shown in Fig. 2A. The overall structure of E146A mutant is very similar to that of the wild type CD38 (PDB code 1YH3) with an RMSD value ranging from 1.08 to 1.78 Å for the Cα atoms of residue T49 to residue R280. Three major structural differences between wild type CD38 and E146A mutant are located at the N-terminus (R45 to Q48), C-terminus (P281 to C296) and a middle loop (G245 to S250), as shown in Fig 2B. The N-terminus of E146A mutant swings about 107° away from that of wild type CD38 due to close crystal packing with the C-terminus of a neighboring molecule. The structure of the C-terminus of E146A mutant is well resolved up to the residue R280. The rest of the residues from P281 to C296 are disordered due to crystal packing. Structural alignment shows that the loop connecting residue H244 and residue R251 is highly flexible, adopting different conformation not only between the mutant and the wild type CD38, but also among all six mutant molecules. Similar flexibility in the N-terminal region is also observed when different molecules of the wild type CD38 are compared 13. The superposition shown in Fig. 2C is based on the main chain atoms of the helix from residue S220 to residue L230. The C domains of these six molecules are well superimposed while there are obvious back-and-forth movements relative to the C domain among the six N domains, especially for the N domains of molecules A and D, indicating a kind of “breathing” action for CD38. These dynamic features of CD38 revealed by the multiple protein-ligand complexes in the asymmetric unit can be important in influencing its catalytic activities, as will be discussed later.
Figure 2.
Overall structure of the E146A mutant. (A) The six molecules (labeled A-F) present in the asymmetric unit of the E146A crystals. Each protein molecule is shown as ribbons and contains one covalent bonded ara-2′ F-NAD that is shown as ball-and-stick models. (B) The Superposition of E146A molecules with wild type CD38 (PDB code 1YH3). Wild type CD38 is shown as red ribbons. Conformational variations are seen at the N-terminus, at a middle loop (G245–S250) and at the C-terminus. (C) The superposition of the six molecules in the asymmetric unit. The molecules are shown as Cα traces with molecules A and D colored in green and blue, respectively. The C domains of these molecules are well superimposed while the helices in the N domains show large variations indicated by the black arrows.
Conformational change of His133
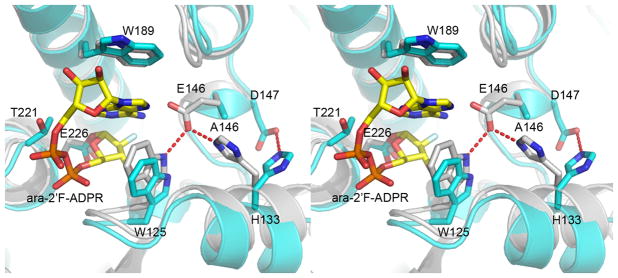
In the wild type CD38, the OE1 atom of residue E146 is at a distance (2.8Ǻ) suitable for forming hydrogen bond with both the NE1 nitrogen atom of residue W125 and the NE2 nitrogen atom of H133 (Fig. 3). This kind of hydrogen network is present in all previously determined CD38 structures, whether in apo form or in complex with a substrate or product. It is interesting that in the structure of the Aplysia ADP-ribosyl cyclase, a CD38 homolog, similar hydrogen bonding is also conserved. In contrast, in the E146A mutant, this hydrogen network is diminished, as the side chain of residue H133 swings away from A146 in an energetically more favorable conformation. In two of the six molecules (E and F) of the E146A mutant, H133 forms a weak hydrogen bond with D147. The flipping away of H133 is only observed in the E146A mutant and is likely the result of the E146A mutation. As will be discussed later, the mutation has major effects on the enzymatic reactions catalyzed by the mutant.
Figure 3.
Stereo view of the superimposed wild type CD38 and E146A mutant at the active site. Wild type CD38 (colored in grey) in complex with ara-2′F-ADPR (PDB code 3I9M) is superimposed onto the E146A mutant (colored in cyan). Residues in the active site including E226, T221, W189, W125, E146/A146, H133 and D147 are shown as ball-and-stick models. Ara-2′F-ADPR in the E146A mutant structure is also shown as ball-and-stick models with yellow carbons. In wild type CD38, E146 forms hydrogen bonds with W125 and H133. In the E146A mutant, H133 swings away from the active site and forms a hydrogen bond with D147 in all six conformations.
Two different conformations for two different catalytic reactions
Human CD38 catalyzes the hydrolysis of NAD to produce a linear product, ADPR (Fig. 1A), which is then released from the active site. For the NAD analogue, ara-2′F-NAD (Fig. 1B), it is also hydrolyzed but the product, ara-2′F-ADPR, is retained in the active site and forms a covalent bond between the C-1′ carbon atom of ara-2′F-ADPR and the OE2 oxygen atom of residue E226 (Fig. 4A and Fig. 4B). The covalent linkage anchors the intermediate at the active site but allows the adenine ring to fold back, as it would during normal cyclization. The covalent linkage also stabilizes the intermediate and blocks the cyclization from completion. Ara-2′F-NAD thus functionally behaves as a catalysis-dependent inhibitor of CD38 20. In the asymmetric unit of the E146A mutant, all six molecules have a covalently bound ara-2′F-ADPR in their active sites (Fig. 2A and Fig. 4C). The interactions of the bound ara-2′F-ADPR with the active site are very similar to those of the previously determined bound substrate 15, reaction intermediate 17 or product 21 of human CD38, all showing that the northern ribose and the diphosphate forming extensive interactions with residues W125, S126, K127, T221 and F222. As shown in Fig. 4C, the C-1′ atom of the northern ribose of ara-2′F-ADPR in E146A mutant structure is 1.6 Ǻ away from the OE2 oxygen of residue E226, indicating that they form a monovalent bond. In all six molecules, the adenine ring of the ara-2′F-ADPR stacks against the side chain of residue W189 through hydrophobic interactions. Fig. 4C shows the superposition of the six molecules with the bound products. The ribose and the diphosphate portions of all the ara-2′F-ADPR molecules are well superimposed. However, the adenine rings of the six ara-2′F-ADPRs are in different conformations and can be divided into two groups with the products in molecule A, B and C in one group and the products in molecule D, E and F in the other group as shown in Fig. 4D and Fig. 4E, respectively. In the first group, the N7 atom of the adenine ring points toward the C-1′ carbon atom of the northern ribose (N7 conformation). This is similar to the conformation of the bound ara-2′F-ADPR seen in the wild type CD38 (PDB code 3I9M) 21. In this orientation, cyclization with the C-1′ of the ribose is unlikely to occur because the N7 of the adenine is known to be very unreactive 22. In striking contrast, in the second group (molecules D-F), it is the N1 atom of the adenine that points toward the C-1′ carbon atom of the ribose (N1 conformation), with the shortest distance of 3.16 Ǻ observed in molecule F. The N1 atom of the adenine is highly reactive and is the site of cyclization as determined in the crystal structure of cADPR 23 (Fig. 1A). It thus represents the cyclizing conformation. In Fig. 4F, the conformation of the bound ara-2′F-ADPR in molecule A of the first group is compared with that in molecule D of the second group. It can be seen that the conformations represents roughly a 180 degree rotation of the adenine ring around the adeninyl-ribose bond.
Figure 4.
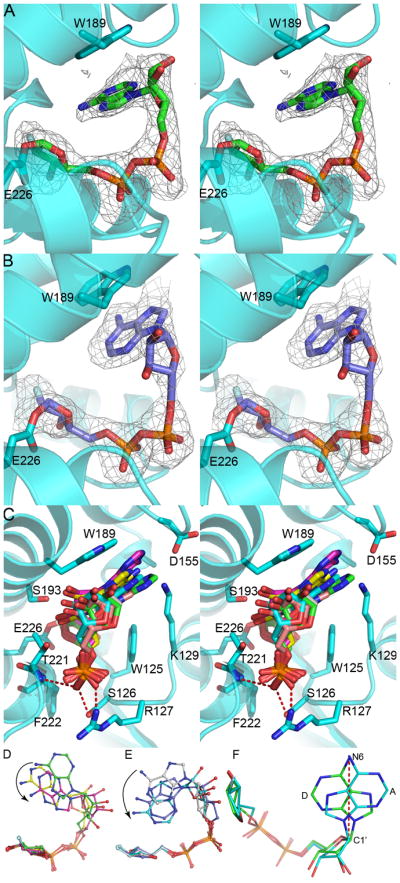
Conformational variations of ara-2′F-ADPR in the active site of the E146A mutant. The protein is shown as cyan ribbons while ara-2′F-ADPR is shown as ball-and-stick models. Stereo view of ara-2′F-ADPR at the active site in N7 conformation (A) and in N1 conformation (B). The Fo-Fc map at 2.0σ is shown as grey mesh. (C) Stereo view of the active site of E146A mutant with ara-2′F-ADPR in the six molecules superimposed. Residues in the active site including W189, S193, E226, F222, T221, W125, S126 and R127 are also shown as ball-and-stick models with cyan carbons. The C-1′ atom is about 1.6Ǻ away from the OE2 oxygen of residue E226. The diphosphate part of ara-2′F-ADPR forms hydrogen bonds with main chain nitrogen atoms of F222 and T221 and side chains of S126 and R127. The adenine ring of ara-2′F-ADPR stacks with the side chain of W189 through hydrophobic interactions. (D) Ara-2′F-ADPR in molecule A, B and C. (E) Ara-2′F-ADPR in molecule D, E and F. The northern ribose and diphosphate part of ara-2′F-ADPR are well superimposed and the adenine rings are in different orientations as shown in (D) and (E). The black arrows indicate the movements of the adenine ring towards the northern ribose. (F) Comparison of conformations: the bound ara-2′F-ADPR in molecule A of the first group and that in molecule D of the second group. The adenine rings of molecules A and D are roughly related by a 180-degree rotation through the N6 atom of adenine and the C1′ atom of southern ribose as indicated by a red dash line.
In each group, the alignment shows the progressive movement of the adenine ring toward the C-1′ carbon atom of the northern ribose, as indicated by the arrows in Fig. 4D and Fig. 4E. The use of multiple protein-ligand complexes in the asymmetric unit presented here thus allows a normally dynamic process of enzymatic catalysis to be captured, in a single crystal, as snapshots of varying conformations of the intermediate. The results provide important insights into the critical role of the residue E146 in the CD38 mediated NAD-cyclization, as will be discussed below.
DISCUSSION
In previous studies 13; 15; 16; 17; 21, we have shown that the crystallographic asymmetric unit generally contains two CD38 molecules. In many cases, we have found each molecule complexes with different ligands or the same ligand with different conformations 15. The observation is not unique to CD38 and is extended to its homolog, the Aplysia ADP-ribosyl cyclase 24. In fact, we have shown that the Aplysia cyclase, under appropriate condition, can form multimeric and microtubular structures 25. This suggests the possibility of capturing various conformations of a bounded ligand in a single crystal, if multiple protein-ligand complexes in the asymmetric unit can be achieved. This is clearly much more preferable than comparing ligand conformations between different crystals.
In this study, crystallization conditions were systemically screened to produce crystals with multimers of CD38(E146A) in an asymmetric unit. The E146A mutation is particularly relevant because it changes the activity of CD38 from hydrolyzing NAD to cyclizing it. Crystals with six molecules in an asymmetric unit were obtained and all of them have a molecule of ara-2′F-ADPR covalently linked to the catalytic residue, E226. More remarkable is that all of them are in different conformations, half of them in the cyclizing (N1) conformation and the other half in the non-cyclizing conformation (N7). In each group, alignment reveals a progressive movement of the adenine ring toward the C-1′ of the ribose, the site of cyclization. That they represent snapshots of the dynamic process of cyclization is more than suggestive.
We have previously shown that ara-2′F-NAD also forms the same covalent intermediate, ara-2′F-ADPR, with the wild type CD38 21. In both molecules in the asymmetric unit, the intermediates are only in the non-cyclizing conformation (N7). The more time the intermediate stays in this non-cyclizing conformation, the higher the chance it would be attacked by bound water observed in the active site 15, resulting in hydrolysis. This is consistent with the fact that the wild type CD38 mainly hydrolyzes NAD to ADPR, and the amount of cADPR formed, the cyclizing product, is vanishingly small 18. In remarkable contrast, here we show that ara-2′F-ADPR in half of the CD38(E146A) molecules in the asymmetric unit are in the cyclizing conformation (N1) instead. By stabilizing the productive conformation of the intermediate, the E146A mutation greatly promotes a cyclizing linkage between N1 of the adenine ring and the C-1′ of the ribose. This, again, is consistent with the catalytic property of the mutant CD38, which forms three times as much cADPR than ADPR 19.
The reason why the E146A mutation would result in stabilizing the N1-conformation is likely related to the change observed in the residue, H133. In CD38(E146A), H133 flipped away from the intermediate (Fig. 3). This would relieve steric hindrance at the active site pocket and allow freer rotation of the adenine ring. Indeed, the N1- and N7- conformations can be interconverted by rotation of the adenine ring around the adeninyl-ribose bond (Fig. 4F).
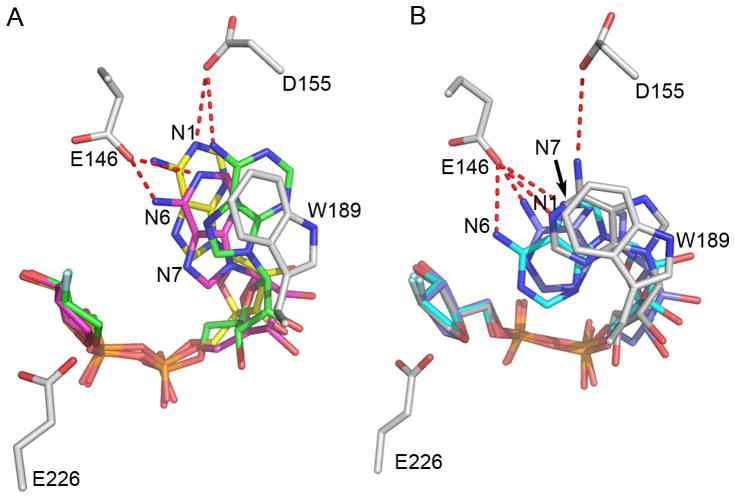
Another important role of the residue, E146, in controlling cyclization verses hydrolysis can be seen in Figure 5. In the wild type CD38, E146 and D155 form extensive hydrogen bonds with N6 and N1 of the adenine ring of the intermediate, which is in the N7-conformation. Superimposed are the three N7-conformations observed in CD38(E146A). Again, extensive hydrogen bonding would be present between E146 and the adenine ring (Fig. 5A). In Fig. 5B, the three N1-conformations observed in CD38(E146A) are superimposed with the wild type CD38. As can be seen, even if the N1 conformation can be formed in the wild type, N1, N6 and N7 of the adenine ring still form extensive hydrogen bonds with E146 and D155 (Fig. 5B). This kind of hydrogen bonding would strongly restrict the ring from folding further back and thus prevent the cyclizing attack of the N1 to the C-1′ of the ribose. The intermediate would thus be stabilized in a non-productive conformation, providing more time for water access and nucleophilic attack of the C-1′. The result is hydrolysis, producing linear product ADPR, as is well documented for the wild type CD38. With a short nonpolar side chain in E146A mutant, this kind of restraint was released and the adenine ring can thus readily fold back, yielding mainly cyclic product. The progressive folding of the adenine ring toward the C1′ of the ribose is shown in Fig. 4D and Fig. 4E where the shortest distance between the N1 atom and the C-1′ atom is about 3.2 Ǻ, a reasonable distance for a nucleophilic attack.
Figure 5.
A local view of two groups of ara-2′F-ADPR in the active site of the E146A mutant with a modeled E146. The wild type CD38 was superimposed onto the E146A mutant and only residues at the active site were shown to make the figure clear. Both ara-2′F-ADPR and residues in the active sites (with grey carbons) including E226, E146, W189 and D155 are shown as ball-and-stick models. In the N7 conformation (A), residues D155 and E146 form hydrogen bonds with the N1 and N6 atoms of the adenine ring of ara-2′F-ADPR while in the N1 conformation (B), they form hydrogen bonds with the N1, N6 and N7 atoms of the adenine ring.
In summary, the multiple protein-ligand complexes in the asymmetric unit described in this study are shown to be helpful in capturing snapshots of the cyclization of NAD catalyzed by CD38, an intrinsically dynamic process. The results provide important insights about the critical role of residue E146 in controlling the process.
Materials and Methods
Protein expression and purification
The E146A mutant of human CD38 was expressed in yeast and produced using a 2L fermentation reactor as described previously14. Briefly, the supernatant of the yeast culture of the E146A mutant was harvested by centrifugation and filtered to remove insoluble materials. The supernatant was adjusted to pH 5.0 with acetate and potassium chloride was added to final concentration of 2M. It was then loaded to a Phenyl Sepharose (GE health care) column and eluted with 50 mM sodium acetate at pH 5.0. The eluted protein was concentrated, desalted and then loaded onto an SP column (GE health care) at pH 5.0. CD38 was eluted with a sodium chloride gradient from 0 to 0.3 M. It was further purified by the Q column (GE health care) at pH8.0 using a sodium chloride gradient from 0 to 0.2M. Final purification was performed using a size-exclusion column, Superdex-75 (GE health care). The purified protein was concentrated to 35 mg/ml for crystallization trials in 20 mM HEPES at pH 7.4. The ara-2′F-NAD compound was synthesized according to established procedures26.
Crystallization, diffraction data collection and structure refinement
The E146A mutant of human CD38 was diluted to 10 mg/ml and mixed with 10 mM ara-2′F-NAD. Crystals were obtained by the hanging droplet vapor diffusion method with the reservoir buffer in 0.1M sodium acetate pH 4.0, 15% PEG 4000, 0.2 M ammonium acetate and 3% isopropanol. They were harvested and soaked in 0.1M sodium acetate pH4.0, 20% PEG 4000 and 15% glycerol and then flash frozen into liquid nitrogen. The diffraction data were collected at 100K at BL17U at the Shanghai Synchrotron Radiation Facility and processed with HKL200027. Molecular replacement was performed using the program Phaser28 from CCP429 and the wild-type human CD38 (PDB code 1YH3) was used as the searching model. The model was refined with Refmac30 and then cycled with rebuilding in Coot31. The catalysis product of ara-2′F-NAD (ara-2′F-ADPR) was built into positive difference electron-density maps after a few restrained refinement runs of the E146A mutant with the stereochemical restraints generated from the program PRODRG32. TLS refinement33 (in 26 groups) was incorporated into the later stages of the refinement process with 11948 atoms independently refined. Solvents were added automatically in Coot and then manually inspected and modified. The final model was analyzed with MolProbity34. Data collection and model refinement statistics are summarized in Table 1.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Space group | P212121 |
|---|---|
| a (Å) | 56.655 |
| b (Å) | 146.662 |
| c (Å) | 191.086 |
| Molecule/asymmetric unit | 6 |
| Wavelength (Å) | 0.97623 |
| Resolution (Å) | 50–2.2 |
| Unique reflections | 79445 (7208) |
| Completeness (%) | 97.7 (89.9) |
| Rsyma (%) | 0.095 (0.354) |
| I/σ(I) | 20.8 (3.6) |
| Redundancy | 7.1 (4.6) |
| Reflections (work/test) | 75411/3969 |
| Rb/Rfree (%) | 20.34/24.96 |
| Ramachandran plot (favored/allowed/outlier %) | 97.3/100/0 |
| Protein Residues/ara-2′F-ADPR/water | 1406/6/392 |
| Average B-factor (Å2) | 18.9 |
| R.m.s.d. from ideal values | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.170 |
Numbers in parentheses are for the highest resolution shell
Rsym=Σhkl|I–<I>|/ΣI, where I is the observed intensity and <I> is the average intensity from observations of symmetry related reflections. A subset of the data (5%) was excluded from the refinement and used to calculate Rfree.
R=Σ||Fo|–|Fc||/Σ|Fo|.
Acknowledgments
This work was supported in part by National Institutes of Health Grant GM061568 (to H. C. L. and Q. H.). This work was also supported by grants HKU 765909M, HKU 769107M and N_HKU 722/08 from the Research Grant Council of Hong Kong and the National Science Foundation of China (to Q. H., H. C. L. and L. H. Zhang). The crystallographic data were collected at the Shanghai Synchrotron Radiation Facility (SSRF).
Abbreviations
- AIDS
Acquired Immune Deficiency Syndrome
- NAD
nicotinamide adenine dinucleotide
- ADPR
adenosine diphosphate ribose
- cADPR
cyclic ADP-ribose
- NIC
nicotinamide
- NADP
nicotinamide adenine dinucleotide phosphate
- NAADP
nicotinic acid adenine dinucleotide phosphate
- ara-2′F-NAD
arabinosyl-2′-fluoro-deoxy-nicotinamide adenine dinucleotide
- HEPES
4-(2-Hydroxyethyl) piperazine-1-ethanesulfonic acid
- ara-2′F-ADPR
arabinosyl-2′-fluoro-deoxy-adenosine diphosphate ribose
- HPLC
high pressure liquid chromatography
- RMSD
root mean square deviation
Footnotes
Accession Number: The coordinates and structure factors of the E146A mutant structure were deposited in the Protein Data Bank with PDB ID: 3OFS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reinherz EL, Kung PC, Goldstein G, Levey RH, Schlossman SF. Discrete stages of human intrathymic differentiation: analysis of normal thymocytes and leukemic lymphoblasts of T-cell lineage. Proc Natl Acad Sci U S A. 1980;77:1588–92. doi: 10.1073/pnas.77.3.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–86. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 3.Ferrero E, Malavasi F. The metamorphosis of a molecule: from soluble enzyme to the leukocyte receptor CD38. J Leukoc Biol. 1999;65:151–61. doi: 10.1002/jlb.65.2.151. [DOI] [PubMed] [Google Scholar]
- 4.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 5.Thornton PD, Fernandez C, Giustolisi GM, Morilla R, Atkinson S, A’Hern RP, Matutes E, Catovsky D. CD38 expression as a prognostic indicator in chronic lymphocytic leukaemia. Hematol J. 2004;5:145–51. doi: 10.1038/sj.thj.6200360. [DOI] [PubMed] [Google Scholar]
- 6.Roussanov BV, Taylor JM, Giorgi JV. Calculation and use of an HIV-1 disease progression score. AIDS. 2000;14:2715–22. doi: 10.1097/00002030-200012010-00011. [DOI] [PubMed] [Google Scholar]
- 7.Antonelli A, Ferrannini E. CD38 autoimmunity: recent advances and relevance to human diabetes. J Endocrinol Invest. 2004;27:695–707. doi: 10.1007/BF03347507. [DOI] [PubMed] [Google Scholar]
- 8.Lee HC, Munshi C, Graeff R. Structures and activities of cyclic ADP-ribose, NAADP and their metabolic enzymes. Mol Cell Biochem. 1999;193:89–98. doi: 10.1007/978-1-4419-8740-2_13. [DOI] [PubMed] [Google Scholar]
- 9.Lee HC. A unified mechanism of enzymatic synthesis of two calcium messengers: cyclic ADP-ribose and NAADP. Biol Chem. 1999;380:785–93. doi: 10.1515/BC.1999.098. [DOI] [PubMed] [Google Scholar]
- 10.Aarhus R, Graeff RM, Dickey DM, Walseth TF, Lee HC. ADP-ribosyl cyclase and CD38 catalyze the synthesis of a calcium-mobilizing metabolite from NADP. J Biol Chem. 1995;270:30327–33. doi: 10.1074/jbc.270.51.30327. [DOI] [PubMed] [Google Scholar]
- 11.Lee HC. NAADP: An emerging calcium signaling molecule. J Membr Biol. 2000;173:1–8. doi: 10.1007/s002320001001. [DOI] [PubMed] [Google Scholar]
- 12.Lee HC. Mechanisms of calcium signaling by cyclic ADP-ribose and NAADP. Physiol Rev. 1997;77:1133–64. doi: 10.1152/physrev.1997.77.4.1133. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q. Crystal structure of human CD38 extracellular domain. Structure. 2005;13:1331–9. doi: 10.1016/j.str.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Munshi C, Aarhus R, Graeff R, Walseth TF, Levitt D, Lee HC. Identification of the enzymatic active site of CD38 by site-directed mutagenesis. J Biol Chem. 2000;275:21566–71. doi: 10.1074/jbc.M909365199. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q. Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis. J Biol Chem. 2006;281:32861–9. doi: 10.1074/jbc.M606365200. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, Kriksunov IA, Moreau C, Graeff R, Potter BV, Lee HC, Hao Q. Catalysis-associated conformational changes revealed by human CD38 complexed with a non-hydrolyzable substrate analog. J Biol Chem. 2007;282:24825–32. doi: 10.1074/jbc.M701653200. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Kriksunov IA, Jiang H, Graeff R, Lin H, Lee HC, Hao Q. Covalent and noncovalent intermediates of an NAD utilizing enzyme, human CD38. Chem Biol. 2008;15:1068–78. doi: 10.1016/j.chembiol.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard M, Grimaldi JC, Bazan JF, Lund FE, Santos-Argumedo L, Parkhouse RM, Walseth TF, Lee HC. Formation and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science. 1993;262:1056–9. doi: 10.1126/science.8235624. [DOI] [PubMed] [Google Scholar]
- 19.Graeff R, Munshi C, Aarhus R, Johns M, Lee HC. A single residue at the active site of CD38 determines its NAD cyclizing and hydrolyzing activities. J Biol Chem. 2001;276:12169–73. doi: 10.1074/jbc.M011299200. [DOI] [PubMed] [Google Scholar]
- 20.Sauve A, Deng H, Angeletti R, Schramm V. A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. J Am Chem Soc. 2000;122:7855–7859. [Google Scholar]
- 21.Liu Q, Graeff R, Kriksunov IA, Jiang H, Zhang B, Oppenheimer N, Lin H, Potter BV, Lee HC, Hao Q. Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase. J Biol Chem. 2009;284:27637–45. doi: 10.1074/jbc.M109.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner GK, Black S, Guse AH, Potter BV. First enzymatic synthesis of an N1-cyclised cADPR (cyclic-ADP ribose) analogue with a hypoxanthine partial structure: discovery of a membrane permeant cADPR agonist. Chem Commun (Camb) 2003:1944–5. doi: 10.1039/b305660k. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Kriksunov IA, Graeff R, Lee HC, Hao Q. Structural basis for formation and hydrolysis of the calcium messenger cyclic ADP-ribose by human CD38. J Biol Chem. 2007;282:5853–61. doi: 10.1074/jbc.M609093200. [DOI] [PubMed] [Google Scholar]
- 24.Graeff R, Liu Q, Kriksunov IA, Kotaka M, Oppenheimer N, Hao Q, Lee HC. Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38. J Biol Chem. 2009;284:27629–36. doi: 10.1074/jbc.M109.030965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Kriksunov IA, Wang Z, Graeff R, Lee HC, Hao Q. Hierarchical and helical self-assembly of ADP-ribosyl cyclase into large-scale protein microtubes. J Phys Chem B. 2008;112:14682–6. doi: 10.1021/jp807990g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sleath P, Handlon A, Oppenheimer N. Pyridine coenzyme analogs. 3. Synthesis of three NAD+ analogs containing a 2′-deoxy-2′-substituted nicotinamide arabinofuranosyl moiety. The Journal of Organic Chemistry. 1991;56:3608–3613. [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology. 1997:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D: Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Schuttelkopf A, Van Aalten D. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica Section D: Biological Crystallography. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 33.Painter J, Merritt E. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 34.Davis I, Leaver-Fay A, Chen V, Block J, Kapral G, Wang X, Murray L, Bryan Arendall W, III, Snoeyink J, Richardson J. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic acids research. 2007 doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]