Abstract
The developmental activity of LIM homeodomain transcription factors (LIM-HDs) is critically controlled by LIM domain-interacting cofactors of LIM-HDs (CLIM, NLI, LDB). CLIM cofactors associate with Single stranded DNA binding proteins (SSDPs, also known as SSBPs) thereby recruiting SSDP1 and/or SSDP2 to LIM-HD/CLIM complexes. Although evidence has been presented that SSDPs are important for the activity of specific LIM-HD/CLIM complexes, the developmental roles of SSDPs are unclear. We show that SSDP1a and SSDP1b mRNAs are widely expressed early during zebrafish development with conspicuous expression of SSDP1b in sensory trigeminal and Rohon-Beard neurons. SSDP1 and CLIM immunoreactivity co-localize in these neuronal cell types and in other structures. Over-expression of the N-terminal portion of SSDP1 (N-SSDP1), which contains the CLIM interaction domain, increases endogenous CLIM protein levels in vivo and impairs the formation of eyes and midbrain-hindbrain boundary. In addition, inhibition of SSDP1 via N-SSDP1 over-expression or SSDP1b knock down impairs trigeminal and Rohon-Beard sensory axon growth. We show that N-SSDP1 is able to partially rescue the inhibition of axon growth induced by a dominant-negative form of CLIM (DN-CLIM). These results reveal specific functions of SSDP in neural patterning and sensory axon growth, in part due to the stabilization of LIM-HD/CLIM complexes.
Introduction
LIM homeodomain transcription factors regulate the development of a variety of different cell types and structures in the nervous system (Bach, 2000; Hobert and Westphal, 2000; Lee and Pfaff, 2001). The association of the LIM domains with the LIM interaction domain (LID) of CLIM cofactors (also known as NLI, Ldb or Chip) is required to exert the in vivo activity of LIM-HD proteins (Matthews and Visvader, 2003). Indeed, disturbing the interaction between CLIM and LIM domains via overexpression of the LID causes phenotypes that are similar/comparable to inhibitions of specific LIM-HD genes (Segawa et al., 2001; Becker et al., 2002; Mukhopadhyay et al., 2003; Thaler et al., 2002).
Single stranded DNA Binding protein 1 (SSDP1) was originally identified by its ability to bind to single-stranded poly-pyrimidine sequences (Bayarsaihan et al., 1998). Later it was found to participate in protein complexes recruited by CLIM cofactors. SSDP1 synergizes with Xlim1/Lhx1 and Ldb1/CLIM2 during gastrulation in Xenopus, and with Apterous and Chip/CLIM during wing development in Drosophila (Chen et al., 2002; van Meyel et al., 2003). Intronic disruption of the SSDP1 gene in mice revealed important roles in the development of late head organizer tissues by activating the Lim1/Lhx1-Ldb1/CLIM complex (Nishioka et al., 2005). SSDP1 exerts parts of its positive activity by preventing the binding of the ubiquitin ligase RLIM to CLIM thereby protecting the LIM-HD/CLIM complex from proteasomal degradation in cells in culture (Gungor et al., 2007; Xu et al., 2007). This is mediated by the N-terminal 94 amino acids that harbor the CLIM interaction domain.
Protein complexes assembled by LIM-HDs are dynamic and vary in different cell types. The formation of tetrameric complexes consisting of two LIM-HD/CLIM dimers appear widespread (van Meyel et al., 1999; Milan and Cohen, 1999). Indeed, the development of specific interneurons requires the formation of a tetrameric complex consisting of Lhx3 bound to CLIM. However, for the differentiation of motor neurons which originate from a common precursor cell, additional expression of the LIM-HD Isl1 and the transient formation of a hexamer complex consisting of each of two Lhx3/Isl1/CLIM molecules is indispensable (Thaler et al., 2002). This illustrates that the composition of LIM-HD multiprotein complexes dictates the developmental read-out. Thus, the correct formation of LIM-HD complexes is critical and cellular stoichiometries of proteins that participate in LIM-HD complexes are crucial for the correct formation of LIM-HD complexes (Fernandez-Funez et al., 1998; Matthews and Visvader, 2003; Hiratani et al., 2003; Song et al., 2009).
To better understand how SSDP1 proteins control these varied and specific functions of LIM-HD complexes in vivo, an analysis of specific SSDP1 phenotypes in relation to CLIM phenotypes is necessary. Although the genes encoding CLIM and SSDP1 cofactors have been mutated/disrupted in several organisms, their functions in conjunction with LIM-HDs and mechanisms of regulation are poorly understood due to the complexity of the knockout/mutant phenotypes and the widespread developmental effects caused by mutation/deletion of SSDP1 and CLIM genes (Mukhopadhyay et al., 2003; van Meyel et al., 2003; Nishioka et al., 2005).
We have used embryonic zebrafish to elucidate the expression and developmental functions of SSDP1 cofactors during neural development in vivo. Inhibiting SSDP cofactors results in specific defects of axon growth of trigeminal as well as Rohon-Beard sensory neurons, and perturbs patterning of the eye and the mid-hindbrain boundary. Morpholino knock down indicate a specific function for SSDP1b in sensory axon growth. The observed phenotypes overlap with those resulting from CLIM perturbation. A truncated form of SSDP1 stabilizes CLIM in zebrafish and rescues motor axon pathfinding errors induced by CLIM perturbation. Thus, our results reveal functions of SSDP1 in instructing axon outgrowth from specific neuronal populations and in neural patterning, exerted at least in part via stabilization of CLIM cofactors.
Material and Methods
Animals
Zebrafish were kept under standard conditions (Kimmel et al., 1995). We used wild-type (wik) and Tg(hb9:gfp) (Flanagan-Steet et al., 2005) embryos.
Cloning
A search of the Ensembl database (www.ensembl.org/Danio_rerio/) predicted ENSDARG00000030155 and ENSDARG00000058237 for SSDP1a and SSDP1b, respectively, to be most closely related to the mouse and human genes. Based on this information, the entire genes could be isolated from cDNA prepared from adult zebrafish brains using PCR with the primers SSDP1a (5’- CAT GTT TCC TAA AGG CAA AAGC -3’; 5’- GAT GCA GGT CAG TCC CAC C -3’) and SSDP1b (5’- ATG TTC GCC AAA GGT AAA GGG -3’; 5’- TCA CAC ACT CAT TGT CAT TGT TG -3’) using the MolTaq DNA polymerase (Vhbio, Newcastle, UK). Sequences were deposited with the GenBank database (SSDP1a: GQ903695.1; SSDP1b: GQ903696.1).
Morpholino and mRNA injection
Morpholinos to SSDP1a (CTAGAATGAGCTTCTTACCTCTGAC) and SSDP1b (GGATAAATATGCAACACACCTCTGA), targeting the exon/intron boundary of exon 2 for both genes, and an inactive control morpholino (GGTCCCTTTACCTTTGGCGAACATG) were purchased from Gene Tools (Philomath, OR). Effectiveness of the morpholinos was tested by PCR using primers in exons 1 and 5 (SSDP-1a forward, exon 1: 5- GCT CCG TCG TGC CAT CG -3, reverse, exon 5: 5- TCC CAT CTC CTG GTG GC -3; SSDP-1b forward, exon1: 5- GGA CCG CGG TGC CAT CG -3; reverse, exon 5: 5- GGT ATA GGG CCG CCT GGC -3).
Messenger RNAs for injection experiments were synthesized as described previously (Becker et al., 2002). We used the N-terminal 92 amino acids of mouse SSDP1 (N-SSDP1), which are highly conserved (94.5% and 97.8% amino acid identity with zebrafish SSDP1a and SSDP1b, respectively), dominant negative CLIM, full length zebrafish SSDP1b, and control RNA, containing a nuclear localization sequence and a myc epitope (NLS-myc; Becker et al., 2002). For injections, rhodamine dextran (0.8%; Invitrogen, Paisley, UK) was added to mRNA or morpholino solutions. A glass micropipette was filled with 1 – 6 µg/µl mRNA and injected at a volume of approximately 1 nl/egg (one-to four-cell stage). Results for mRNA injections were pooled because no systematic differences were observed in phenotypes between different concentrations. Morpholino solutions were injected in the same way at concentrations of 0.5 and 1 mM as described previously (Feldner et al., 2005).
In situ hybridization and RT-qPCR
Full length SSDP1a and SSDP1b probes were labeled with digoxigenin using the Megascript kit (Ambion, Warrington, UK) and used on 2 to 24 h post fertilization (hpf) whole-mounted embryos as described (Feldner et al., 2005). Additional probes to label head structures have been described (Becker et al., 2002). RT-qPCR was performed as reported previously (Gungor et al., 2007).
Western blots
Western blots on total extracts of embryonic zebrafish (24 hpf) and αT3 cells, a cell line derived from gonadotrope cells of the mouse pituitary (Windle et al., 1990), were carried out essentially as described previously (Becker et al., 2002; Tursun et al., 2005). Blots were incubated with antibodies directed against SSDP1 (Abnova), Myc (Santa Cruz), GAPDH (Chemicon), α-tubulin (Sigma), GFP (Chemicon) and CLIM (Ostendorff et al., 2002). Protein A horseradish peroxidase (HRP; BioRad) was used to visualize specific bands. Infections with retrovirus carrying GFP-N-SSDP1 or GFP-SSDP1 of αT3 cells were performed as described (Gungor et al., 2007).
Immunohistochemistry
Whole-mount immunohistochemistry was performed as described previously (Becker et al., 2002). Ventral motor axons were labeled with a monoclonal antibody against acetylated tubulin (6-11B-1; Sigma-Aldrich, Poole, UK). The 412 monoclonal antibody to the HNK-1 epitope labels cell surface and Golgi apparatus of most neurons of the early nervous system (Becker et al., 2001). The antibody 3A10 to a neurofilament-associated antigen, which labels commissural primary ascending interneurons in the spinal cord and Mauthner axons, was obtained from the Developmental Studies Hybridoma Bank maintained by The University of Iowa, Department of Biological Sciences (Iowa City, IA). Antibodies to SSDP1 and CLIM were the same as for Western blots. Secondary Cy2, Cy3 or HRP-coupled antibodies were from Jackson Laboratories (Newmarket, UK).
Quantification of axons
Axons were counted as previously described (Becker et al., 2002). Briefly, axon fascicles, axons or branches were counted within one cell diameter away from the ganglion on the upper ganglion in laterally-mounted, anti-tubulin immuno-labeled embryos. Rohon-Beard axons or branches were counted in the most dorsal aspect of the upper epidermis. Primary motor axons were counted on both sides of the embryos in segments 7–14. Control and treatment groups were always processed in parallel from the same clutch of eggs to avoid differences in overall development from influencing results. The observer was blinded to the treatments. Counts are collectively expressed as number of axons, comprising single axons, fascicles of axons and axonal branches.
Results
SSDP1 cofactors co-localize with CLIM during zebrafish nervous system development
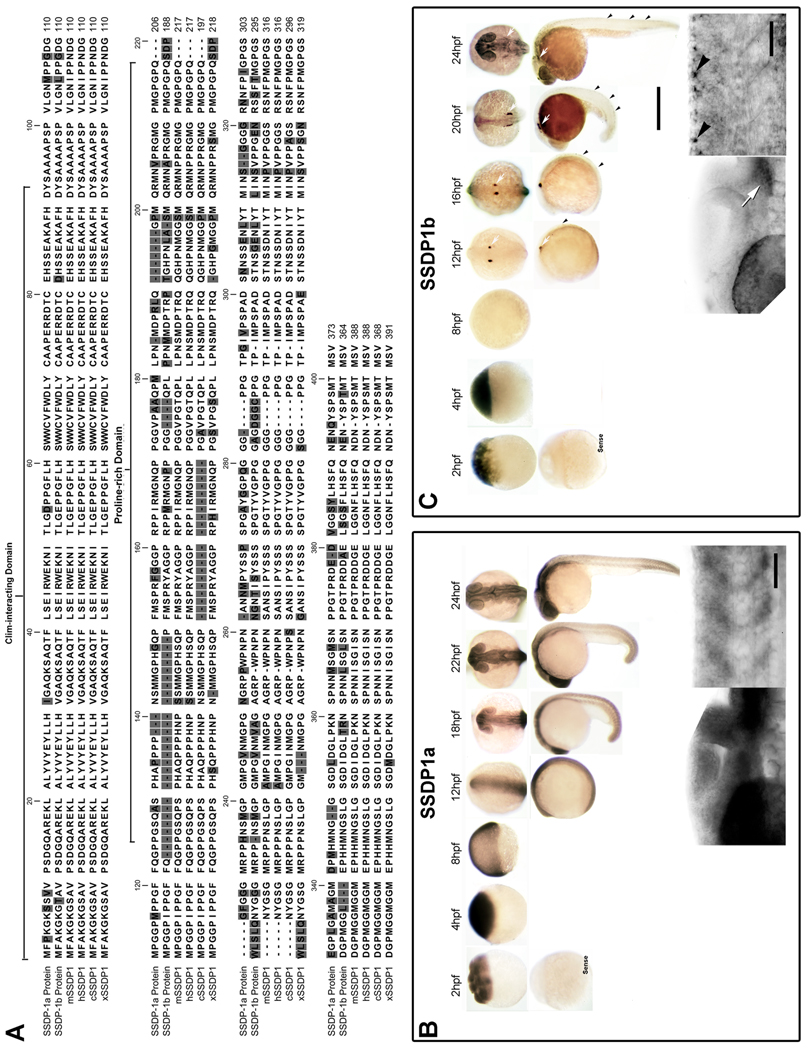
We cloned two SSDP1 genes from the zebrafish genome, SSDP1a (bankit1264174 GQ903695) and SSDP1b (bankit1264294 GQ903696), encoding proteins of 373 and 364 amino acids (aa), respectively (Fig. 1A). Sequence comparisons reveal that SSDP1a and SSDP1b share 70% aa identity. The predicted overall aa identities with human and mouse SSDP1 are 78% and 77% for SSDP1a, respectively, and 76% and 81% for SSDP1b, respectively (Fig. 1A; Suppl. Fig. 1). These high sequence conservations suggest important functions of both zebrafish SSDP1s.
Fig. 1.
SSDP1a and SSDP1b protein sequences are conserved and mRNAs are expressed during embryonic development in zebrafish. A: The CLIM- interacting domain is most strongly conserved between zebrafish SSDP1s and the mouse (mSSDP1), human (hSSDP1), chick (cSSDP1) and Xenopus (xSSDP1) protein. B, C: Whole-mount in situ hybridization between 2 and 24 hpf indicates that SSDP1a is ubiquitously expressed with stronger expression in the head, whereas SSDP1b mRNA is most strongly expressed in trigeminal (white arrows) and Rohon-Beard (black arrowheads) neurons. Higher magnification pictures in lower row show lateral views of the head (left) and trunk (right). Upper scale bar in C = 500 µm for low magnification pictures; lower scale bar = 50 µm for high magnification pictures.
To investigate developmental functions of SSDP1 genes we first localized gene expression by in situ hybridization. This showed that SSDP1a mRNA was ubiquitously detectable in the developing embryo from 2 to 12 hours post-fertilization (hpf) (Fig. 1B). At 18–24 hpf the expression of SSDP1a was still widespread but concentrated more towards anterior embryonic regions including brain and eye (Fig. 1B). Similar to SSDP1a, mRNA encoding SSDP1b was also highly and ubiquitously present at 2 to 4 hpf (Fig. 1C). However, in contrast to SSDP1a, we detected only very low SSDP1b mRNAs levels at 8 hpf. Strong SSDP1b mRNA expression was detected in Rohon-Beard neurons and in trigeminal neurons from 12 hpf onwards (Fig. 1C). These results suggest that the mRNA detectable at 2–4 hpf is maternally derived and zygotic expression of SSDP1b only starts at 12 hpf in sensory neurons. In zebrafish, zygotic mRNA synthesis starts at approximately 3 hpf (Kane and Kimmel, 1993). Our data show a highly dynamic expression pattern of SSDP1 mRNAs and suggest functions of SSDP1b for the development of trigeminal and Rohon-Beard neurons.
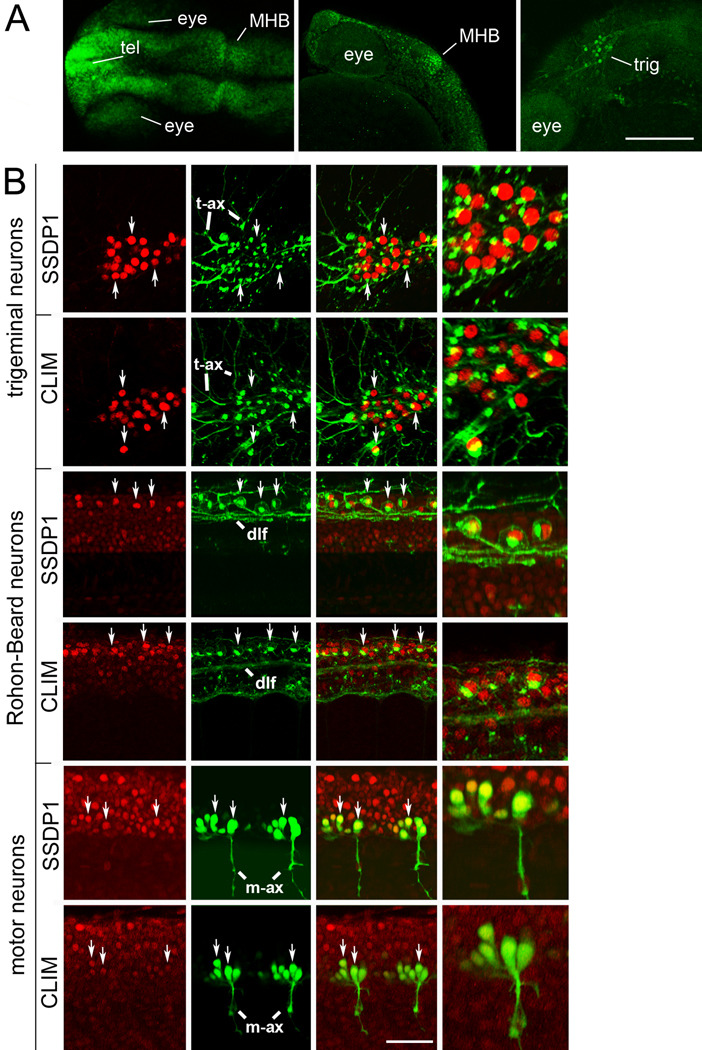
To further elucidate expression of SSDP1 in conjunction with the LIM-HD/CLIM complex we compared the expression profile of SSDP1 and CLIM cofactors early during zebrafish development at the protein level, as LIM-HD recruited protein complexes are regulated by the proteasome (Gungor et al., 2007). We used a specific SSDP1 antibody that recognizes predominantly zebrafish SSDP1b. This was indicated by recognition of a protein band in zebrafish tissue that was identical in size to that found in mouse cells in Western blot analysis and by reduced immunohistochemical labeling after selective morpholino knock down of SSDP1a and SSDP1b in embryos (Suppl. Fig. 2A,B). We found widespread expression of the antigen in embryos at 24 hours post-fertilization (hpf) in various neuronal structures including telencephalon and midhindbrain boundary (MHB) (Fig. 2A). Double-labeling experiments with antibodies directed against the HNK-1 epitope, recognizing neuronal cell membranes and Golgi apparatus (Becker et al., 2001), identified Rohon-Beard and trigeminal neurons as cells with particularly high expression of SSDP1 protein (Fig. 2B). This was expected because of the strong expression of SSDP1b mRNA in these cell types and the preferential labeling of SSDP1b by the antibody. CLIM was also strongly expressed in Rohon-Beard and trigeminal neurons. Antibody labeling of SSDP1 and CLIM in transgenic fish, in which motor neurons express green fluorescent protein under the motor neuron specific promoter HB9 (Flanagan-Steet et al., 2005), revealed lower expression of SSDP1 and CLIM in primary motor neurons (Fig. 2B). All trigeminal and Rohon-Beard cells, defined by HNK-1 immunohistochemistry, and all motor neurons, defined by HB9:GFP transgene, were immuno-positive for SSDP1 and CLIM in their nuclei, indicating co-localization of the antigens. These results are consistent with the expression profile of SSDP1 mRNAs and show that cofactors SSDP1 and CLIM co-localize in specific neuronal cell types including trigeminal neurons, Rohon-Beard neurons and primary motor neurons.
Fig. 2.
SSDP1 protein expression corresponds to in situ hybridization patterns and SSDP1 is co-expressed with CLIM protein in neuronal cell types. A: Dorsal (left) and lateral (middle, right) views of the head labeled with SSDP1 antibodies at 24 hpf are shown. Strong immuno-reactivity is detectable in the telencephalon (tel), in the MHB and in the trigeminal ganglion (trig). The right panel is a selective z-stack of the epidermal region. Scale bar: 200 µm. B: Neuronal localization of SSDP1 and CLIM immunoreactivity (red) is revealed by co-labeling with an HNK-1 antibody (trigeminal and Rohon-Beard neurons in green) or by transgenic GFP expression in HB9:GFP transgenic embryos (motor neurons in green) at 24 hpf. The right outer column gives a higher magnification of double-labeled neurons. HNK-1 is strongly labeled on axonal membranes and Golgi-apparatus next to nuclear labeling of SSDP1 and CLIM in trigeminal and Rohon-Beard neuron. In GFP expressing motor neurons, nuclear labeling is found inside the somata. Some double-labeled neurons are indicated by arrows. Trigeminal (t-ax) and motor axons (m-ax) as well as the dorsal longitudinal fascicle (dlf) are indicated for orientation. Scale bar: 50 µm for low and 100 µm for high magnification.
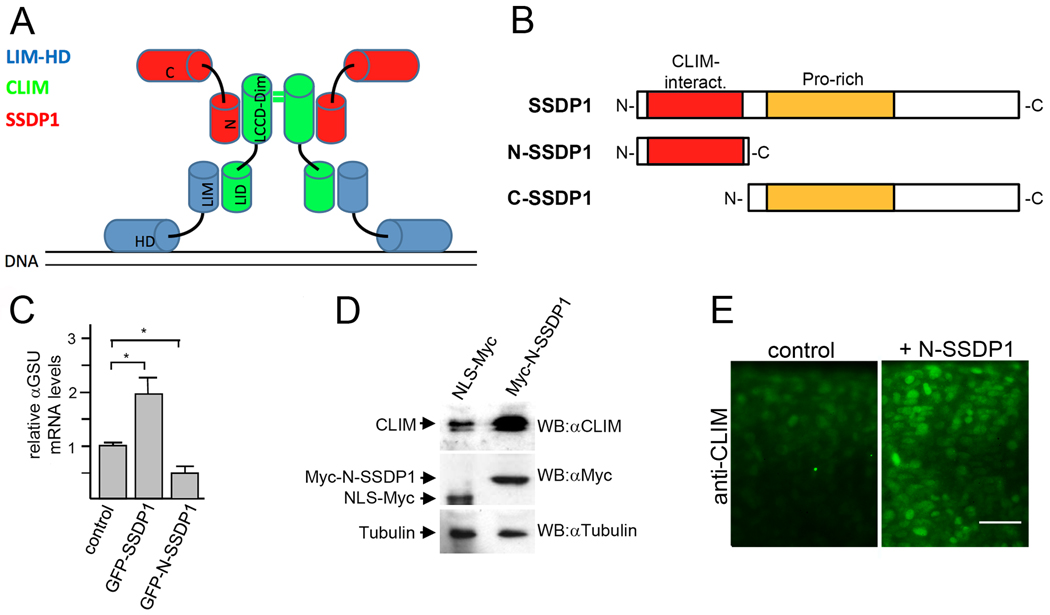
An N-terminal fragment of SSDP1 does not transactivate LIM-HDs, but protects CLIM in vivo
SSDP1 forms complexes with CLIM and LIM-HDs on DNA (Fig. 3A). We have previously generated a dominant-negative CLIM (DN-CLIM) molecule that contains the LIM interaction domain (LID) of CLIM, able to compete with endogenous CLIM for binding to LIM domains (Bach et al., 1999). When ectopically over-expressed early during zebrafish development, DN-CLIM induces specific phenotypes that resemble phenotypes of targeted LIM-HD gene deletions in mice (Becker et al., 2002). To evaluate whether such an approach is also feasible for the elucidation of functions of SSDP1 we examined the potential of the N-terminal 92 amino acids of SSDP1 (N-SSDP1), which contains the CLIM-interaction domain (van Meyel et al., 2003), but lacks a proline-rich domain (Enkhmandakh et al., 2006) as well as a C-terminal transactivation domain (Wu, 2006) (Fig. 3B). We used the pituitary αT3 cell line (Windle et al., 1990) to test effects of N-SSDP1 on the transcriptional activations mediated by LIM-HD proteins. This cell line endogenously expresses the LIM network proteins Lhx3, CLIM and SSDP1 and the Lhx3 target gene αGSU (Bach et al., 1995). Indeed, cofactors CLIM and SSDP1 associate with the αGSU promoter critically regulating αGSU transcription in these cells (Bach et al., 1997; Gungor et al., 2007). Overexpression of SSDP1 in full length fused to GFP via lentiviral infection led to increased endogenous αGSU mRNA levels in αT3 cells. In contrast, overexpression of GFP-N-SSDP1 (Suppl. Fig. 2C) resulted in significantly decreased αGSU mRNA levels when compared to control (Fig. 3C).
Fig. 3.
N-SSDP1 inhibits LIM-HD transcriptional complex in vitro and stabilizes CLIM in vivo. A: Model of transcriptional complexes consisting of LIM-HDs (blue), CLIM (green) and SSDP1 (red) on DNA. LIM domain (LIM); homeodomain (HD); LIM interaction domain (LID); Ldb/Chip conserved domain (LCCD); dimerization domain (dim); N- and C-terminal portions of SSDP1 (N and C, respectively) are indicated. B: N-SSDP1 contains the CLIM interacting domain, but not the proline-rich domain. C-SSDP1 lacks the CLIM interacting domain of SSDP1. C: Overexpression of full-length SSDP1 in alphaT3 cells significantly increases transcriptional activity of LIM-HD factors in these cells whereas GFP-N-SSDP1 decreases it, as indicated by relative levels of the target alphaGSU mRNA measured by RT-qPCR. D: Western Blot analysis of embryos (24 hpf) after over-expression of myc-tagged N-SSDP1 shows increased CLIM protein levels as compared to embryos injected with control mRNA for myc-tagged NLS. E: N-SSDP1 mRNA injection increases CLIM immunofluorescence in embryos, confirming Western Blot results. Lateral views of the trunk region of whole-mounted embryos (24 hpf) are shown. Scale bar = 25 µm.
Another activity of SSDP1 is its ability to stabilize CLIM cofactors in cells in culture (Gungor et al., 2007; Xu et al., 2007). To test the ability of N-SSDP1 to stabilize CLIM cofactors in vivo we injected mRNA encoding Myc-N-SSDP1 into zebrafish embryos at the one-to-two cell stage and tested endogenous CLIM levels. The over-expression of N-SSDP1 resulted in 3.2× increased CLIM levels in Western blots of extracts prepared from 24 hpf embryos when compared with NLS-myc control mRNA injected animals (Fig. 3D). In addition, although non-quantitative, the immunoreactivity of CLIM antibodies in immunohistochemical experiments was stronger in embryos injected with N-SSDP1 when compared to control (Fig. 3E). These results show that while N-SSDP1 has lost its ability for enhancing the transcriptional activity of LIM-HD complexes in vitro, it still protects CLIM co-factors from proteasomal turn-over in zebrafish embryos. Thus, the overexpression of N-SSDP1 in cells is predicted to inhibit functions mediated by C-terminal sequences of SSDP1 and SSDP2 including their transactivation potentials, but not their CLIM-stabilizing functions mediated by N-terminal sequences.
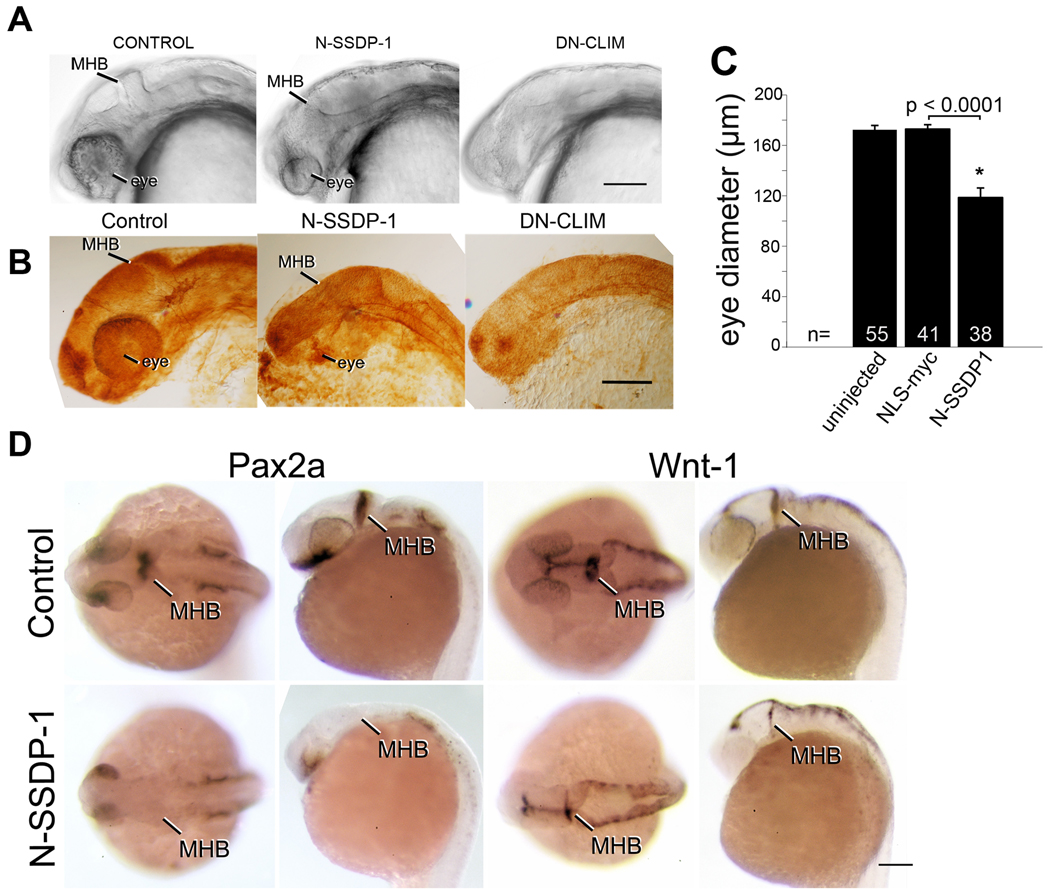
N-SSDP1 over-expression inhibits eye and MHB development
To elucidate functions of SSDP1 in conjunction with LIM-HDs, we compared phenotypes induced by ectopic over-expression of N-SSDP1 and DN-CLIM in developing zebrafish. We have previously shown that DN-CLIM over-expression inhibited the development of eyes, the MHB, and peripheral axonal projections of Rohon-Beard neurons, trigeminal neurons and primary motor neurons (Becker et al., 2002). Indeed, differential interference contrast (DIC) on live (Fig. 4A) and fixed embryos (Fig. 4B) revealed that the eye size of N-SSDP1 and DN-CLIM over-expressing animals was significantly reduced (Fig. 4C). Over-expression of full length zebrafish SSDP1b also significantly reduced eye size (Suppl. Fig. 4). However, N-SSDP1 and SSDP1b overexpression only reduced eye size, whereas upon DN-CLIM overexpression eyes were usually lost completely.
Fig. 4.
N-SSDP impairs eye and MHB development. A,B: Lateral views of the heads of living (A) or fixed, anti-tubulin immuno-labeled (B) 24 hpf embryos are shown. N-SSDP1 mRNA injection leads to reduced eye and MHB size compared to control embryos. DN-CLIM mRNA completely inhibits eye development and more strongly affects the MHB. C: Eye size is significantly reduced after N-SSDP1 mRNA over-expression compared to NLS-myc control mRNA over-expression. D: Dorsal (left columns) and lateral views (right columns) of whole-mounted 24 hpf embryos reveal loss of Pax2a and reduction of Wnt-1 mRNA expression in the MHB after N-SSDP1 mRNA over-expression. Scale bars = 200 µm (A, B and D).
In situ hybridization for Pax2a revealed that the development of the MHB was also affected by N-SSDP1 over-expression at 24 hpf (Fig. 4D). However, Wnt-1 and engrailed transcript levels in the MHB were reduced but still detectable (Fig. 4D; Suppl. Fig. 2E), indicating that this phenotype was milder when compared to DN-CLIM over-expression, which completely abolishes expression of both mRNAs at this stage (Becker et al., 2002).
To determine the influence of N-SSDP1 and DN-CLIM on hindbrain differentiation, we analyzed differentiation of the Mauthner neurons, a pair of early differentiating neurons with descending axons to the spinal cord, which are situated in the hindbrain caudal to the MHB. Immunohistochemistry with the 3A10 antibody indicated that in DN-CLIM mRNA injected embryos these neurons did not form, whereas N-SSDP1 over-expression had no influence on their differentiation (Suppl. Fig. 3A). This again indicates a greater influence of DN-CLIM than N-SSDP1 on embryonic differentiation.
In situ hybridization for Otx2, Pax6a, NeuroD and Six3a showed that the overall brain patterning, except for eyes and MHB, appeared grossly normal in N-SSDP1 mRNA injected embryos. However, N-SSDP1 overexpression resulted in generally smaller structures (Suppl. Fig. 2E). This is very similar to DN-CLIM overexpression (Becker et al., 2002). Because embryos over-expressing NLS-Myc protein were indistinguishable from wild-type animals, phenotypes induced by N-SSDP1 were specific. Furthermore, we did not detect an effect on zebrafish eye and MHB development upon overexpression of the C-terminal 294 aa of SSDP1 (Suppl. Fig. 2D). Strong resemblance of N-SSDP1 and DN-CLIM (Becker et al., 2002) patterning phenotypes in eye and MHB, but not hindbrain, suggests overlapping functions for SSDP1 and CLIM in brain patterning.
SSDP1 regulates axon outgrowth of trigeminal and Rohon-Beard neurons but not motor neurons
Next, we examined the effects of N-SSDP1 over-expression on the development of peripheral axons of trigeminal neurons, Rohon-Beard neurons and primary motor neurons, as we have shown previously that DN-CLIM inhibits the development of these projections (Becker et al., 2002). Indeed, over-expression of N-SSDP1 resulted in significantly decreased numbers of peripheral trigeminal as well as Rohon-Beard axons, but not primary motor axons in anti-tubulin immunohistochemistry (Figs. 5 A, B). These effects were specific as axons in NLS-Myc or C-SSDP1 over-expressing animals appeared indistinguishable from uninjected embryos (Fig. 5B; Suppl. Fig. 5). The reduction in the number of peripheral Rohon-Beard axons was not due to cell loss, as we observed 40.3 ± 0.71 Rohon-Beard neurons in NLS-myc injected embryos (n = 30) and 41.9 ± 0.99 RHB neurons in N-SSDP1 injected embryos, which was not statistically different (P = 0.24). Trigeminal neurons were also present but could not be quantified due to their dense packing. This is very similar to DN-CLIM injected embryos, in which somata of sensory neurons are also retained (Becker et al., 2002). Moreover, we did not find any effect of N-SSDP1 or DN-CLIM over-expression on the differentiation of a spinal intrinsic neuron, the commissural primary ascending interneuron (CoPA). CoPAs differentiate in the dorsal spinal cord. Their axons course ventrally, cross the midline at the floor plate and ascend in the contra-lateral side of the spinal cord in the dorsal longitudinal fascicle. Immunohistochemistry with the 3A10 antibody selectively labeled these neurons in N-SSDP1 and DN-CLIM mRNA injected embryos in a pattern that was indistinguishable from uninjected embryos (Suppl. Fig. 3B). This suggests the absence of general effects of N-SSDP1 and DN-CLIM on spinal floor plate differentiation and axon growth inside the spinal cord. Thus phenotypes of SSDP manipulations in sensory neurons strongly resemble those observed after DN-CLIM overexpression (Becker 2002), in agreement with overlapping functions of SSDP1 and CLIM.
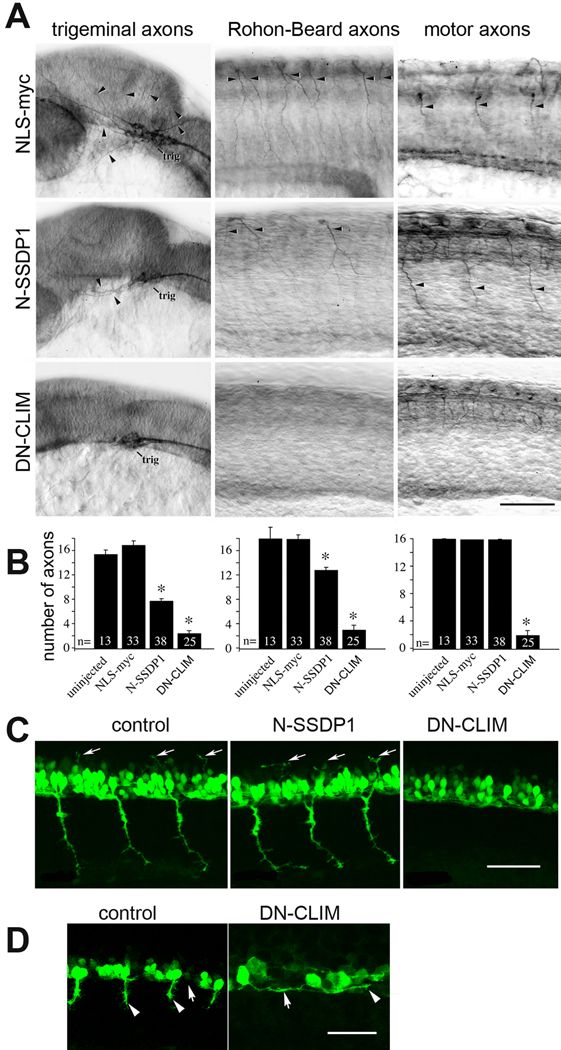
Fig. 5.
Phenotypes induced by N-SSDP over-expression partially overlap with those induced by DN-CLIM over-expression. A: Lateral views of anti-tubulin immuno-labeled whole-mounted 24 hpf embryos are shown. Injection of N-SSDP1 mRNA leads to loss of peripheral axons of trigeminal (trig) and Rohon-Beard neurons, but not of ventral motor axons (NLS-myc = control mRNA). DN-CLIM mRNA injections affect all three types of axons. Arrows indicate axons. B: Quantification confirms significant loss of peripheral sensory but not motor axons upon N-SSDP1 over-expression, whereas DN-CLIM over-expression affects also motor axons. C: DN-CLIM, but not N-SSDP1 over-expression inhibits growth of dorsal MiP axons (arrows), as shown in lateral trunk views of HB9:GFP transgenic embryos at 28 hpf. D: Higher magnification of the ventral border of the caudal spinal cord (arrows) at 24 hpf shows that HB9:GFP+ motor neurons grow axons (arrowheads) ventrally out of the spinal cord in controls. In DN-CLIM mRNA injected embryos axons grow along the ventral border of the spinal cord. Scale bars = 100 µm.
The absence of an effect of N-SSDP1 overexpression on the development of primary motor axons in anti-tubulin immunohistochemistry was surprising because over-expression of DN-CLIM strongly inhibited primary motor axon development (Becker et al., 2002) and it is well established that LIM-HD proteins play crucial roles for the development of motor axons in many vertebrates including zebrafish (Lee and Pfaff, 2001; Hutchinson and Eisen, 2006). Therefore, we decided to examine the effects of N-SSDP1 and DN-CLIM over-expression on primary motor axon development in greater detail. We used HB9:GFP transgenic animals, in which primary motor neurons and their axons can be readily visualized. At 31 hpf, ventrally projecting CaP axons and dorsally projecting MiP axons are detectable in these embryos. Both types of projections require the activity of LIM-HDs of the Isl class (Hutchinson and Eisen, 2006). The use of HB9:GFP embryos revealed that DN-CLIM inhibited both the ventral projection by CaP and the dorsal projection by MiP, whereas N-SSDP1 had no effect on either projection (Fig. 5C). In contrast to unperturbed primary motor axons, GFP-positive axons in DN-CLIM over-expressing embryos extended inside the spinal cord along the rostro-caudal axis at the ventral border of the spinal cord (Fig. 5D), indicating that CLIM cofactors are required for primary motor axons to exit the spinal cord.
To elucidate possible mechanisms of SSDP1 action, we also injected mRNAs encoding zebrafish SSDP1b in full length (zf SSDP1b). Indeed, similar to phenotypes observed for N-SSDP1, embryos overexpressing zf SSDP1b developed smaller eyes and the development of trigeminal and Rohon-Beard axons was inhibited (Suppl. Fig. 4). These phenotypes were weaker when compared to the ones induced by N-SSDP1 (compare Figs. 4, 5 and Suppl. Fig. 4), probably due to lower protein stability in cells.
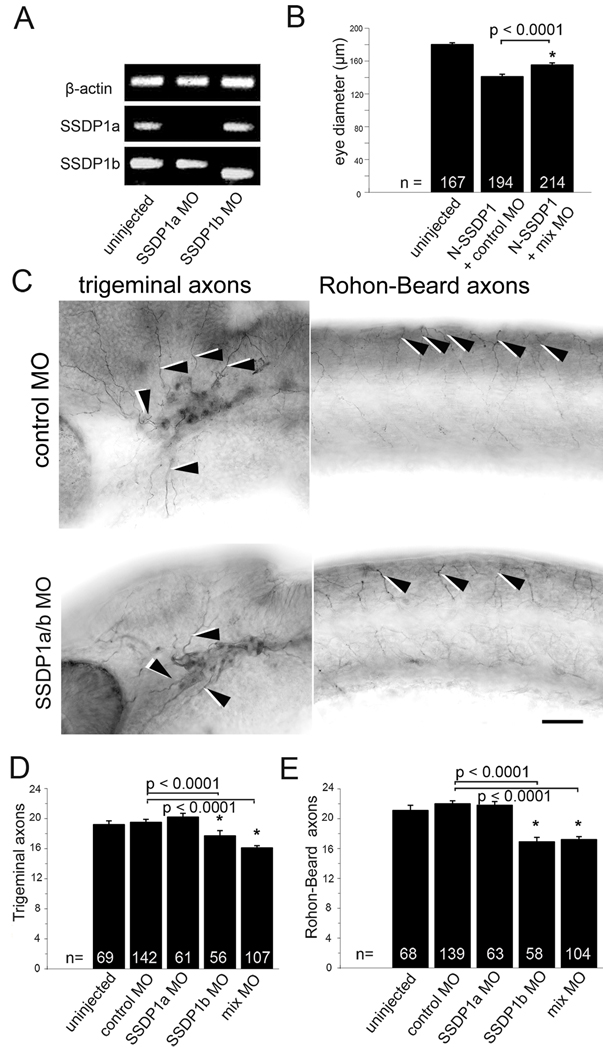
To confirm the roles of SSDP1a and b for zebrafish development we generated specific SSDP1 morpholino oligonucleotides to knock down levels of SSDP1 early during zebrafish development. RT-PCR experiments confirmed the functionality of our morpholinos: Whereas the injection of SSDP1a morpholino (MO SSDP1a) resulted in undetectable mRNA levels, probably due to non-sense mediated decay (Bill et al., 2009), the morpholino against SSDP1b (MO SSDP1b) induced a shift in the size of the amplicon from 327 bp to 254bp in the morphant. This led to a frame shift thereby introducing an early stop codon at the beginning of exon 3, such that it is highly unlikely that functional protein was transcribed (Fig. 6A). Next, we examined the developmental effects caused by individually or simultaneously knocking down SSDP1a and SSDP1b. These experiments revealed that the knockdown of SSDP1b, but not SSDP1a, significantly inhibited the development of peripheral axons of trigeminal and Rohon-Beard neurons (Figs. 6C–E). This is consistent with conspicuous expression of SSDP1b, but not SSDP1a mRNA in these neuronal cell types (see Fig. 1). To exclude that effects of morpholinos were a consequence of retarded overall development we determined the segmental position of the lateral line primordium, an indicator of embryonic developmental stage (Kimmel et al., 1995), and found no differences between any of the morpholino injected groups and uninjected embryos (data not shown). This suggests specificity of the axonal phenotypes in morphants.
Fig. 6.
Knock-down of SSDP1b, but not SSDP1a partially inhibits growth of peripheral axons of trigeminal and Rohon-Beard neurons. A: Morpholino knock-down reduces levels of correctly spliced SSDP1a and SSDP1b to levels that are undetectable by PCR. An ectopic band is amplified after SSDP1b knock-down, which contains premature stop codons and, therefore, cannot lead to functional protein. Beta-actin was used to equalize cDNA concentrations. B: Morpholinos to SSDP1 partially rescue reduced eye size induced by N-SSDP1 over-expression, suggesting specificity of the morpholinos. C: Lateral views of anti-tubulin immuno-labeled whole-mounted 24 hpf embryos injected with Morpholinos against SSDP1a/b are shown. Arrowheads point to trigeminal and Rohon-Beard axons, respectively. Scale bar = 50 µm. D,E: Quantifications indicate significant loss of peripheral axons of trigeminal and Rohon-Beard neurons after Morpholino knock-down of SSDP1b.
However, we did not detect significant effects on eye and MHB development nor on PMN axons by knocking down SSDP1a, b or both (data not shown). These results indicate functions of SSDP1b for developing peripheral axons of trigeminal and Rohon-Beard neurons. The fact that eye and MHB development was inhibited upon overexpression of N-SSDP1 (Fig. 4) but not in our SSDP1 knockdown experiments (data not shown) suggests that members of the SSDP family other than SSDP1 may participate in LIM-HD/CLIM complexes in these structures and act redundantly with SSDP1a,b.
The eye phenotype caused by N-SSDP1 over-expression was partially rescued by simultaneously knocking down endogenous SSDP1a and b (Fig. 6B). Because there is only a 6 out of 25 morpholino bases overlap with N-SSDP1, and morpholinos lose all activity when more than 4 bases are mismatched it is highly likely that neither of the morpholino oligonucleotides recognize mouse-derived N-SSDP1. This may be explained by opposing actions of N-SSDP1 and SSDP1 morpholinos on CLIM stability. This rescue experiment also suggests specific action of the morpholinos.
SSDPs stabilize CLIM during PMN development
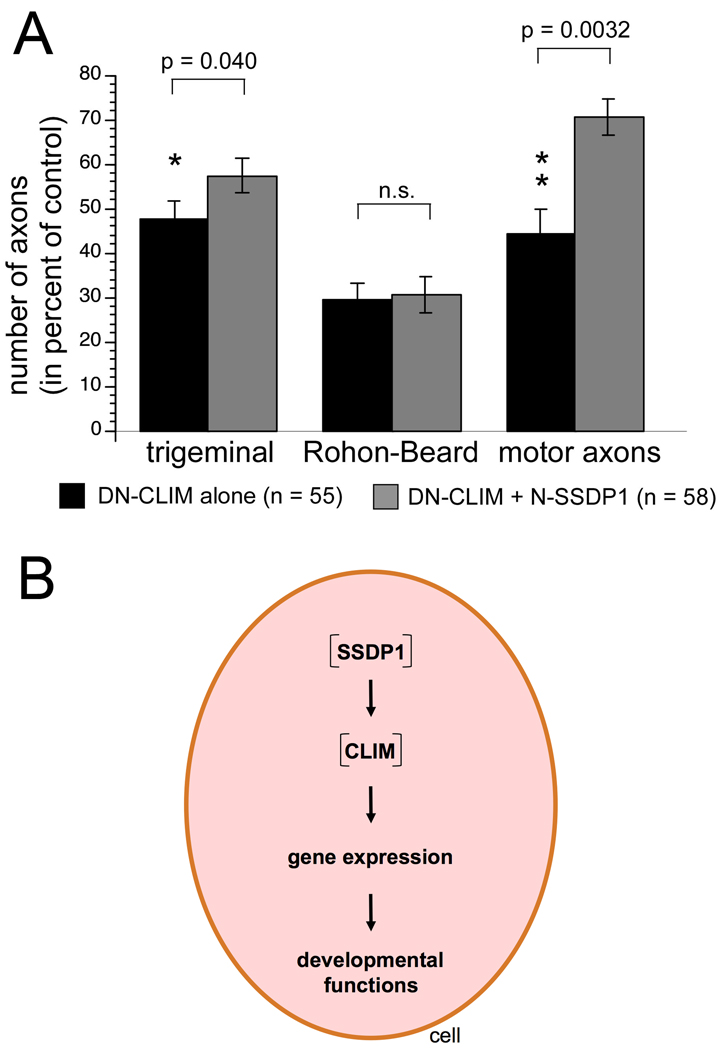
The finding that in contrast to DN-CLIM, overexpression of N-SSDP1 or zebrafish SSDP1b did not affect peripheral axons of primary motor neurons (Fig. 5; data not shown) showed that either SSDPs are dispensable for primary motor neuron development or that only N-terminal sequences of SSDPs are important for a correct formation and functioning of LIM-HD complexes. To examine if SSDP1 exerts functions via CLIM stabilization we co-injected DN-CLIM together with N-SSDP1. In these co-injection experiments we titrated the amount of DN-CLIM mRNA (75%) to only moderately affect axon growth. Indeed, co-injection of N-SSDP1 (25%) partially rescued the effect of DN-CLIM on PMN axons as well as trigeminal axons in a significant manner, whereas no effect was detected on DN-CLIM-mediated inhibition of RHB peripheral axon formation (Fig. 7A). Although the average eye size was 45% larger in N-SSDP1/DN-CLIM co-injected embryos when compared to DN-CLIM injected embryos, this difference was not significant. The MHB could not be quantitatively assessed. Because N-SSDP1 is able to stabilize CLIM (Fig. 3) and overexpression of N-SSDP1 or knockdown of SSDP1 does not affect PMN axons (Fig. 5, not shown), the rescue in this cell type is likely due to a stabilization of endogenous CLIM cofactors by N-SSDP1. Thus, these results reveal a function for SSDPs for the stabilization of LIM-HD multiprotein complexes in primary motor neurons.
Fig. 7.
A: N-SSDP1 over-expression partially rescues phenotypes induced by DN-CLIM. Significantly fewer axons of trigeminal neurons and motor neurons, but not of Rohon-Beard neurons, are lost when N-SSDP1 mRNA is co-injected with DN-CLIM mRNA, compared to DN-CLIM mRNA injection alone. B: Schematic representation of cellular functions of SSDP1.
Discussion
We show previously unknown and highly specific functions of SSDP1 co-factors in the development of peripheral sensory axons and in patterning of the anterior CNS. Our results provide evidence that these functions are mediated at least in part by stabilization of CLIM cofactors.
Specific functions of SSDP1b in sensory axon growth
Expression analysis of SSDP1a and SSDP1b mRNAs indicated widespread expression throughout embryonic development for both genes, in agreement with multiple functions of these co-factors. However, SSDP1b showed specific accumulation in sensory trigeminal and Rohon-Beard neurons. This is reminiscent of the expression of CLIM cofactors with wide expression of CLIM2 and a more restricted expression pattern of CLIM1 (Bach et al., 1997; Bach et al., 1999; Ostendorff et al., 2006). Because cofactor levels are under tight proteasomal regulation (Gungor et al., 2007; Xu et al., 2007) we also analyzed the protein distribution of SSDP1 and CLIM. This confirmed specific accumulation of both SSDP1 and CLIM in trigeminal and Rohon-Beard neurons, suggesting specific roles of these co-factors here. Interfering with SSDP1 function, either by over-expression of N-SSDP1, SSDP1b in full length or by morpholino knock down of SSDP1b, impaired outgrowth of peripheral trigeminal and Rohon-Beard axons. This phenotype strongly resembles that of over-expression of a dominant-negative form of the CLIM co-factor, DN-CLIM. Effects of SSDP1 manipulations and DN-CLIM were highly cell type-specific, because another spinal cell type, the CoPA neuron, showed an unaffected axonal trajectory inside the spinal cord. Interestingly, the LIM-HD islet-1/-2 is strongly expressed in trigeminal and Rohon-Beard neurons (Becker et al., 2002), suggesting that SSDP1 and CLIM could participate in islet-1/-2 transcriptional complexes in these sensory neurons. Taken together, these observations are consistent with functions of SSDP1b/CLIM interactions in the formation of sensory axons during early development.
SSDP1/CLIM interactions may be involved in motor axon pathfinding
Extending on previous findings we show here that DN-CLIM overexpression affects ventral and dorsal motor axon growth. Instead of inhibiting axon formation altogether, as for sensory axons, axons of HB9:GFP positive cells still grow, but are unable to exit the spinal cord. Axons grow along the ventral edge of the spinal cord, close to the floor plate. This phenotype is similar to that observed after knock down of the LIM-HDs islet-1 and islet-2 (Hutchinsen and Eisen, 2006). The fact that CoPA neurons showed correct growth inside the spinal cord, including commissural growth at the floor plate, indicated that patterning of the spinal cord was largely unaffected by DN-CLIM overexpression. This suggests that LIM-HD complexes could be involved in the expression of specific guidance receptors or proteins in only motor neurons that lead their axons to their specific targets (Lee et al., 2008). For example we have shown for another cell type, trigeminal neurons, that expression of the axonal recognition molecule Contactin1 is dramatically reduced when DN-CLIM is overexpressed (Gimnopoulos et al., 2002). The lack of a motor axon phenotype by manipulations of SSDP1 was the only non-overlapping finding with those of DN-CLIM overexpression. However, N-SSDP1 overexpression partially rescued the axon pathfinding defect elicited by DN-CLIM. This suggests that SSDP1/CLIM interactions may also play roles in motor axon pathfinding.
SSDP1 is involved in patterning of the anterior CNS
Patterning defects induced by N-SSDP1 over-expression in the eye and MHB were also similar to those observed after DN-CLIM over-expression. We show that SSDP1 and CLIM proteins are co-expressed in these structures. However, DN-CLIM generally had stronger effects. Reduction of eye size and loss of MHB marker expression was more severe in DN-CLIM overexpressing embryos compared to N-SSDP1 or SSDP1b overexpressing embryos. N-SSDP1 over-expressions lead to complete loss of pax2 and a reduction of engrailed and wnt-1 labeling in the MHB, whereas after DN-CLIM injection all three markers are lost (Becker et al., 2002). Here we show that DN-CLIM inhibits differentiation of the Mauthner neurons in the hindbrain, whereas N-SSDP1 over-expression was not sufficient to inhibit differentiation of this cell type.
The fact that knock down of SSDP1a and b had no apparent effect on eyes and MHB could be explained by the fact that at least five different SSDP and four CLIM proteins are present in zebrafish (zfin.org), which may act redundantly. Indeed, SSDP2 has been shown to participate in LIM-HD complexes (Cai et al., 2008) and to be necessary to maintain levels of CLIM protein (Wang et al., 2010). Overall, patterning effects of manipulations of SSDP1 and CLIM cofactors on CNS patterning resemble those observed in mouse mutants for SSDP1 and CLIM, in which genetically linked smaller or absent head structures have been reported (Mukhopadhyay et al., 2003; van Meyel et al., 2003; Nishioka et al., 2005; Enkhmandakh et al., 2006). Axonal phenotypes have not been demonstrated in mice. We show here highly specific functions of SSDPs in axon formation and pathfinding.
Evidence that effects of SSDP1 manipulation are mediated by CLIM in vivo
Too low or too high CLIM levels inhibit LIM-HDs (Fernandez-Funez et al., 1998; Matthews and Visvader, 2003). The inhibition of LIM-HD/CLIM transcriptional complexes via overexpression of DN-CLIM in zebrafish (Becker et al., 2002) is reminiscent of the phenotypes obtained by disturbing SSDP1 via overexpression or knockdown. Thus, because our overexpression of SSDP1 lead to an increase and the knockdown of SSDP1 via morpholino to a decrease in endogenous CLIM levels (Fig. 7B; Gungor et al., 2007), most of the phenotypes observed by disturbing SSDP1 might be explained by induced changes in cellular CLIM/LIM-HD stoichiometries leading to a functional inhibition of these complexes.
Further support for the significance of the CLIM stabilization of SSDP1 stems from our observation that both N-SSDP1 and SSDP1 in full length lead to increased cellular CLIM levels when overexpressed (Gungor et al., 2007) and that the induced phenotypes are very similar (Figs 4, 5 and Suppl. Fig. 4). These results argue against dominant-negative effects of N-SSDP1. In addition, overexpression of N-SSDP1 that contains the CLIM interaction domain rescues phenotypes induced by DN-CLIM in motor axon pathfinding and formation of trigeminal axons. This is consistent with a scenario in which N-SSDP1 stabilizes endogenous CLIM, such that it can compete with DN-CLIM to form functional LIM-HD complexes. In Rohon-Beard neurons, for which no rescue of peripheral axon growth was observed, N-SSDP1 overexpression might not have stabilized CLIM proteins to functional levels.
Taken together, stabilization of CLIM by SSDP1 in vivo, rescue of DN-CLIM induced motor axon phenotypes by DN-CLIM and overlapping phenotypes of DN-CLIM overexpression and SSDP1 manipulations are consistent with in a role of SSDP1 in the control of the transcriptional activity of LIM-HD complexes by stabilizing and protecting CLIM cofactors from proteasomal degradation (Gungor et al., 2007; Xu et al., 2007). However, as overexpression of N-SSDP1 inhibits endogenous alphaGSU expression in pituitary cells in vitro (Fig. 3C) and a proline-rich region that is deleted in N-SSDP1 appears important for at least some of its functions in LIM-HD complexes (Enkhmandakh et al., 2006), we cannot exclude the possibility that dominant-negative actions contribute to the phenotype observed in embryos overexpressing N-SSDP1. Finally, it has to be noted that SSDP1/CLIM interactions may have functions independent of LIM-HDs, as CLIM co-factors have been shown to interact with several non-LIM domain-containing transcription factors (Bach et al., 1997; Torigoi et al., 2000; Johnsen et al., 2009; Heitzler et al., 2003).
In summary, overlapping expression patterns and perturbation phenotypes of SSDP1 and CLIM, as well as in vivo stabilization of CLIM and rescue of DN-CLIM phenotypes by N-SSDP1 indicate important regulatory functions of SSDP1/CLIM interactions for CNS patterning, as well as growth and pathfinding of specific axon types.
Research Highlights
-
-
SSDP cofactors regulate neural patterning and sensory axon growth
-
-
Functions of SSDP cofactors are mediated via stabilization of CLIM/Ldb/NLI cofactors
Supplementary Material
Suppl. Fig. 1 Phylogenetic analysis confirms identities of the cloned SSDP1a and SSDP1b in zebrafish. Zebrafish SSDP1a and SSDP1b co-segregate with SSDP1 genes of other vertebrate species. The phylogenetic tree is constructed by the fast minimum evolution method (Desper R and Gascuel O, Mol Biol Evol 21:587-98, 2004 PMID: 14694080) based on Grishin tree distance (Grishin NV, J Mol Evol, 41:675-79, 1995 PMID: 18345592).
Suppl. Fig. 2 A: Western blot analysis indicates that the SSDP1 antibody detects a band of the same molecular weight in protein extracts of alphaT3 cells and zebrafish embryos, suggesting specific reactivity of the antibody in zebrafish embryos. B: Immunodetectability of SSDP1 is slightly reduced by SSDP1a and strongly reduced by SSDP1b morpholino injection, indicating that the morpholino predominantly recognizes SSDP1b. Lateral trunk views of whole-mounted embryos are shown. The strongly immunoreactive spinal cord level is indicated by brackets. C: Western blot analysis indicates that GFP-tagged N-SSDP1 and full length SSDP1 can be expressed in alphaT3 cells. D: Lateral views of anti-tubulin labeled embryos at 24 hpf are shown. Over-expression of C-SSDP1, which lacks the CLIM-interacting domain, does not induce phenotypes in eye or MHB. E: Lateral views of the heads of 24 hpf embryos after in situ hybridization are shown. The indicated markers retain their general expression patterns when N-SSDP1 is over-expressed. Scale bar in B = 50 µm; bars in D,E = 200 µm.
Suppl. Fig. 3 DN-CLIM but not N-SSDP1 over-expression induces patterning defects specifically in the hindbrain. A: Dorsal views of the hindbrain (rostral is up) of 24 hpf embryos immuno-labelled with the 3A10 antibody are shown. Development of Mauthner neurons (arrows) and crossing of their axons at the brain midline (arrowhead) is not affected by N-SSDP1 over-expression. In contrast, DN-CLIM over-expression leads to complete or partial loss of Mauthner neurons. Note that in those cases in which only one Mauthner neuron is present its axonal trajectory is not altered (ov = otic vesicle). B: Lateral views of the spinal cord show that 3A10 immuno-labelled CoPA neurons (arrowheads) and their commissural axons (arrows) are not affected by N-SSDP1 or DN-CLIM over-expression. Scale bars = 50 µm.
Suppl. Fig. 4 Full length zebrafish SSDP1b over-expression has similar effects to N-SSDP1. Lateral views of anti-tubulin labeled whole-mounted 24 hpf embryos are shown. Arrowheads indicate axons. Eye size as well as axon numbers of trigeminal and Rohon-Beard neurons are significantly reduced by SSDP1b over-expression compared to NLS-myc control. Scale bar = 50 µm.
Suppl. Fig. 5 C-SSDP1 overexpression does not affect peripheral trigeminal or Rohon-Beard axons. Lateral views of head (for trigeminal axons) and trunk (for Rohon-Beard axons) are shown. Some of the axons are indicated by arrows. Scale bar = 100 µm.
Acknowledgments
We thank Bénédicte Autin for excellent in situ hybridization and fish care. Core resources supported by the Diabetes Endocrinology Research Center grant DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases were used. This work has been supported by NIH grants 5 P30 DK32520 (NIDDK) and 1R01CA131158 (NCI) to I.B. and the College of Medicine and Veterinary Medicine and the University of Edinburgh Scottish Overseas Research Students Award Scheme (to Z.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bach I. The LIM domain: regulation by association. Mech. Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse RV, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat. Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Soto RJ, Lukens LN. Cloning and characterization of a novel sequence-specific single-stranded-DNA-binding protein. Biochem. J. 1998;331(Pt 2):447–452. doi: 10.1042/bj3310447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Becker CG, Schachner M, Bernhardt RR. Antibody to the HNK-1 glycoepitope affects fasciculation and axonal pathfinding in the developing posterior lateral line nerve of embryonic zebrafish. Mech. Dev. 2001;109:37–49. doi: 10.1016/s0925-4773(01)00504-4. [DOI] [PubMed] [Google Scholar]
- Becker T, Ostendorff HP, Bossenz M, Schlüter A, Becker C, Peirano RI, Bach I. Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech. Dev. 2002;117:75–85. doi: 10.1016/s0925-4773(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Cai Y, Xu Z, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance and function of the LIM-homeodomain transcription factor LHX2 in pituitary cells. Biochem. Biophys. Res. Commun. 2008;373:303–308. doi: 10.1016/j.bbrc.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, Ogryzko VV, Dawid IB, Westphal H. Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhmandakh B, Makeyev AV, Bayarsaihan D. The role of the proline-rich domain of Ssdp1 in the modular architecture of the vertebrate head organizer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11631–11636. doi: 10.1073/pnas.0605209103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldner J, Becker T, Goishi K, Schweitzer J, Lee P, Schachner M, Klagsbrun M, Becker CG. Neuropilin-1a is involved in trunk motor axon outgrowth in embryonic zebrafish. Dev. Dyn. 2005;234:535–549. doi: 10.1002/dvdy.20520. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P, Lu CH, Rincon-Limas DE, Garcia-Bellido A, Botas J. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 1998;17:6846–6853. doi: 10.1093/emboj/17.23.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Vanolst L, Biryukova I, Ramain P. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev. 2003;17:591–596. doi: 10.1101/gad.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Yamamoto N, Mochizuki T, Ohmori SY, Taira M. Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development. 2003;130:4161–4175. doi: 10.1242/dev.00621. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development. 2006;133:2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- Johnsen SA, Gungor C, Prenzel T, Riethdorf S, Riethdorf L, Taniguchi-Ishigaki N, Rau T, Tursun B, Furlow JD, Sauter G, Scheffner M, Pantel K, Gannon F, Bach I. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69:128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 2001;4 Suppl:1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Nagano S, Nakayama R, Kiyonari H, Ijiri T, Taniguchi K, Shawlot W, Hayashizaki Y, Westphal H, Behringer RR, Matsuda Y, Sakoda S, Kondoh H, Sasaki H. Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1-Ldb1 complex. Development. 2005;132:2535–2546. doi: 10.1242/dev.01844. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Peirano RI, Peters MA, Schlüter A, Bossenz M, Scheffner M, Bach I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, Bach I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev. Dyn. 2006;235:786–791. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Song MR, Sun Y, Bryson A, Gill GN, Evans SM, Pfaff SL. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136:2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Torigoi E, Bennani-Baiti IM, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Schluter A, Peters MA, Viehweger B, Ostendorff HP, Soosairajah J, Drung A, Bossenz M, Johnsen SA, Schweizer M, Bernard O, Bach I. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19:2307–2319. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel DJ, O'Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol. Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klumpp S, Amin HM, Liang H, Li J, Estrov Z, Zweidler-McKay P, Brandt SJ, Agulnick A, Nagarajan L. SSBP2 is an in vivo tumor suppressor and regulator of LDB1 stability. Oncogene. 2010 doi: 10.1038/onc.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol. Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- Wu L. Structure and functional characterization of single-strand DNA binding protein SSDP1: carboxyl-terminal of SSDP1 has transcription activity. Biochem. Biophys. Res. Commun. 2006;339:977–984. doi: 10.1016/j.bbrc.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1 Phylogenetic analysis confirms identities of the cloned SSDP1a and SSDP1b in zebrafish. Zebrafish SSDP1a and SSDP1b co-segregate with SSDP1 genes of other vertebrate species. The phylogenetic tree is constructed by the fast minimum evolution method (Desper R and Gascuel O, Mol Biol Evol 21:587-98, 2004 PMID: 14694080) based on Grishin tree distance (Grishin NV, J Mol Evol, 41:675-79, 1995 PMID: 18345592).
Suppl. Fig. 2 A: Western blot analysis indicates that the SSDP1 antibody detects a band of the same molecular weight in protein extracts of alphaT3 cells and zebrafish embryos, suggesting specific reactivity of the antibody in zebrafish embryos. B: Immunodetectability of SSDP1 is slightly reduced by SSDP1a and strongly reduced by SSDP1b morpholino injection, indicating that the morpholino predominantly recognizes SSDP1b. Lateral trunk views of whole-mounted embryos are shown. The strongly immunoreactive spinal cord level is indicated by brackets. C: Western blot analysis indicates that GFP-tagged N-SSDP1 and full length SSDP1 can be expressed in alphaT3 cells. D: Lateral views of anti-tubulin labeled embryos at 24 hpf are shown. Over-expression of C-SSDP1, which lacks the CLIM-interacting domain, does not induce phenotypes in eye or MHB. E: Lateral views of the heads of 24 hpf embryos after in situ hybridization are shown. The indicated markers retain their general expression patterns when N-SSDP1 is over-expressed. Scale bar in B = 50 µm; bars in D,E = 200 µm.
Suppl. Fig. 3 DN-CLIM but not N-SSDP1 over-expression induces patterning defects specifically in the hindbrain. A: Dorsal views of the hindbrain (rostral is up) of 24 hpf embryos immuno-labelled with the 3A10 antibody are shown. Development of Mauthner neurons (arrows) and crossing of their axons at the brain midline (arrowhead) is not affected by N-SSDP1 over-expression. In contrast, DN-CLIM over-expression leads to complete or partial loss of Mauthner neurons. Note that in those cases in which only one Mauthner neuron is present its axonal trajectory is not altered (ov = otic vesicle). B: Lateral views of the spinal cord show that 3A10 immuno-labelled CoPA neurons (arrowheads) and their commissural axons (arrows) are not affected by N-SSDP1 or DN-CLIM over-expression. Scale bars = 50 µm.
Suppl. Fig. 4 Full length zebrafish SSDP1b over-expression has similar effects to N-SSDP1. Lateral views of anti-tubulin labeled whole-mounted 24 hpf embryos are shown. Arrowheads indicate axons. Eye size as well as axon numbers of trigeminal and Rohon-Beard neurons are significantly reduced by SSDP1b over-expression compared to NLS-myc control. Scale bar = 50 µm.
Suppl. Fig. 5 C-SSDP1 overexpression does not affect peripheral trigeminal or Rohon-Beard axons. Lateral views of head (for trigeminal axons) and trunk (for Rohon-Beard axons) are shown. Some of the axons are indicated by arrows. Scale bar = 100 µm.