Abstract
A series of new zinc binding groups (ZBGs) has been evaluated kinetically on 13 carbonic anhydrase (CA) isoforms. The fragments show affinity for all isoforms with IC50 values in the range of 2–11 µm. The crystal structure of hCA II in complex with one such fragment reveals a bidentate binding mode with a trigonal-bipyramidal coordination geometry at the Zn2+ center. The fragment also interacts with Thr199 and Thr200 through hydrogen bonding and participates in a water network. Further development of this ZBG should increase the binding affinity leading to a structurally distinct and promising class of CA inhibitors.
Keywords: carbonic anhydrase, crystal structures, drug discovery, inhibitors, metalloenzymes, zinc binding group
Introduction
Carbonic anhydrases (CAs, EC 4.2.1.1) constitute one of the most extensively studied groups of metalloenzymes.[1, 2] They belong to a superfamily of ubiquitous metalloproteins present in prokaryotes and eukaryotes and are encoded by five distinct evolutionarily unrelated gene families: α class (present in vertebrates, bacteria, algae, and cytoplasm of green plants), β class (predominantly in bacteria, algae, and chloroplasts), γ class (mainly in Archaea spp. and some bacteria), and δ and ξ classes present in marine diatoms. They catalyze the reversible hydration of carbon dioxide into hydrogen carbonate and a proton. The catalytically relevant metal ion, which is Zn2+ in the α-CAs, but may be Fe2+ in γ- or Cd2+ in ξ-CAs, among others, is located (for the α class enzymes) at the bottom of the 15 Å deep active site, coordinated by three histidine residues—His94, His96, and His119—and a water molecule to form a tetrahedral geometry at the active site.[1, 2]
Sixteen different isoforms have been described in mammals, each differing by their relative hydrase activity, their subcellular localization, and their susceptibility to inhibition. Because these CAs are involved in several physiological processes, it is not surprising that carbonic anhydrase inhibitors (CAIs) have been developed for the treatment of various pathologies such as glaucoma, neurological disorders, and osteoporosis.[3, 4] The recent discovery of CA isoforms that are involved in cancer or obesity has demonstrated the need for potent, novel classes of CAIs. However, the large number of diverse α-CAs leads to a lack of inhibitor selectivity, especially for first-generation CAIs, i.e., sulfonamides and their bioisosteres. To minimize unwanted side effects of CAI therapy, the design of novel inhibitors must discriminate between the different isozymes.[1, 2, 5, 6]
So far, most rationally designed CAIs are based on a zinc binding group (ZBG) that interacts with the Zn2+ ion of the enzyme and Thr199. Linked to this ZBG is a diversely substituted aromatic ring, heterocycle, aliphatic group, or sugar moiety that interacts with hydrophobic and hydrophilic pockets neighboring the active site. The sulfonamide group (Figure 1) is the predominant ZBG present in CAIs; this is in contrast to other Zn2+ metalloenzymes, such as the matrix metalloproteinases (MMPs), for which hydroxamic acids serve as the dominant ZBG (Figure 1).[7] The sulfonamide moiety coordinates to the Zn2+ ion with its terminal deprotonated nitrogen atom (2.0 Å) and participates in a hydrogen bonding network with Thr199 (2.8 Å) and Glu106 (2.7 Å). The latter amino acids are referred to as the “door-keepers” in the CA active site.[8] In addition, one sulfonamide oxygen atom forms a hydrogen bond with the backbone NH group of Thr199 (3.0 Å). With this interaction pattern, many highly potent sulfonamide CAIs have been developed.[ 1, 2, 9, 10]
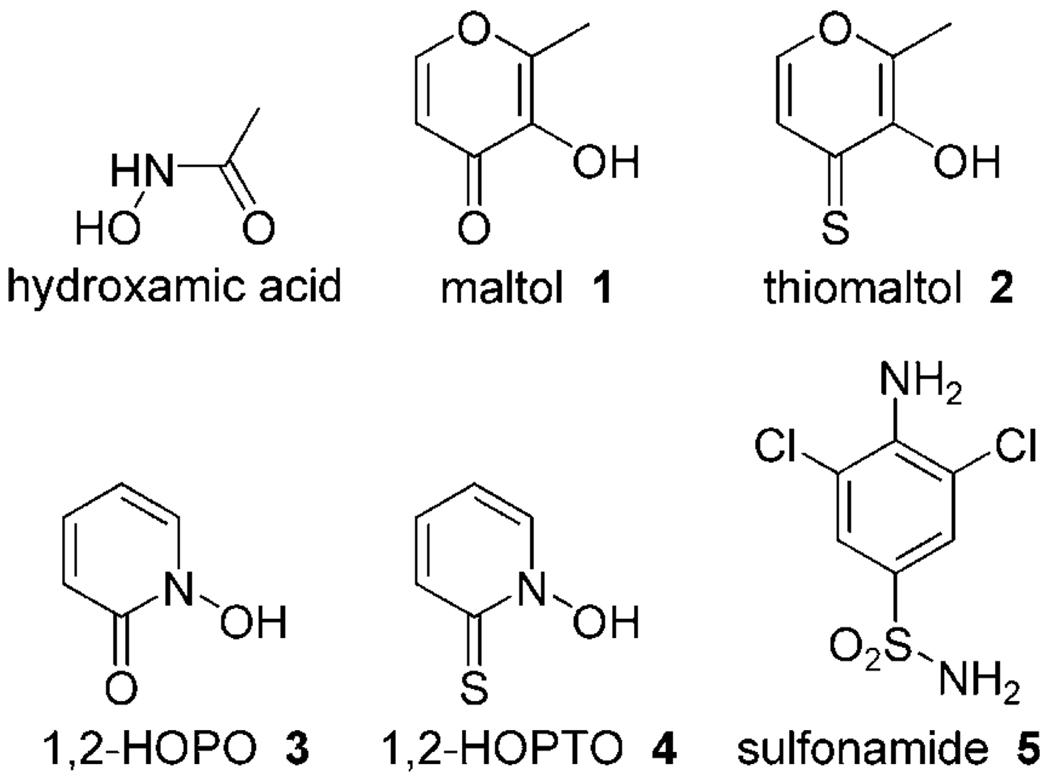
Figure 1.
Zinc binding groups (ZBGs) used in this study as CA inhibitors.
The side effects of sulfonamide CAIs have led to new strategies for selective inhibitor design, such as modifications of the inhibitor backbone.[1–4] One attractive approach to new CAIs is the use of non-sulfonamide ZBGs. A range of ZBGs have been tested so far, including sulfamates, sulfamides, substituted sulfonamides, Schiff bases, ureas and hydroxyureas, as well as hydroxamic acids. These studies revealed sulfamates and sulfamides, structurally related to sulfonamides, to be amongst the most promising alternative ZBGs.[11–13]
To find new ZBGs for use in CAIs, ligands from other Zn2+-dependent metalloenzymes are of particular interest. Recently, Cohen and colleagues developed a series of new ZBGs, which include hydroxypyr(thi)ones and hydroxypyridine(thi)ones, that have been examined as alternative ZBGs for MMP inhibitors (Figure 1). These ZBGs show potent affinity, are nontoxic, biocompatible, and show promise for incorporation into a new family of MMP inhibitors.[14, 15] The studies reported herein examine the use of these alternative ZBGs as warheads for CAIs; we present the investigation of four ligands from this series. Compounds 1–4 were tested in a kinetic enzyme assay on all catalytically active mammalian CAs (Figure 1). The crystal structure of CA II in complex with compound 4 was determined and revealed an intriguing bidentate binding mode. Fragment 4 coordinates the active site Zn2+ ion in a chelating, bidentate mode, and also interacts with Thr199 and Thr200. In addition, the compound participates in a hydrogen bonding network that is translated via water molecules within the entire active site of CA II.
Results
Inhibition data
The inhibition data for compounds 1–4 against 13 CA isoforms (hCA–mCA XV) are listed in Table 1. All of these compounds show affinities against the tested isoforms in the range of 2.8–10.8 µm. The inhibition of each isoform with compounds 1–4 is generally very similar (<2.5 µm). The best inhibition was obtained with maltol 1 against mCA XV, with a Ki value of 2.8 µm; the weakest inhibition was observed for 1,2-HOPTO 4 against hCA XIII, with a Ki value of 10.8 µm. Isoforms hCA I, III, IX, XIII, and XIV show generally weaker inhibition, with Ki values ranging from 7.5 to 10.8 µm, in comparison with all other isoforms, for which Ki values from 2.8 to 4.9 µm were found.
Table 1.
hCA I–mCA XV inhibition data with compounds 1–4.
| Isoform | Ki [µm][a] | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| hCA I | 9.1 | 8.3 | 9.7 | 9.4 | 7.0 |
| hCA II | 3.6 | 4.0 | 3.9 | 4.1 | 50 nm |
| hCA III | 9.5 | 8.4 | 9.1 | 7.5 | - |
| hCA IV | 4.3 | 4.6 | 3.9 | 4.4 | - |
| hCA VA | 4.6 | 4.2 | 4.4 | 4.0 | - |
| hCA VB | 4.3 | 4.0 | 4.4 | 3.8 | - |
| hCA VI | 3.5 | 3.2 | 3.9 | 3.6 | - |
| hCA VII | 3.4 | 3.6 | 3.8 | 3.6 | - |
| hCA IX | 9.5 | 10.0 | 9.3 | 9.2 | 27 nm |
| hCA XII | 4.0 | 4.5 | 4.7 | 4.8 | - |
| hCA XIII | 10.4 | 10.4 | 10.8 | 10.8 | - |
| hCA XIV | 7.2 | 6.7 | 6.8 | 7.1 | - |
| mCA XV | 2.8 | 3.1 | 4.9 | 4.7 | - |
Errors in the range of ± 10 % of the reported value from three different assays.
X-ray crystallography
The crystal structure of CA II in complex with fragment 4 (1,2-HOPTO) was determined at a resolution of 1.35 Å. The difference electron density of 4 is very well defined, and the orientation of the fragment can be unambiguously assigned due to the higher diffraction power of the sulfur atom and the shape of the ring system, which is clearly visible in the difference electron density and refined B values. The positioning of the oxygen atom was ambiguous at the beginning of the refinement. However, repeated refinement cycles using various fragment orientations (ring flip) and considerations about a consistent hydrogen bond network render the assigned binding mode as most likely. The B values for 4 are somewhat larger than those of the protein residues which is probably related to weak fragment affinity and thus a decreased occupancy. The B values are: main chain 14.1 Å2, side chain 18.2 Å2, water 27.1 Å2, ligand 4 24.1 Å2, and Zn2+ ion 8.4 Å2.
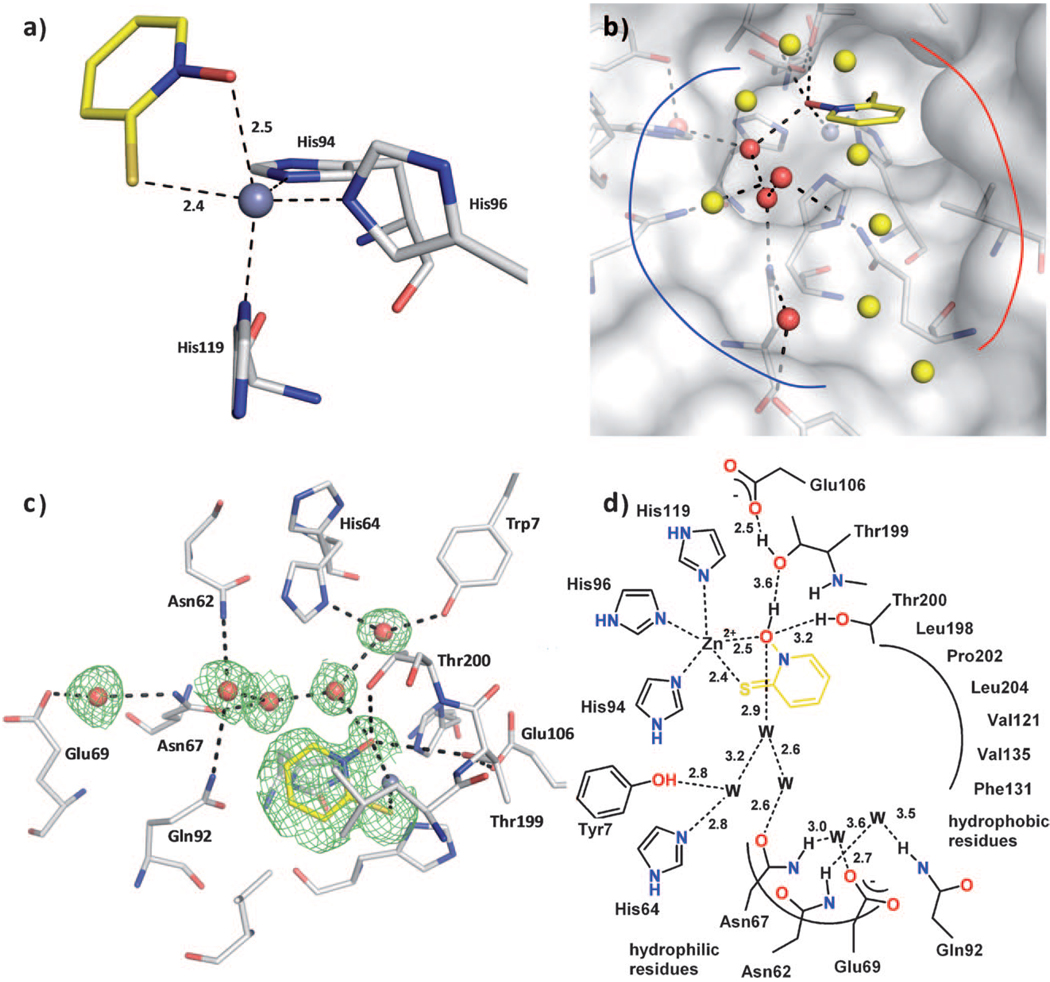
As expected, fragment 4 binds to CA II by coordinating to the active site Zn2+ ion. The exocyclic sulfur and oxygen atoms participate in the coordination of the metal ion at distances of 2.4 (S) and 2.5 Å (O) (Figure 2 a). Importantly, the coordination of the Zn2+ ion changes upon binding of 4 from a four-coordinate tetrahedral geometry (including the bound OH− ion, PDB ID: 3D92)[16] to a distorted, five-coordinate trigonal-bipyramidal geometry. Two nitrogen atoms (Nε2-His94 and Nε2-His96) and the sulfur atom of 4 form the equatorial plane, while Nδ1 of His119 and the oxygen atom of 4 reside at the axial positions. A similar coordination change from four- to five-coordinate has been proposed for the binding of fragment 4 to MMPs.[17]
Figure 2.
Compound 4 binding to CA II. The protein residues are shown in stick representation (protein:  ; ligand:
; ligand:  ), and water molecules are shown as red and yellow spheres. The solvent accessible surface is represented in white in panel b). a) Coordination geometry of the active site Zn2+ ion upon binding of 4. The tetrahedral geometry is changed to distorted trigonal-bipyramidal arrangement with Nε2-His94 (2.0 Å), Nε2-His96 (2.0 Å), and the sulfur atom of 4 (2.4 Å) in the equatorial plane, whereas Nδ1-His119 (2.1 Å) and the oxygen atom of 4 (2.5 Å) are in the axial positions. The angles in the plane are 143.5° (S-Zn-His96), 109.5° (S-Zn-His94), and 101.7° (His94-Zn-His96). The two axial ligands and the Zn2+ ion form an angle of 161.2°. b) The binding pocket of CA II can be divided into a hydrophobic (red line) and a hydrophilic binding region (blue line). The fragment is stabilized by a hydrogen bond network which is mediated by five water molecules (red spheres) addressing the typical hydrophilic residues in CA II. The remaining waters in the binding pocket (yellow spheres) are connected to the hydrogen bond network as well, however not performing any further strong interactions to the protein. c) Detailed binding mode. Hydrogen bond interactions are indicated by the dashed lines. The equivalent distances are shown in the schematic representation of the binding mode in panel d). The difference electron density (Fo—Fc) for the ligands and directly interacting water molecules is shown at a σ level of 2.0. Atomic distances are shown in Å. The fragment coordinates with its sulfur and oxygen atom to the active site Zn2+ ion. Hydrogen bonds to Thr200 and Thr199 as well as a water network, which is spread over the entire binding pocket, linking the fragment to the common hydrophilic residues can be observed. d) Binding mode in schematic representation.
), and water molecules are shown as red and yellow spheres. The solvent accessible surface is represented in white in panel b). a) Coordination geometry of the active site Zn2+ ion upon binding of 4. The tetrahedral geometry is changed to distorted trigonal-bipyramidal arrangement with Nε2-His94 (2.0 Å), Nε2-His96 (2.0 Å), and the sulfur atom of 4 (2.4 Å) in the equatorial plane, whereas Nδ1-His119 (2.1 Å) and the oxygen atom of 4 (2.5 Å) are in the axial positions. The angles in the plane are 143.5° (S-Zn-His96), 109.5° (S-Zn-His94), and 101.7° (His94-Zn-His96). The two axial ligands and the Zn2+ ion form an angle of 161.2°. b) The binding pocket of CA II can be divided into a hydrophobic (red line) and a hydrophilic binding region (blue line). The fragment is stabilized by a hydrogen bond network which is mediated by five water molecules (red spheres) addressing the typical hydrophilic residues in CA II. The remaining waters in the binding pocket (yellow spheres) are connected to the hydrogen bond network as well, however not performing any further strong interactions to the protein. c) Detailed binding mode. Hydrogen bond interactions are indicated by the dashed lines. The equivalent distances are shown in the schematic representation of the binding mode in panel d). The difference electron density (Fo—Fc) for the ligands and directly interacting water molecules is shown at a σ level of 2.0. Atomic distances are shown in Å. The fragment coordinates with its sulfur and oxygen atom to the active site Zn2+ ion. Hydrogen bonds to Thr200 and Thr199 as well as a water network, which is spread over the entire binding pocket, linking the fragment to the common hydrophilic residues can be observed. d) Binding mode in schematic representation.
The equatorial plane shows Zn2+ distances of 2.0 Å for Nε2-His94, 2.0 Å for Nε2-His96, and 2.4 Å for the sulfur atom with angles of 143.5° (S–Zn–His96), 109.5° (S–Zn–His94), and 101.7° (His94–Zn–His96). The Zn2+ ion is positioned 0.25 Å away from the plane formed by the equatorial coordinating ligands. The axial oxygen atom forms a distance of 2.5 Å and Nδ1-His199 is positioned 2.1 Å away from the metal ion. The two axial coordinating ligand atoms and the Zn2+ ion form an angle of 161.2°.
In addition to metal coordination, compound 4 also participates in three hydrogen bonds (Figure 2b–d). The Oγ1 atom of Thr200 forms a hydrogen bond to the oxygen in 4 at a distance of 3.2 Å. Furthermore, the same oxygen in 4 acts as the donor in a hydrogen bond network via Oγ1 of Thr199 (3.6 Å) to Oε1 of Glu106 (2.5 Å). This network is well known for typical sulfonamide inhibitors; however, the distance to Thr199 is increased, as the fragment most likely cannot move closer to Thr199. Interestingly, fragment 4 participates in a hydrogen bonding network that is spread over the entire binding pocket, which provides additional stabilization in the active site of CA II (Figure 2 b). The interactions are mediated by five water molecules, which address all residues in the hydrophilic pocket of CA II, i.e., Tyr7, Asn62, His64, Asn67, Glu69, and Gln92 (Figure 2 c, d). Fragment 4 is linked via two water molecules to the terminal side chain OH group of Tyr7 and Nδ1 of the proton shuttle His64 in the “in” conformation. In addition, 4 is connected via two water molecules to Asn67 that forms an additional hydrogen bond network to Glu69 at the exit of the binding pocket mediated by a water molecule. Nε2 of Gln92 and Nδ2 of Asn62 also participate in this network via a water molecule (Figure 2b–d). In addition to these direct interactions with water molecules, several other water molecules close to the exit of the binding pocket are involved in this network (Figure 2 b–d). The distribution of B values of the participating water molecules illustrates the well-coordinated water chain (20.5, 14.8, 26.3, 32.8, and 17.1 Å2). The conjugated ring system of 4 also experiences lipophilic interactions, as it points toward a hydrophobic binding pocket. In total, 26 van der Waals interactions can be identified.
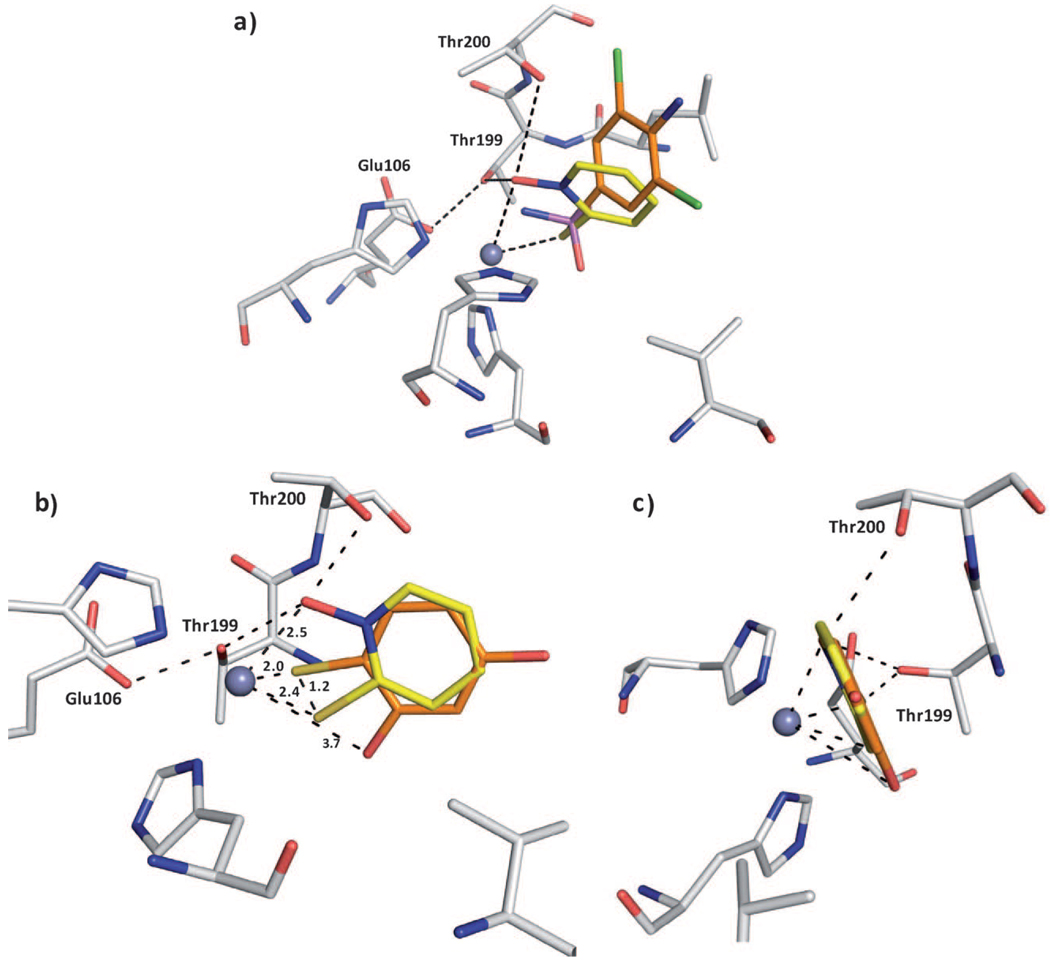
The superposition of 4 with a sulfonamide inhibitor 5 (PDB ID: 1ZGE) reveals that the sulfur atom of 4 adopts a position shifted by 0.9 Å relative to the sulfonamide sulfur atom (Figure 3 a).[18] This shift depends on the fact that the sulfur atom in 4 coordinates the Zn2+ ion, which is not the case for 5. Furthermore, the ring system is shifted relative to the phenyl group of the sulfonamide inhibitor. Because the oxygen atom is coordinated to the Zn2+ ion and stabilized by Thr199 and Thr200, the ring system is slightly rotated and shifted toward the Zn2+ ion in comparison with the sulfonamide fragment.
Figure 3.
Superpositions of compound 4. a) Superposition with sulfonamide inhibitor 5 in complex with CA II; the protein residues and ligands are shown in stick representation (protein:  ; compound 4:
; compound 4:  ; sulfonamide 5:
; sulfonamide 5:  ; PDB ID: 1ZGE). The sulfur atoms are located at different positions. Fragment 4 forms an additional coordination to the Zn2+ ion. The ring system of 4 is shifted relative to the sulfonamide inhibitor which is dependent on the hydrogen bonds of 4 to the oxygen atoms of Thr199 and Thr200. b) Superposition of 4 with a structurally related fragment 7 from a crystal structure PDB ID: 2OSF (
; PDB ID: 1ZGE). The sulfur atoms are located at different positions. Fragment 4 forms an additional coordination to the Zn2+ ion. The ring system of 4 is shifted relative to the sulfonamide inhibitor which is dependent on the hydrogen bonds of 4 to the oxygen atoms of Thr199 and Thr200. b) Superposition of 4 with a structurally related fragment 7 from a crystal structure PDB ID: 2OSF ( ). The sulfur atoms are shifted by 1.2 Å. The ring system of the superimposed ligands is rotated by 180°. In contrast to this ligand, 4 can form hydrogen bonds to Thr199 and Thr200. c) Image in panel b) rotated by 90°.
). The sulfur atoms are shifted by 1.2 Å. The ring system of the superimposed ligands is rotated by 180°. In contrast to this ligand, 4 can form hydrogen bonds to Thr199 and Thr200. c) Image in panel b) rotated by 90°.
Discussion
The inhibition of 1–4 with respect to the considered CA isoforms is not exceptionally strong; however, they reveal a new and promising anchoring motif for CA inhibitors. Crystallographic analysis of the CA II–4 complex reveals a bidentate binding mode to the Zn2+ ion with interactions to Thr199, which itself donates a hydrogen bond to Glu106. Such binding has been observed earlier only in the hCA II–N-hydroxyurea complex, in which the Zn2+ ion is coordinated by the N and O atoms of the inhibitor (which is presumably deprotonated).[19] In addition, 4 accepts a hydrogen bond from Thr200. The weaker affinity toward some of the isoforms can easily be understood, because these show variable amino acid compositions within the active site, for instance, at position 200. For example, a valine in hCA XIII and a histidine in hCA I is present at this position (and many others, mainly toward the exit of the cavity). These residues cannot produce a similar interaction with 4, which leads to a greater than twofold decrease in affinity against these isoforms.
From inhibition, crystallographic, and previously reported modeling data, we can also infer the binding mode of compounds 1–3.[17] As the inhibition profile of all four compounds is the same against each isoform, and their metal complexes with a tris(histidine) active site model complex are all similar, the binding modes of 1–3 are most likely nearly identical to that of 4.[17] Indeed, in a previously reported tris(pyrazolyl)borate Zn2+ model complex, fragment 4 was shown to bind in a bidentate fashion to the Zn2+ center, generating a five-coordinate, distorted trigonal-bipyramidal coordination geometry at the Zn2+ ion. In this model complex, the Zn—S distance was 2.32 Å, and the Zn—O distance was 2.08 Å.[17] In the structure reported herein, the Zn—S distance is ~2.4 Å, very close to that observed in the model complex. The Zn–O distance in the complex of 4 with CA is longer than that found in the model complex, at 2.5 Å. The oxygen atom is arranged in this increased distance to the metal ion by additional interactions with Thr199 and Thr200. These favorable hydrogen bonds are also formed at rather long distances, which shows that the oxygen atom in 4 adopts a position from which interactions to all three groups are possible. Nonetheless, it is likely that the related fragments 1, 2, and 3 also coordinate in a chelating fashion with oxygen and/or sulfur atoms to the Zn2+ ion in bidentate manner.
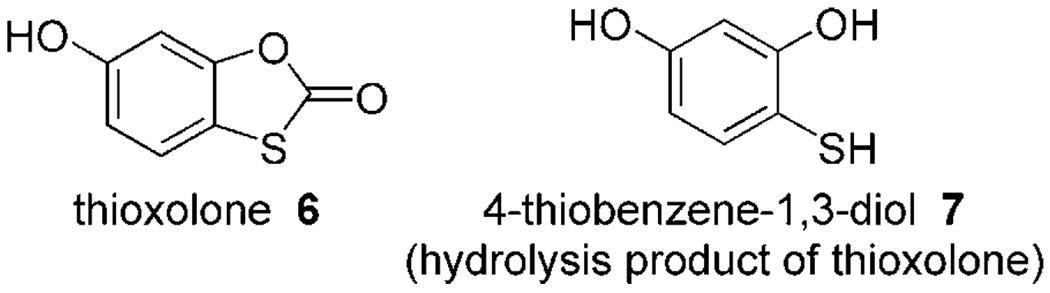
A similar CA II complex has been determined crystallographically and was recently published by Barrese et al.[20] Investigating the inhibition profile and binding mode of thioxolone 6 in CA II, they observed that the inhibitor, most likely a prodrug, is hydrolyzed upon binding to form the fragment 4-thiobenzene-1,3-diol 7 (Ki=148 µm)[20] (Figure 4). However, superposition of 7 with 4 shows significant differences in the binding mode, with 7 binding in a monodentate fashion (Figure 3b, c). Although both inhibitor ring systems are similarly substituted with oxygen and sulfur donor atoms, the interaction pattern with CA II is completely different. The ring systems are rotated by 180° relative to each other, and in 7 the oxygen donor atom does not interact with Thr199 and Thr200. Moreover, the sulfur atom in 7 resides in a different position, in total shifted by 1.2 Å. It appears that the nitrogen atom in the heterocycle of 4, which is not present in the compared fragment 7, as well as the thione (C=S) nature of the donor functionality in 4 influences the binding mode and improves the Zn2+ coordination in CA II. The trigonal-bipyramidal coordination of 4 and the interactions with Thr199 and Thr200 result in an excellent set of interactions of fragment 4 with the active site Zn2+ ion.
Figure 4.
Thioxolone 6 and its hydrolysis fragment 7 from PDB ID: 2OSF.
The binding affinities of 1–4 to CA II are significantly better than those of other alternative ZBGs such as the sulfamide (1130 µm) and sulfamic acid (390 µm) groups.[13] The observed bidentate binding mode of 4 to CA II is very promising and rarely observed in CAIs. The substitution of 4 with additional functional groups should notably increase the affinity toward CAs, as shown previously for sulfamides, for example.[12] Lipophilic moieties can be attached to interact with the hydrophobic cavities found inside CAs. In addition, hydrophilic residues such as Asn62, Asn67, Glu69, and Gln92 should be targeted for hydrogen bonding. Further development of this ZBG could lead to a new, promising class of CAIs.
Experimental Section
Chemistry and CA II inhibition
CA inhibition
An Applied Photophysics stopped-flow instrument was used for assays of the CA-catalyzed CO2 hydration activity.[21] Phenol red (at a concentration of 0.2 mm) was used as an indicator, working at the absorbance maximum of λ 557 nm, with 20 mm HEPES (pH 7.5) as buffer, and 20 mm Na2SO4 (for maintaining a constant ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mm for determination of the kinetic parameters and inhibition constants (five different substrate concentrations were used). For each inhibitor, at least six traces of the initial 5–10% of the reaction were used for determining the initial velocity. The uncatalyzed rates were determined by the same protocol and subtracted from the total observed rates. Stock solutions of inhibitor (0.1 mm) were prepared in distilled deionized H2O, and dilutions up to 0.01 nm were done thereafter with distilled deionized H2O. Experiments were performed with six different inhibitor concentrations, varying from 100 µm to 0.1 nm. Inhibitor and enzyme solutions were pre-incubated for 15 min–24 h: at room temperature for 15 min, or at 4 °C for all other incubation times prior to assay, in order to allow formation of the enzyme–inhibitor complex or for the eventual active-site-mediated hydrolysis of the inhibitor. Data reported in Table 1 show the inhibition after 15 min incubation, as there were no differences in inhibitory potency if the enzyme and inhibitors were incubated for longer periods.[1] The inhibition constants were obtained by nonlinear least-squares methods using PRISM 3, as reported earlier,[22–26] and represent the mean from at least three different determinations. CA isozymes were prepared as reported previously.[27–33]
Synthesis
Maltol 1, 1,2-HOPO 3, and 1,2-HOPTO 4 were obtained from commercial sources (Aldrich). Thiomaltol 2 was synthesized as previously described.[34]
Crystallization, X-ray data collection, and refinement
Crystals of hCA II (from Sigma–Aldrich) in complex with 4 were obtained by the sitting drop technique, using 10 mg mL−1 protein with 2 mm inhibitor in 50 mm Tris·HCl buffer (pH 8.0). The drops consisted of 5 µL of the enzyme–inhibitor solution and 5 µL of the precipitant solution containing 3.0 m (NH4)2SO4 in 100 mm Tris·HCl (pH 8.0) and 0.15 mm p-chloromercuribenzoic acid to promote the growth of highly oriented crystals. The drops were equilibrated by vapor diffusion against the precipitant solution at 18°C, and crystals appeared after 1–4 days. The data were collected at the synchrotron BESSY II in Berlin (Germany) on PSF beamline 14.2. The experiment was performed at 100 K using synchrotron radiation (λ 0.91841 Å) on a MAR CCD 165 mm detector. The crystals were monoclinic P21 with the following cell parameters: a=42.3 Å, b= 41.4 Å, c=72.4 Å, β=104.48. The data collection parameters and refinement statistics are listed in Table 2.
Table 2.
Crystallographic data for PDB ID: 3M1K.
| Parameter | Value |
|---|---|
| A. Data collection and processing: | |
| beamline | BESSY 14.2 |
| no. crystals used | 1 |
| λ [Å] | 0.91841 |
| space group | P21 |
| Unit cell parameters: | |
| a, b, c [Å] | 42.3, 41.4, 72.4 |
| β [°] | 104.4 |
| B. Diffraction data: | |
| resolution range [Å][a] | 25–1.35 (1.37–1.35) |
| unique reflections[a] | 50925 (2347) |
| R(I)sym [%][a] | 5.1 (25.4) |
| completeness [%][a] | 95.2 (88.8) |
| redundancy[a] | 2.2 (1.8) |
| I/σ(I)[a] | 15.1 (2.9) |
| C. Refinement: | |
| program used for refinement | SHELXL |
| resolution range [Å] | 10–1.35 |
| reflections used in refinement | 49131 |
| Rfree [Fo; Fo > 4 σFo] | 18.6; 17.6 |
| Rwork [Fo; Fo > 4 σFo] | 12.8; 12.1 |
| No. atoms (non hydrogen): | |
| protein atoms | 2080 |
| water molecules | 239 |
| ligand atoms | 18 |
| RMSD, angle [°] | 2.2 |
| RMSD, bond [Å] | 0.012 |
| Ramachandran plot: | |
| most favoured regions [%] | 88.4 |
| additionally allowed regions [%] | 11.1 |
| generously allowed regions [%] | 0.5 |
| disallowed regions [%] | 0 |
| Mean B factor [Å2]: | |
| main chain | 14.1 |
| side chain | 18.2 |
| water molecules | 27.1 |
| ligand atoms | 24.1 |
| zinc atom | 8.4 |
| Matthews’ coefficient [Å3 Da−1] | 2.2 |
| solvent content [%] | 43.4 |
Highest-resolution shell.
Data were processed and scaled with Denzo and Scalepack as implemented in HKL2000.[35] The complex structures were analyzed by difference Fourier techniques, using the PDB file 2OQ5 as starting model for refinement. Refinement was continued with CNS[36] and SHELXL-97.[37] For each refinement step, at least 10 cycles of conjugate minimization were performed, with restraints on bond distances, angles, and B values. For each crystal, the 2 Fo—Fc and Fo—Fc maps were calculated, and the inhibitor binding mode assigned from the difference electron density. Intermittent cycles of model building were done with the program COOT.[38] The coordinates have been deposited in the RCSB Protein Data Bank (PDB; http://www.rcsb.org/pdb/) with the reference code 3M1K.
Acknowledgements
This research was supported financially by the DFG (KL1204/10-1) and by two EU projects to C.T.S. (DeZnIT, 6th FP, and METOXI, 7th FP), which are gratefully acknowledged. S.M.C. was supported by a grant from the National Institutes of Health (R01 HL00049-01). We acknowledge support from the beamline staff at Bessy II, Berlin and a travel grant from the Helmholtz Zentrum Berlin.
References
- 1.Supuran CT, Scozzafava A, Conway J, editors. Carbonic Anhydrase: Its Inhibitors and Activators. Boca Raton, FL: CRC Press; 2004. [Google Scholar]
- 2.Supuran CT, Scozzafava A, Casini A. Med. Res. Rev. 2003;23:146–189. doi: 10.1002/med.10025. [DOI] [PubMed] [Google Scholar]
- 3.Supuran CT. Nat. Rev. Drug Discovery. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 4.Scozzafava A, Mastrolorenzo A, Supuran CT. Expert Opin. Ther. Pat. 2006;16:1627–1664. doi: 10.1517/13543776.16.1.27. [DOI] [PubMed] [Google Scholar]
- 5.Scozzafava A, Mastrolorenzo A, Supuran CT. Expert Opin. Ther. Pat. 2004;14:667–702. [Google Scholar]
- 6.Pastorekova S, Parkkila S, Pastorek J, Supuran CT. J. Enzyme Inhib. Med. Chem. 2004;19:199–229. doi: 10.1080/14756360410001689540. [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen JA, Major Jourden JL, Miller MT, Cohen SM. Biochim. Biophys. Acta Mol. Cell Res. 2010;1803:72–94. doi: 10.1016/j.bbamcr.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Christianson DW, Fierke CA. Acc. Chem. Res. 1996;29:331–339. [Google Scholar]
- 9.Supuran CT, Scozzafava A. Expert Opin. Ther. Pat. 2002;12:217–242. [Google Scholar]
- 10.Supuran CT, Scozzafava A. Expert Opin. Ther. Pat. 2000;10:575–600. [Google Scholar]
- 11.Winum J-Y, Scozzafava A, Montero J-L, Supuran CT. Med. Res. Rev. 2005;25:186–228. doi: 10.1002/med.20021. [DOI] [PubMed] [Google Scholar]
- 12.Winum J-Y, Scozzafava A, Montero J-L, Supuran CT. Med. Res. Rev. 2006;26:767–792. doi: 10.1002/med.20068. [DOI] [PubMed] [Google Scholar]
- 13.Winum JY, Scozzafava A, Montero J-L, Supuran CT. Cur. Top. Med. Chem. 2007;7:835–848. doi: 10.2174/156802607780636771. [DOI] [PubMed] [Google Scholar]
- 14.Puerta DT, Lewis JA, Cohen SM. J. Am. Chem. Soc. 2004;126:8388–8389. doi: 10.1021/ja0485513. [DOI] [PubMed] [Google Scholar]
- 15.Puerta DT, Griffin MO, Lewis JA, Romero-Perez D, Garcia R, Villarreal FJ, Cohen SM. J. Biol. Inorg. Chem. 2006;11:131–138. doi: 10.1007/s00775-005-0053-x. [DOI] [PubMed] [Google Scholar]
- 16.Domsic JF, Avvaru BS, Kim CU, Gruner SM, Agbandje-McKenna M, Silverman DN, McKenna R. J. Biol. Chem. 2008;283:30766–30771. doi: 10.1074/jbc.M805353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puerta DT, Cohen SM. Inorg. Chem. 2003;42:3423–3430. doi: 10.1021/ic026029g. [DOI] [PubMed] [Google Scholar]
- 18.Ilies MA, Vullo D, Pastorek J, Scozzafava A, Ilies M, Caproiu MT, Pastorekova S, Supuran CT. J. Med. Chem. 2003;46:2187–2196. doi: 10.1021/jm021123s. [DOI] [PubMed] [Google Scholar]
- 19.Temperini C, Innocenti A, Scozzafava A, Supuran CT. Bioorg. Med. Chem. Lett. 2006;16:4316–4320. doi: 10.1016/j.bmcl.2006.05.068. [DOI] [PubMed] [Google Scholar]
- 20.Barrese AA, Genis C, Fisher SZ, Orwenyo JN, Kumara MT, Dutta SK, Phillips E, Kiddle JJ, Tu C, Silverman DN, Govindasamy L, Agbandje-McKenna M, McKenna R, Tripp BC. Biochemistry. 2008;47:3174–3184. doi: 10.1021/bi702385k. [DOI] [PubMed] [Google Scholar]
- 21.Khalifah RG. J. Biol. Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 22.Briganti F, Mangani S, Orioli P, Scozzafava A, Vernaglione G, Supuran CT. Biochemistry. 1997;36:10384–10392. doi: 10.1021/bi970760v. [DOI] [PubMed] [Google Scholar]
- 23.Temperini C, Scozzafava A, Puccetti L, Supuran CT. Bioorg. Med. Chem. Lett. 2005;15:5136–5141. doi: 10.1016/j.bmcl.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 24.Temperini C, Scozzafava A, Vullo D, Supuran CT. Chem. Eur. J. 2006;12:7057–7066. doi: 10.1002/chem.200600159. [DOI] [PubMed] [Google Scholar]
- 25.Temperini C, Innocenti A, Scozzafava A, Mastrolorenzo A, Supuran CT. Bioorg. Med. Chem. Lett. 2007;17:628–635. doi: 10.1016/j.bmcl.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 26.Temperini C, Innocenti A, Scozzafava A, Supuran CT. Bioorg. Med. Chem. 2008;16:8373–8378. doi: 10.1016/j.bmc.2008.08.043. [DOI] [PubMed] [Google Scholar]
- 27.Innocenti A, Vullo D, Scozzafava A, Supuran CT. Bioorg. Med. Chem. Lett. 2008;18:1583–1587. doi: 10.1016/j.bmcl.2008.01.077. [DOI] [PubMed] [Google Scholar]
- 28.Innocenti A, Hilvo M, Scozzafava A, Parkkila S, Supuran CT. Bioorg. Med. Chem. Lett. 2008;18:3593–3596. doi: 10.1016/j.bmcl.2008.04.077. [DOI] [PubMed] [Google Scholar]
- 29.Innocenti A, Vullo D, Scozzafava A, Supuran CT. Bioorg. Med. Chem. 2008;16:7424–7428. doi: 10.1016/j.bmc.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Bayram E, Senturk M, Irfan Kufrevioglu O, Supuran CT. Bioorg. Med. Chem. 2008;16:9101–9105. doi: 10.1016/j.bmc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Maresca A, Temperini C, Vu H, Pham NB, Poulsen S-A, Scozzafava A, Quinn RJ, Supuran CT. J. Am. Chem. Soc. 2009;131:3057–3062. doi: 10.1021/ja809683v. [DOI] [PubMed] [Google Scholar]
- 32.Maresca A, Temperini C, Pochet L, Masereel B, Scozzafava A, Supuran CT. J. Med. Chem. 2010;53:335–344. doi: 10.1021/jm901287j. [DOI] [PubMed] [Google Scholar]
- 33.Temperini C, Innocenti A, Scozzafava A, Parkkila S, Supuran CT. J. Med. Chem. 2010;53:850–854. doi: 10.1021/jm901524f. [DOI] [PubMed] [Google Scholar]
- 34.Lewis JA, Puerta DT, Cohen SM. Inorg. Chem. 2003;42:7455–7459. doi: 10.1021/ic0347135. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W, Carter CW., Jr . Methods in Enzymology. Vol. 276. London: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick GM, Schneider TR, Charles WC, Jr, Robert MS. Methods in Enzymology. Vol. 277. London: Academic Press; 1997. pp. 319–343. [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]