Active translation of HIF-1α requires association and trafficking of HIF mRNA on dynamic microtubules, whereas microtubule disruption represses HIF-1α translation by releasing its mRNA from polysomes and sequestering it to P-bodies.
Abstract
The hypoxia inducible factor 1α (HIF-1α) is overexpressed in solid tumors, driving tumor angiogenesis and survival. However, the mechanisms regulating HIF-1α expression in solid tumors are not fully understood. In this study, we find that microtubule integrity and dynamics are intricately involved in orchestrating HIF-1α translation. HIF-1α messenger RNA (mRNA) traffics on dynamic microtubules when it is actively translated. Microtubule perturbation by taxol (TX) and other microtubule-targeting drugs stalls HIF-1α mRNA transport and releases it from polysomes, suppressing its translation. Immunoprecipitation of the P-body component Argonaute 2 (Ago2) after microtubule disruption shows significant enrichment of HIF-1α mRNAs and HIF-targeting microRNAs (miRNAs). Inhibition of HIF-repressing miRNAs or Ago2 knockdown abrogates TX’s ability to suppress HIF-1α translation. Interestingly, microtubule repolymerization after nocodazole washout allows HIF-1α mRNA to reenter active translation, suggesting that microtubule dynamics exert tight yet reversible control over HIF-1α translation. Collectively, we provide evidence for a new mechanism of microtubule-dependent HIF-1α translation with important implications for cell biology.
Introduction
The majority of solid tumors contain regions of hypoxia as a result of their poor local vasculature (Pouysségur et al., 2006). The hypoxia inducible factor 1 (HIF-1) is the master regulator of the cell’s response to low-oxygen tension that controls the transcription of >100 genes essential for hypoxic adaptation, angiogenesis, invasion, and tumor progression (Semenza and Wang, 1992; Semenza, 2003). HIF-1α, the oxygen-regulated subunit of HIF-1, is found overexpressed in >70% of all human tumors, and its expression is correlated with poor prognosis and resistance to therapy. Therefore, inhibition of HIF-1α represents an attractive strategy for anticancer therapy.
The availability of HIF-1α protein under different environmental conditions is tightly controlled though protein synthesis and degradation. The oxygen-dependent proteasomal degradation of HIF-1α, mediated primarily by the von Hippel Lindau protein (pVHL), is a well-characterized pathway, and it is widely accepted that inhibition of pVHL results in HIF-1α protein accumulation during hypoxia (Jaakkola et al., 2001). In contrast to the inhibition of global protein synthesis that occurs in the absence of oxygen (Guppy et al., 2005), HIF-1α translation continues unobstructed under hypoxia, contributing to the increase in HIF-1α protein and transcriptional activity. Although the pathways involved in HIF-1α protein stability and degradation are well characterized, little is known about the regulation of HIF-1α translation, hindering the potential therapeutic exploitation of the latter mechanism.
In this study, we report a new mechanism of HIF-1α translational regulation that requires the presence of functional microtubules. The microtubule cytoskeleton is an extensive network of filaments that undergoes constant remodeling according to its role in diverse cellular functions, ranging from cell division to intracellular trafficking. These features make microtubules attractive targets for cancer therapy, as indicated by the broad clinical use of existing microtubule-targeting drugs (MTDs), such as taxanes and vinca alkaloids, and the large number of new MTDs in development (Jordan and Wilson, 2004; Kavallaris, 2010). Unlike conventional thinking supporting inhibition of mitosis as the predominant mechanism of MTD action, the slow growth of human tumors (Skipper, 1971) together with the increasing body of literature showing mitosis-independent cell kill by MTDs (Giannakakou et al., 2000; Mabjeesh et al., 2003; Gascoigne and Taylor, 2008) suggest that the antitumor activity of this class of drugs can also be attributed to their effects in interphase. However, the pathways regulated by interphase microtubule dynamics in epithelial cancer biology are poorly understood. Understanding these pathways may help elucidate mechanisms of resistance to MTDs and provide important insights into the selection of tumor types and/or individual patients most likely to benefit from this class of chemotherapeutics.
In this study, we provide evidence that taxol (TX) and other MTDs shift the association of HIF-1α mRNA from actively translating polysomes to translationally inactive ribosomal subunits. This change in HIF-1α polysome association occurs downstream of microtubule disruption, as the presence of a tubulin mutation in either the taxane- or 2-methoxyestradiol (2ME2)–binding site prevented the drug-induced changes. Using molecular beacons (MBs) to visualize endogenous HIF-1α mRNA in living cells, we showed that HIF-1α translation requires active transport of HIF-1α mRNA on interphase microtubules. Microtubule perturbation targeted HIF-1α mRNA to the P-body component Ago2, where HIF-repressing microRNAs (miRNAs) were also recruited. Microtubule repolymerization reversed this process and enabled HIF-1α mRNA to reenter active translation. Moreover, inhibition of endogenous HIF-targeting miRNAs abrogated the ability of TX to suppress HIF-1α, suggesting an important role for miRNA machinery in the microtubule-dependent regulation of HIF-1α translation. In addition, knockdown (KD) of Ago2 also counteracted TX’s effect on HIF translation, highlighting the importance of the P-body component for this mechanism of action. Further elucidation of this mechanism will not only provide important insights into the role of microtubules in the regulation of protein translation in epithelial cell biology but will also provide opportunities to specifically target HIF-1α translation for cancer therapy.
Results
MTDs inhibit HIF-1α translation
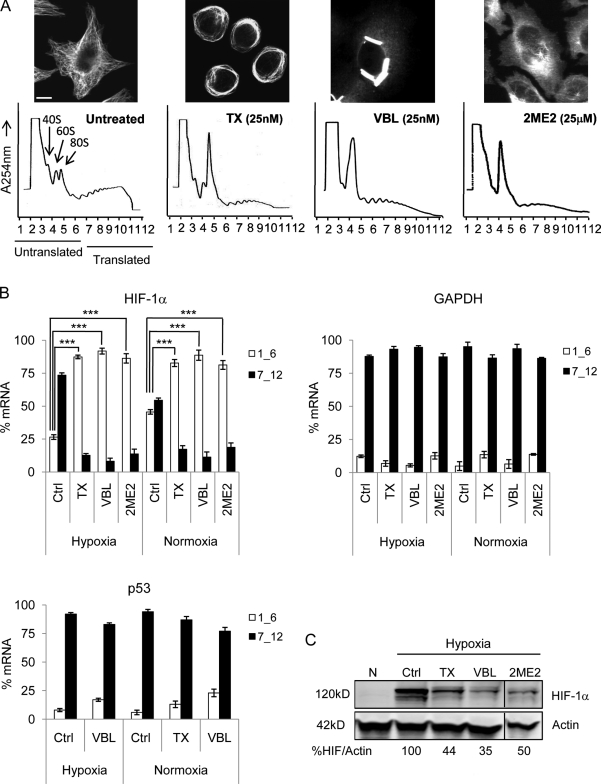
We have previously shown that TX, 2ME2, and other MTDs exert their antiangiogenic effects by inhibiting HIF-1α protein expression and transcriptional activation of HIF target genes, including VEGF (Mabjeesh et al., 2003; Escuin et al., 2005). However, the mechanism underlying the drug-induced HIF-1α inhibition was unknown. To investigate whether microtubule disruption inhibited HIF-1α translation, we treated MCF7 cells with TX, vinblastine, or 2ME2 and assessed the polysome association profile of HIF-1α after sucrose gradient centrifugation. In this assay, we divided each gradient into 12 equal-volume fractions and monitored the position of nontranslating ribosomal subunits (1–6) or translationally active polysomes (7–12) with continuous A254 measurements (Fig. 1 A). The amount of HIF-1α mRNA in each fraction was quantified by quantitative RT-PCR (qRT-PCR), and its distribution between translating and nontranslating fractions is shown in Fig. 1 B. In untreated cells, the majority of HIF-1α mRNA cofractionated with actively translating polysomes under both hypoxia (∼74%) and normoxia (∼55%). Treatment with all of the MTDs resulted in a shift of HIF-1α mRNA from active to inactive translation, which is consistent with the ability of each MTD to efficiently disrupt cellular microtubules (Fig. 1 A, top) and down-regulate HIF-1α protein (Fig. 1 C). No shift was observed in the translation status of unrelated messages such as GAPDH (Fig. 1 B), HIF-1β, actin (not depicted), or the transcription factor p53 (Fig. 1 B). The latter is an important control, as we have previously shown that p53 protein utilizes microtubules for its nuclear targeting; however, in contrast to HIF-1α, microtubule disruption had no effect on p53 protein expression (Giannakakou et al., 2000, 2002). The polysome profile of HIF-1α was also altered by MTDs in MDA-MB-231 breast cancer cells and PC3 prostate cancer cells (unpublished data), suggesting that the drug-induced inhibition of HIF-1α translation is not cell type dependent.
Figure 1.
MTDs inhibit HIF-1α translation. (A) Polysome association profile of MCF7 cells after overnight treatment with 25 nM TX, 25 nM vinblastine (VBL), or 25 µM 2ME2 visualized after sucrose gradient centrifugation. The 40, 60, and 80S ribosomal subunits and polysomes were fractionated and monitored with continuous A254 measurements. Representative profiles and microtubule images after each drug treatment are shown. Bar, 10 µm. (B) RNA was extracted from each fraction, and HIF-1α, GAPDH, and p53 expression was quantified by qRT-PCR. The distribution of each mRNA between nontranslating (1_6) and translating (7_12) fractions is plotted (mean ± SEM; n = 3–10; ***, P < 0.001). (C) Immunoblot of HIF-1α and actin from MCF7 cells treated as in A and exposed to normoxia (N) or 4-h hypoxia to visualize HIF-1α protein. Black lines indicate that intervening lanes have been spliced out.
Inhibition of HIF-1α translation occurs downstream of microtubule disruption
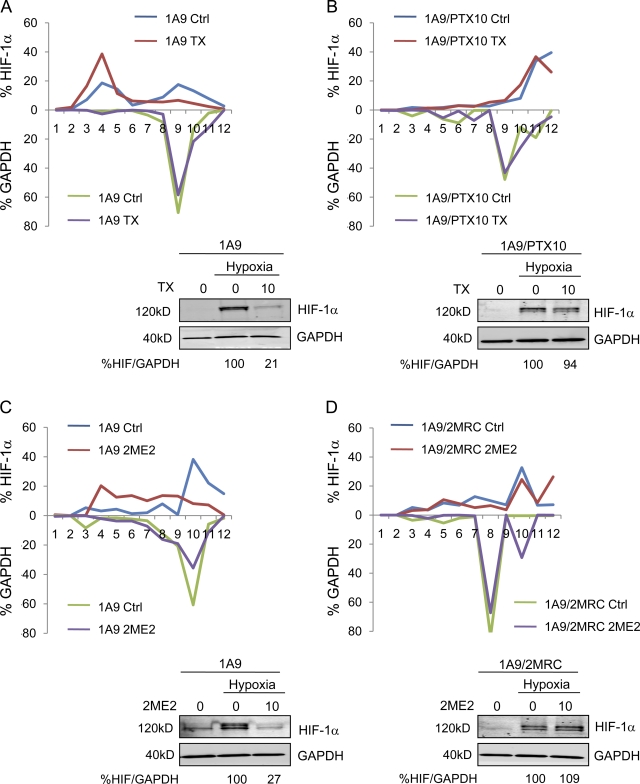
To further investigate whether the inhibition of HIF-1α translation is dependent on chemomechanics of the microtubule cytoskeleton, we used the TX-resistant human ovarian carcinoma cell line 1A9/PTX10, which was previously established in our laboratory. These cells harbor an acquired β-tubulin mutation (Fβ270V) at the taxane-binding site (Giannakakou et al., 1997) that prevents drug–target interaction. We treated parental 1A9 and 1A9/PTX10 cells with TX and assessed the polysome association profile of HIF-1α as described in Fig. 1. TX treatment led to the association of 71% of HIF-1α mRNA with the nontranslating ribosomal subunits consistent with the TX-induced inhibition of HIF-1α protein (Fig. 2 A, bottom) in 1A9 cells. In contrast, TX treatment did not affect HIF-1α translation in 1A9/PTX10 cells, as >90% of HIF-1α mRNA was associated with translating polysomes in both untreated and treated cells (Fig. 2 B and Table I). These results were corroborated by the lack of TX-induced microtubule stabilization (Fig. S1 A) and HIF-1α protein inhibition in 1A9/PTX10 cells (Fig. 2 B). Collectively, these findings indicate that efficient drug–tubulin interaction is required for the inhibition of HIF-1α translation.
Figure 2.
Microtubule disruption is required for drug-induced inhibition of HIF-1α translation. (A and B) Parental 1A9 (A) and 1A9/PTX10 (B) cells treated with 10 nM TX overnight and subjected to sucrose gradient fractionation. The percentage of HIF-1α or GAPDH mRNA/fraction is plotted for each cell line and drug treatment. (bottom) HIF-1α and GAPDH protein levels in 1A9 (A) and 1A9/PTX10 (B) cells treated with 10 nM TX overnight assessed by immunoblotting after 4-h hypoxia. (C and D) Similar experiments performed using 1A9 (C) or 1A9/2MRC (D) cells to assess the HIF-1α polysome profile after 10 µM 2ME2 treatment. (bottom) Immunoblot of HIF-1α and GAPDH after 4-h hypoxia (Table I).
Table I.
HIF-1α inhibition requires microtubule disruption
| Cell line | 1A9 | 1A9/PTX10 | 1A9 | 1A9/2MRC | ||||
| Ctrl | TX | Ctrl | TX | Ctrl | 2ME2 | Ctrl | 2ME2 | |
| HIF-1α | ||||||||
| Untranslated | 41.3 ± 0.6 | 71.0 ± 2.1 | 6.3 ± 1.5 | 4.5 ± 1.6 | 14.4 ± 1.5 | 52.9 ± 4.3 | 16.9 ± 2.2 | 17.5 ± 1.1 |
| Translated | 58.7 ± 1.6 | 29.0 ± 2.1 | 93.7 ± 1.5 | 95.5 ± 1.6 | 85.6 ± 1.5 | 47.1 ± 4.3 | 83.1 ± 2.2 | 82.5 ± 1.1 |
| GAPDH | ||||||||
| Untranslated | 2.5 ± 1.1 | 6.8 ± 2.9 | 18.8 ± 2.5 | 15.5 ± 4.9 | 12.3 ± 1.5 | 12.6 ± 4.1 | 10.3 ± 1.1 | 4.5 ± 1.8 |
| Translated | 97.5 ± 1.1 | 93.2 ± 2.9 | 81.2 ± 2.5 | 84.5 ± 4.9 | 87.7 ± 1.5 | 87.4 ± 4.1 | 89.7 ± 1.1 | 95.5 ± 1.8 |
Ctrl, control. Polysome association of HIF-1α and GAPDH mRNA in 1A9, 1A9/PTX10, and 1A9/2MRC cells. The percentage of translated versus untranslated mRNA is displayed as mean ± SEM (n = 2).
To rule out the possibility that these results were applicable to TX alone or to the specific pair of parental and drug-resistant cell lines, we developed a cell line resistant to the microtubule-depolymerizing drug 2ME2 (1A9/2MRC). The 1A9/2MRC cells acquired a different β-tubulin mutation (Vβ236I) that impairs 2ME2 binding and renders these cells >80-fold resistant to 2ME2 (Escuin et al., 2009). Treatment with 2ME2 inhibited HIF-1α translation in 1A9 cells (Fig. 2 C) but had no effect in 1A9/2MRC cells (Fig. 2 D and Table I), which is consistent with 2ME2’s inability to inhibit HIF-1α protein in these cells (Fig. 2 D). This result is consistent with the lack of 2ME2-mediated microtubule depolymerization in 1A9/2MRC cells (Fig. S1 A; Escuin et al., 2009) and further supports the notion that microtubule disruption is required for and precedes HIF-1α translation inhibition. No effect on the protein levels or polysome association of GAPDH (Fig. 2) or p53 (not depicted) was observed after MTD treatment in any cell line. Collectively, our results show that both microtubule stabilization and depolymerization act as triggers for HIF-1α translational repression.
HIF-1α mRNA colocalizes with the microtubule cytoskeleton
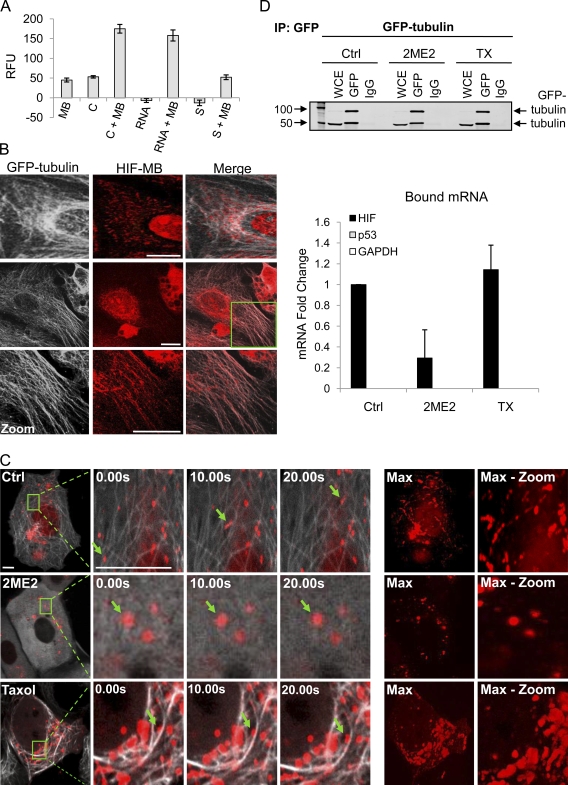
The inhibition of HIF-1α translation after MTD treatment prompted us to investigate the spatial relationship between HIF-1α mRNA and cytoplasmic microtubules. To visualize HIF-1α mRNA, we designed HIF-1α–specific MBs, single-stranded oligonucleotides (ODNs) labeled with a 5′ Texas red fluorophore and a 3′ quencher. In the absence of target mRNA, MBs form a hairpin and are devoid of fluorescence because of the proximity of the fluorophore and quencher. Upon hybridization with its target, the hairpin opens, resulting in fluorescence (Fig. S2, A and B; Tyagi and Kramer, 1996; Bratu et al., 2003). We first examined the specificity of our MBs in vitro and observed a more than threefold increase in fluorescence intensity after coincubation with a complementary sequence or total RNA from HIF-1α–expressing cells. In contrast, only background fluorescence was observed upon coincubation with a sense DNA ODN (Fig. 3 A). We next evaluated the subcellular localization of HIF-1α mRNA by hybridizing the MB with fixed MCF7 cells stably expressing EGFP–α-tubulin (MCF7–GFP-tub). Interestingly, we observed colocalization of the MBs with microtubules in untreated cells (Fig. 3 B). This intriguing result prompted us to examine the spatial and temporal distribution of HIF-1α mRNA after MB transfection in MCF7–GFP-tub cells. Live cell confocal microscopy revealed that HIF-1α MBs were dispersed throughout the cytoplasm and exhibited clear association with microtubules (Fig. 3 C). Furthermore, time-lapse imaging and particle tracking revealed microtubule-dependent movements of the MBs (Fig. 3 C and Videos 1 and 2). After 2ME2 or TX treatment, the HIF-1α MBs were redistributed into larger cytoplasmic clusters and exhibited more constrained, nondirectional movements rather than the long-range trafficking observed in untreated cells (Fig. 3 C and Videos 3 and 4). Notably, some HIF-1α MBs remained associated with TX-induced microtubule bundles; however, their movement was significantly compromised.
Figure 3.
HIF-1α mRNA associates with cellular microtubules. (A) HIF-1α MBs incubated at 37°C for 1 h with a complementary (C) or sense (S) DNA ODN or total RNA from MCF7 cells. Relative fluorescence units (RFU) are represented as mean ± SEM. (B) MCF7–GFP-tub (white) cells fixed and processed for MB (red) hybridization. (zoom) Higher magnification views of the boxed area are shown. Two representative untreated cells are shown. (C) MCF7–GFP-tub (white) cells transfected with 100 nM HIF-1α MB (red) overnight, treated with 100 µM 2ME2 or 100 nM TX (1 h) and imaged continuously for 60 s. Arrows show inhibition of HIF-1α movement. (max) Stack arithmetic shows trajectory of MB movement. (max-zoom) Higher magnification view of max from boxed areas in the first column (Videos 1–4). (D) Anti-GFP was used to immunoprecipitate lysates from MCF7–GFP-tub cells treated overnight with 25 µM 2ME2 or 25 nM TX. HIF-1α, p53, and GAPDH mRNA expression are shown as mean fold change ± SEM (n = 3). Bars, 10 µm.
To further investigate the specificity of HIF-1α MB association with microtubules, we transfected MCF7–GFP-tub cells with a GAPDH-specific MB or a scrambled MB. No microtubule colocalization or microtubule-dependent trafficking was observed for either MB (Fig. S2 C and Video 5). Collectively, these results reveal that HIF-1α mRNA associates with and traffics on microtubules in epithelial cancer cells.
HIF-1α mRNA coprecipitates with tubulin
To further confirm the interaction between tubulin and HIF-1α mRNA, we performed coprecipitations using MCF7–GFP-tub cells or MCF7–GFP mock cells as a negative control. Cell lysates treated with 2ME2, TX, or DMSO were subjected to anti-GFP immunoprecipitation. Bound RNA was extracted from each condition, and HIF-1α, p53, and GAPDH mRNA expression was quantified by qRT-PCR. Our results showed that HIF-1α mRNA was specifically bound to tubulin, as neither p53 nor GAPDH mRNA coprecipitated with microtubules (Fig. 3 D), and no bound mRNAs were identified in immunoprecipitates from MCF7–GFP mock cells (Fig. S2 F). Microtubule depolymerization by 2ME2 at concentrations that effectively inhibited HIF-1α protein levels (Fig. S2 D) resulted in dissociation of HIF-1α mRNA from tubulin, confirming our earlier observations using HIF-1α MBs (Fig. 3 D). In contrast, HIF-1α mRNA remained bound to microtubules after TX treatment (Fig. 3 D), which is consistent with our results in Fig. 3 C, where HIF-1α MBs remained associated with TX-induced microtubule bundles. No change in total RNA levels of HIF-1α, p53, or GAPDH was observed after MTD treatment (Fig. S2 E). Our results suggest that mRNA–tubulin binding alone is not sufficient to regulate HIF-1α translation but, rather, that the integrity and dynamics of the microtubule cytoskeleton are important for regulation of HIF-1α translation.
Microtubule disruption induces P-body formation
To understand how microtubule integrity regulates HIF-1α translation, we further examined the significance of the increased 80S monosomal peak that was consistently observed in the polysome profiles after MTD treatment (Fig. 1 A). We showed that this MTD-induced 80S increase was absent in traces from the two tubulin mutant cell lines (1A9/2MRC and 1A9/PTX10), suggesting that efficient microtubule disruption was causal to this effect (Fig. S1 C). This increase in 80S peak has been previously associated with inhibition of translation initiation after exposure to translation inhibitors, glucose starvation, and heat shock (Ashe et al., 2000; Low et al., 2005; Teixeira et al., 2005; Luke et al., 2007; Sivan et al., 2007). Furthermore, these 80S peaks are known to contain protein components of P-bodies and stress granules, both of which represent distinct cytoplasmic sites of translational repression (Ashe et al., 2000; Low et al., 2005). These data, together with our own observations, prompted us to investigate the potential involvement of P-bodies in the microtubule-dependent repression of HIF-1α translation.
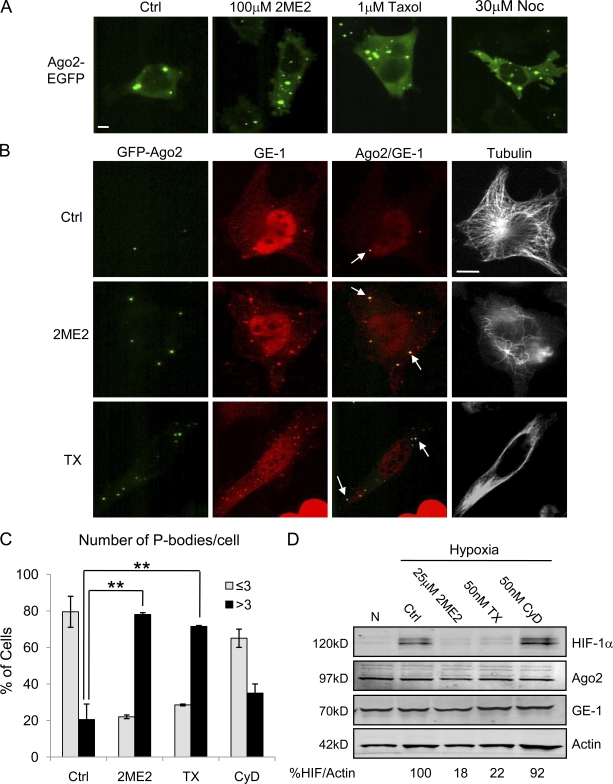
To visualize P-body formation in vivo, we used HeLa cells stably expressing the Argonaute family protein Ago2 fused to EGFP (HeLa–GFP-Ago2; Leung et al., 2006). Argonaute proteins are major components of the miRNA machinery that localize to P-bodies (Liu et al., 2005b; Sen and Blau, 2005). Live cell imaging of HeLa–GFP-Ago2 cells revealed distinct fluorescent particles of heterogeneous size in the cytoplasm of untreated cells. After treatment with the microtubule-depolymerizing agents 2ME2 or nocodazole (Noc) or the stabilizing drug TX, we observed a rapid and robust increase in the number of Ago2 foci per cell (Fig. 4 A). We confirmed that these Ago2 foci were P-bodies by immunostaining fixed samples with an antibody against GE-1/Hedls (human enhancer of decapping large subunit), a protein found exclusively in P-bodies, as opposed to other types of RNA-containing granules such as stress granules (Yu et al., 2005; Kedersha and Anderson, 2007). Visualization of cytoplasmic GE-1 and Ago2 revealed that 100% of Ago2 foci colocalized with GE-1 (Fig. 4 B, arrows). Importantly, P-body quantification revealed a significant increase after MTD treatment at concentrations that effectively disrupted microtubules and inhibited HIF-1α protein expression (Fig. 4, B and D). Specifically, 80% of untreated cells contained three or less P-bodies, whereas >70% of cells contained more than three P-bodies upon 2ME2 or TX treatment (Fig. 4 C). Although we arbitrarily used a cutoff of three P-bodies per cell, many MTD-treated cells contained >40 P-bodies. This change in P-body number is not because of an increase in Ago2 or GE-1 protein levels (Fig. 4 D).
Figure 4.
MTDs induce P-body formation. (A) Live cell imaging of HeLa–GFP-Ago2 cells treated with 100 µM 2ME2, 1 µM TX, or 30 µM Noc for 1 h. (B) HeLa–GFP-Ago2 (green) cells treated with 25 µM 2ME2 or 50 nM TX overnight, fixed, and stained for GE-1 (red) and α-tubulin (white). Arrows show colocalization of Ago2 and GE-1. (C) The number of Ago2 foci per cell was quantified (100 cells/condition) and plotted as percentage of cells with either three or less or more than three P-bodies (mean ± SEM; n = 3; **, P < 0.01). (D) Immunoblot of HeLa–GFP-Ago2 cells treated as in B and followed by 4-h hypoxia. Bars, 10 µm.
To determine whether the increase in P-body formation is specific to disruption of the microtubule cytoskeleton or whether it could result from more general cytoskeletal stress, we used the actin-depolymerizing agent cytochalasin D. Actin depolymerization did not affect P-body number (Fig. 4 C), suggesting that this effect was specific to disruption of microtubules. Consistent with this result, cytochalasin D treatment had no effect on HIF-1α protein levels (Fig. 4 D).
HIF-1α mRNA reversibly accumulates in P-bodies after microtubule disruption
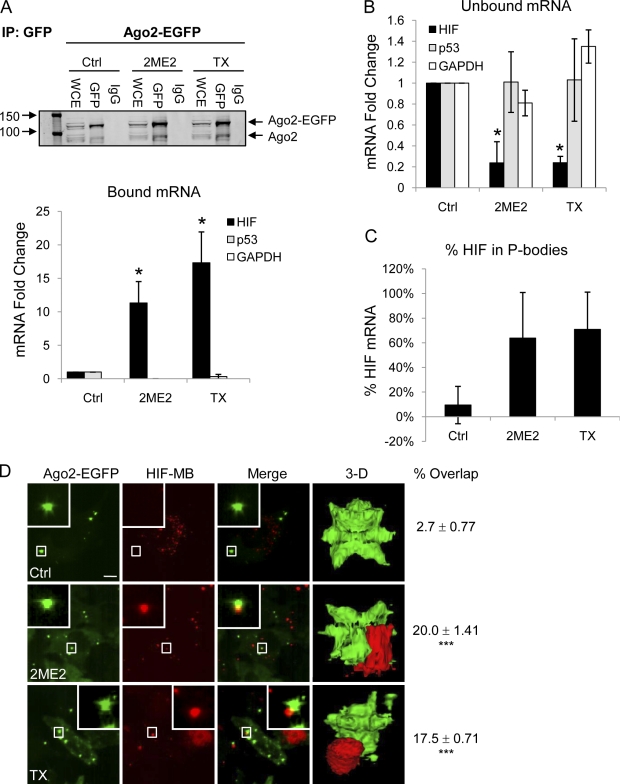
Thus far, we have shown that disruption of microtubule dynamics inhibits HIF-1α translation, redistributes HIF-1α MBs into larger cytoplasmic foci, and stimulates P-body formation (Figs. 1, 3, and 5). To examine whether microtubule stabilization or depolymerization sequesters HIF-1α mRNA into P-bodies, we immunoprecipitated Ago2 protein, extracted bound mRNAs, and performed qRT-PCR for HIF-1α, GAPDH, and p53. TX and 2ME2 treatment resulted in 17-fold and 12-fold increases, respectively, in the amount of HIF-1α mRNA associated with Ago2 compared with untreated cells (Fig. 5 A, bottom). A concomitant decrease in HIF-1α mRNA isolated from the non-Ago2–bound fraction was also observed (Fig. 5 B), further confirming the significant enrichment of HIF-1α mRNA in P-bodies after MTD treatment. More specifically, we found that 9% of total HIF-1α mRNA localized to P-bodies in control cells, whereas 64% and 70% of HIF-1α mRNA were found in P-bodies after treatment with 2ME2 or TX, respectively (Fig. 5 C). No association with Ago2 was seen for GAPDH or p53 mRNA (Fig. 5 A), and no change in the amount of total HIF-1α, p53, or GAPDH mRNA was observed upon MTD treatment (Fig. S3 B) at concentrations that effectively down-regulated HIF-1α protein levels (Fig. S3 A). To confirm the redistribution of HIF-1α mRNA into cytoplasmic P-bodies after MTD treatment in living cells, we transfected HeLa–GFP-Ago2 cells with HIF-1α or GAPDH-specific MBs and performed confocal microscopy. MTD treatment induced significant colocalization of HIF-1α MBs with Ago2-containing P-bodies, as we observed an ∼20% overlap of HIF-1α mRNA and Ago2 after 1 h or overnight treatment with 2ME2 or TX compared with 1% colocalization in untreated cells (P < 0.001; Fig. 5 D). In contrast, no overlap was observed between Ago2 and GAPDH mRNA in either control or MTD-treated cells (unpublished data). 3D rendering of HIF-1α mRNA and Ago2 further revealed their overlap (Fig. 5 D and Videos 6 and 7). Together, these results demonstrate that disruption of microtubule dynamics triggers the accumulation of HIF-1α mRNA into cytoplasmic P-bodies for translational repression, suggesting that microtubule integrity is required for active translation of HIF-1α.
Figure 5.
HIF-1α mRNA is sequestered to P-bodies upon microtubule disruption. (A) HeLa–GFP-Ago2 cells treated with 25 µM 2ME2 or 25 nM TX overnight, lysed, and immunoprecipitated (IP) using an anti-GFP antibody. Ago2 protein was detected by immunoblotting. Ago2-bound RNA was extracted, and HIF-1α, p53, and GAPDH expression is displayed as mean fold change ± SEM (n = 3; *, P < 0.05). (B) RNA from Ago2 unbound lysates was processed as in A (n = 3; *, P < 0.05). (C) Bar graph showing the percentage of HIF-1α mRNA localized to P-bodies after the indicated drug treatments. (D) HeLa–GFP-Ago2 cells transfected with 100 nM HIF-1α MB (red) and treated with 100 nM TX or 100 µM 2ME2 for 1 h. Bar, 10 µm. Right column shows the percentage of overlap (mean ± SEM; n = 3) between Ago2 foci and HIF-1α MB (***, P < 0.001; Videos 6 and 7).
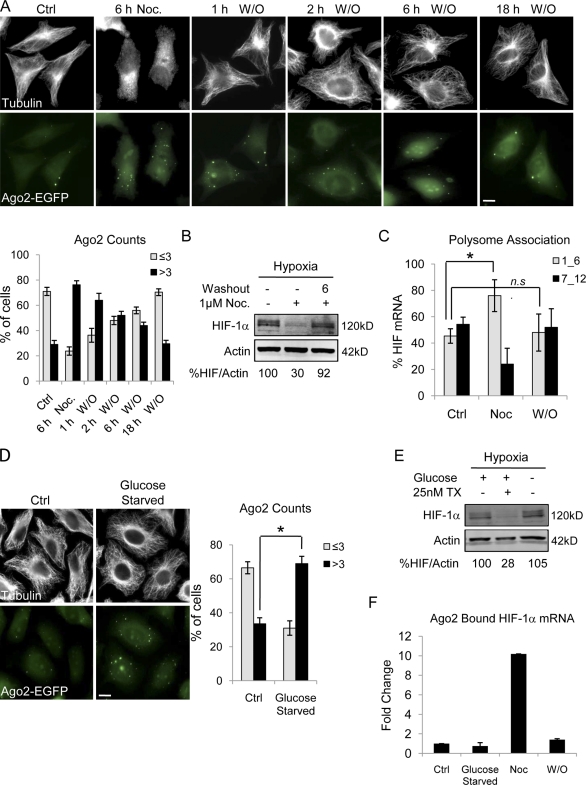
We next investigated whether restoration of microtubules can relieve the translational repression of HIF-1α. To do so, we used the reversible microtubule depolymerizer Noc and monitored P-body number after drug treatment and subsequent washout. Noc treatment resulted in near-complete depolymerization accompanied by a profound increase in P-body number (Fig. 6 A). Noc was then removed, and cells were incubated in drug-free media for the indicated times. After drug washout, microtubule polymers were quickly reformed (within 1 h), and P-body numbers gradually decreased to the level observed in untreated cells (Fig. 6 A). Similar results were obtained in a parallel experiment in which single HeLa–GFP-Ago2 cells were followed over time though live cell imaging (Fig. S4 A). Again, a decrease in P-body number was evident after drug removal, suggesting that P-bodies disassemble and release repressed mRNAs once microtubules are reformed. To test this hypothesis, we monitored HIF-1α protein expression after Noc washout. As expected, Noc treatment inhibited HIF-1α protein, whereas drug removal restored HIF-1α protein (Fig. 6 B). To further confirm that HIF-1α mRNA returns to active translation after microtubule repolymerization, we assessed HIF-1α’s polysome association profile. We observed that HIF-1α translation was significantly inhibited in Noc-treated cells and that this effect was reversed after drug washout and microtubule repolymerization (Fig. 6 C). These results are further corroborated by the accumulation of HIF-1α mRNA into P-bodies after Noc treatment and its subsequent release from Ago2 after drug washout (Fig. 6 F). Together, these results indicate that, in concert with the microtubule cytoskeleton, P-bodies regulate HIF-1α translation and that HIF-1α mRNA can return to a translationally active state once the cellular stress of microtubule disruption is relieved.
Figure 6.
HIF-1α translation inhibition is reversed after microtubule repolymerization. (A) HeLa–GFP-Ago2 (green) cells treated with 10 µM Noc for 6 h followed by drug washout (W/O). Cells were fixed at the indicated times and immunostained for α-tubulin (white). The percentage of cells with either three or less or more than three P-bodies is plotted (mean ± SEM; n = 10 fields of view/condition). (B) HIF-1α and actin protein levels in HeLa–GFP-Ago2 cells treated with 1 µM Noc overnight and lysed after 6-h washout (6) or continued drug treatment (−) with 4-h hypoxia. (C) The percentage of HIF-1α mRNA associated with untranslating fractions (1_6) or actively translating polysomes (7_12) after 1 µM Noc treatment overnight and 6 h drug washout is plotted (mean ± SEM; n = 3; *, P < 0.05). (D) HeLa–GFP-Ago2 cells were incubated in glucose-free media for 1 h and stained for α-tubulin (white). The percentage of cells with three or less or more than three P-bodies (mean ± SEM; n = 3; *, P < 0.05) is plotted. (E) Immunoblot of HIF-1α and actin after 1-h glucose starvation and 4-h hypoxia. (F) Ago2-bound RNA extracted from control (Ctrl), glucose-starved (1 h) cells, or cells treated overnight with 1 µM Noc (Noc) and followed by 6-h washout. HIF-1α expression was quantified by qRT-PCR (mean ± SEM; n = 2). Bars, 10 µm.
To investigate the contribution of the P-body number increase to HIF-1α translation repression, we used glucose starvation to increase P-body number independently of disruption of microtubule dynamics (Teixeira et al., 2005). As expected, glucose starvation (1 h or overnight) induced a rapid increase in P-body number without affecting microtubules (Fig. 6 D). However, there was no inhibition of HIF-1α protein (Fig. 6 E and Fig. S4 B) nor any change in the amount of HIF-1α mRNA bound to Ago2 after glucose starvation (Fig. 6 F). Together, these results demonstrate that P-body formation alone is not sufficient to inhibit HIF-1α translation but, rather, that perturbation of the microtubule cytoskeleton is required to trigger the accumulation of HIF-1α mRNA into these cytoplasmic sites of translational repression. Importantly, we show that microtubules exert a tight, reversible control over these processes.
MTD treatment enhances the binding of HIF-1α–targeting miRNAs to Ago2
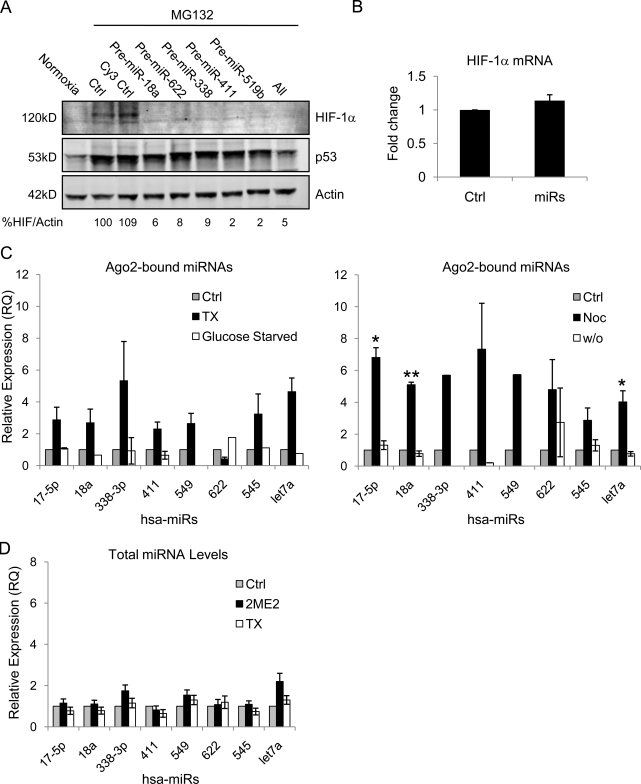
P-bodies are known hubs for miRNAs, which are short, noncoding regulatory RNAs that repress translation upon binding to their target mRNA sequence (Pillai, 2005; Tomari and Zamore, 2005). Though the exact mechanism of miRNA-mediated translational repression is not well elucidated, several studies have shown that Argonaute proteins, miRNAs, and their target mRNAs all accumulate in P-bodies (Liu et al., 2005b; Pillai et al., 2005; Sen and Blau, 2005). Thus, we hypothesized that sequestration of HIF-1α mRNA to Ago2 after stabilization or depolymerization of microtubules facilitates its interaction with specific miRNAs that, in turn, inhibit its translation. Using the miRBase Targets database (Sanger Institute), we identified nine miRNAs predicted to target the 3′ UTR of human HIF-1α. Transfection of specific premiRs (miRNA mimics) into HeLa or MCF7 cells (not depicted) resulted in down-regulation of HIF-1α protein (Fig. 7 A) without affecting HIF-1α transcription (Fig. 7 B). This result is consistent with the predicted role of miRNAs in the repression of HIF-1α translation.
Figure 7.
Role of HIF-1α–targeting miRNAs in the microtubule-dependent translation of HIF-1α. (A) HIF-1α immunoblot of MCF7 cells transfected with each pre-miR, a combination of all four, or a Cy3-labeled negative control (Cy3 Ctrl) and treated with 10 µM proteasome inhibitor MG-132 for 4 h. (B) HIF-1α mRNA expression quantified by qRT-PCR after transfection of all HIF-targeting miRNAs (miRs) in combination (mean ± SEM; n = 2). (C) Relative expression of Ago2-bound miRNAs quantified by qRT-PCR from HeLa–GFP-Ago2 cells treated overnight with 25 nM TX, glucose starvation (left; mean ± SEM; n = 3), or 1 µM Noc followed by a 6-h drug washout (right; mean ± SEM; n = 2, *, P < 0.05; **, P < 0.01). (D) Endogenous miRNA expression was assessed by qRT-PCR in HeLa cells after overnight treatment with 25 nM TX or 25 µM 2ME2.
To examine whether MTD treatment affects the P-body localization of HIF-targeting miRNAs, we immunoprecipitated Ago2 and quantified the relative abundance of associated miRNAs. This analysis revealed an increase in the amount of Ago2-bound HIF-targeting miRNAs after either TX or Noc treatment (Fig. 7 C). Repolymerization of microtubules after Noc washout resulted in dissociation of HIF-targeting miRNAs from Ago2, further implicating these miRNAs in the regulation of HIF-1α translation (Fig. 7 C). In addition, glucose starvation did not induce miRNA association with Ago2, suggesting that the mere increase in P-body number is not sufficient to recruit HIF-targeting miRNAs to Ago2 (Fig. 7 C). Finally, there was no significant increase in the level of total miRNAs observed after drug-induced microtubule stabilization or depolymerization (Fig. 7 D).
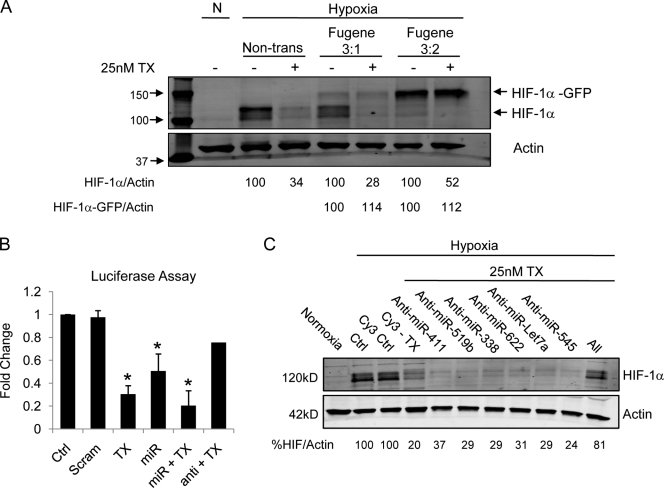
Our data showing miRNA enrichment to P-bodies suggested that the 3′ UTR of HIF-1α plays an important role in its translational regulation. To test this hypothesis, we transfected HeLa cells with a construct encoding GFP-tagged HIF-1α cDNA lacking its UTRs. TX treatment had no effect on the exogenous HIF-1α protein, whereas it efficiently down-regulated endogenous HIF-1α protein (Fig. 8 A). This result indicated that the TX-induced suppression of HIF-1α requires the presence of HIF’s UTRs. Next, we transfected cells with a HIF-1α 3′ UTR luciferase reporter and monitored TX’s effect on its activity. TX treatment inhibited HIF 3′ UTR luciferase activity, as did cotransfection of the HIF-targeting miRs both alone and together with TX (Fig. 8 B). Importantly, inhibition of the endogenous HIF-targeting miRNAs after transfection with HIF-specific anti-miRs counteracted TX’s effect and restored luciferase activity (Fig. 8 B). To examine the effect of HIF-specific anti-miRs on HIF-1α protein expression, we transfected cells with a combination of anti-miRs and assessed HIF-1α by immunoblotting. Although transfection of each individual anti-miR did not significantly affect TX’s activity, transfection of all four HIF-specific anti-miRs together almost completely abrogated TX’s ability to inhibit HIF-1α protein (Fig. 8 C). Interestingly, inhibition of two miRNAs not predicted to target HIF-1α, namely hsa-miR-Let7a and hsa-miR-545, had no effect on the TX-induced translational inhibition of HIF-1α (Fig. 8 C), although MTD treatment enhanced their binding to Ago2 (Fig. 7 C). Together, these results demonstrate that miRNA targeting of HIF-1α’s 3′ UTR plays a critical role in the microtubule-dependent regulation of HIF-1α.
Figure 8.
TX requires the 3′ UTR of HIF-1α to regulate its translation. (A) HeLa–GFP-Ago2 cells transfected (3:1 or 3:2 Fugene/DNA ratio) with a GFP-tagged HIF-1α construct lacking both the 5′ and 3′ UTR (HIF-1α–GFP) and treated with 25 nM TX overnight. (B) HeLa–GFP-Ago2 cells transfected with a HIF-1α 3′ UTR luciferase expression vector either alone or cotransfected with four HIF-targeting miRNAs (miR), four HIF-specific anti-miRs (anti), or a scrambled miRNA-like sequence (Scram). Luciferase activity was measured in untreated or TX-treated cells as indicated. *, P < 0.05. Error bars indicate mean ± SEM. (C) HIF-1α immunoblot of HeLa–GFP-Ago2 cells transfected with HIF-specific anti-miRs (622, 338, 411, and 519b), a combination of all four, or Cy3 control for 24 h followed by overnight 25 nM TX treatment. Transfection of two non-HIF–related anti-miRs (let7a and 545) was used as a control.
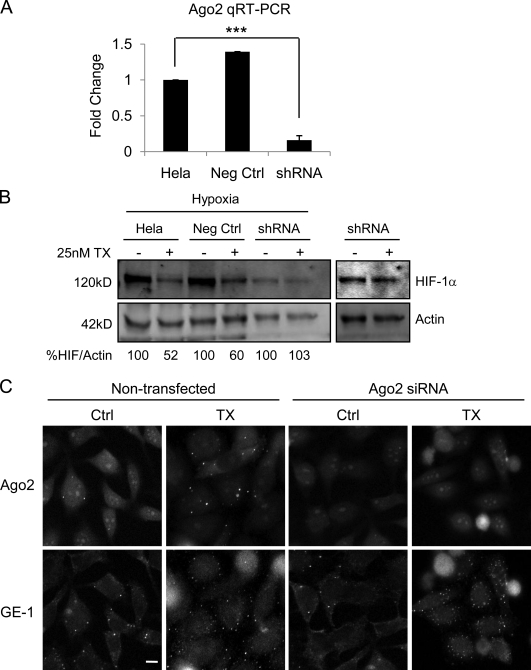
We next sought to elucidate the role of Ago2 in the regulation of HIF-1α translation. To do so, we generated HeLa cells stably expressing Ago2 shRNA and confirmed KD by qRT-PCR (Fig. 9 A) and immunofluorescence staining (Fig. 9 C, top). Interestingly, TX treatment was unable to inhibit HIF-1α protein in Ago2 KD cells in contrast to the effective inhibition observed in control or mock-transfected cells (Fig. 9 B). This result was confirmed by transient KD of Ago2 using Ago2-specific siRNAs (unpublished data). Notably, although HIF-1α was not affected by TX treatment in Ago2 KD cells, P-body number (detected by GE-1 immunostaining) was still increased (Fig. 9 C).
Figure 9.
Ago2 is necessary for MTD-induced inhibition of HIF-1α translation. (A) Ago2 mRNA expression by qRT-PCR after stable expression of Ago2 (shRNA; mean ± SEM; n = 2; ***, P < 0.001) or control shRNA (Neg Ctrl). (B) Immunoblot of HeLa cells transfected as in A and treated with 25 nM TX overnight. (right) A more intense scan of lanes 5 and 6 is depicted as a result of sample underloading. (C) Ago2-GFP is visualized in fixed untreated or TX-treated HeLa–GFP-Ago2 cells transfected with Ago2 siRNA (top) and stained for GE-1 (bottom). Bar, 10 µm.
Discussion
The importance of HIF-1α in the pathophysiology of cancer, cardiovascular disease, and other ischemic conditions has placed considerable interest in understanding HIF-1α regulation. Although most studies address the pathways involved in HIF-1α protein degradation, the mechanisms regulating HIF-1α translation remain unclear. In addition, current knowledge of HIF-1α translation stems from studies focusing on the mechanisms that enable continued translation of HIF-1α under hypoxia (Lang et al., 2002; Schepens et al., 2005); however, mechanisms involved in the repression of HIF-1α translation have not been identified.
In this study, we report a new mechanism of cytoskeletal-mediated repression of HIF-1α translation. Our studies demonstrate that microtubule integrity is critical for efficient translation of HIF-1α, as HIF mRNA binds to and traffics on dynamic microtubules, presumably to sites of active translation (Fig. 3 and Videos 1–4). Our results are the first to show that the fate of an mRNA encoding a transcription factor is tightly regulated by the microtubule cytoskeleton. We show that collapse of the microtubule network by MTDs is detrimental for HIF-1α translation, as it triggers the release of HIF-1α mRNA from polysomes and recruitment to P-bodies (Figs. 1 and 5) where translationally silenced mRNA is stored and/or degraded (Anderson and Kedersha, 2002; Kedersha and Anderson, 2002; Cougot et al., 2004; Andrei et al., 2005; Brengues et al., 2005; Kedersha et al., 2005; Teixeira et al., 2005). However, the signals that lead to recruitment of specific messages to P-bodies are not well understood. Our data reveal dynamic shuttling of HIF-1α mRNA from polysomes to P-bodies in a microtubule-dependent manner (Figs. 5 and 6). A similar dynamic exchange between P-bodies and polysomes has been reported for CAT-1 mRNA where glucose starvation released CAT-1 mRNA from P-bodies and enabled its recruitment to polysomes (Bhattacharyya et al., 2006). Although the role of the microtubule network was not addressed in that study, it would be interesting to investigate its involvement in the exchange of mRNAs between P-bodies and polysomes. It is currently unknown how different types of cellular stress regulate translation by recruitment or release of specific messages to and from P-bodies and which additional mRNAs are susceptible to microtubule control.
We also showed that microtubule disruption led to an increase in the number of P-bodies, in agreement with previous studies in yeast and mammalian cells (Sweet et al., 2007; Aizer et al., 2008). Other cellular stressors such as nutrient deprivation and glucose starvation have been associated with an increase in P-body number. Although this increase could be interpreted as a cell’s defense mechanism to cope with environmental stress by shutting down translation, we demonstrate that the mere increase in P-body number had no effect on HIF-1α translation, suggesting that disruption of microtubule dynamics is the critical initiating step in the repression of HIF translation. In spite of these data, what remains puzzling is the exact signaling cascade that links microtubule stabilization or destabilization with recruitment of HIF-1α mRNA and HIF-targeting miRNAs to P-bodies. In this study, we show that one important component of this cascade is Ago2, as its depletion prevented TX’s effect on HIF translation (Fig. 9), although microtubules were efficiently stabilized (not depicted). TX also led to an increase in P-body number in Ago2 KD cells as a result of microtubule hyperstabilization.
P-bodies are known hubs for miRNAs, which function in association with Argonaute family proteins and other components of RNA-induced silencing complexes (Liu et al., 2005a; Meister et al., 2005; Sen and Blau, 2005) to suppress translation. However, the contribution of different P-body components in this process is not well understood. Moreover, it remains unclear whether HIF-1α mRNA interacts with miRNAs after its entry into P-bodies or is first targeted by miRNAs in the cytoplasm. Previous studies have proposed two general models of P-body–mediated translational repression. The first proposes that formation of the mRNA–miRNA complex occurs before P-body localization (Liu et al., 2005b), whereas in the second model, mRNAs accumulate in P-bodies regardless of prior miRNA coupling, and translation is suppressed as the result of ribosome exclusion (Andrei et al., 2005; Teixeira et al., 2005). Our Ago2 KD results support the first model whereby Ago2 mediates the association between HIF-1α mRNA and HIF-targeting miRNAs in the cytoplasm after microtubule disruption and subsequently targets these complexes into P-bodies for translational repression.
The findings presented in this study, together with recent data from our laboratory showing that HIF-1α protein also binds microtubules (unpublished data), raise the possibility that HIF-1α translation may physically occur on microtubules. In support of this hypothesis, many proteins that regulate translation, including eukaryotic initiation and elongation factors (eEF-1α, eEF-1-2, eIF-2, eIF-3, eIF-4a, eIF-4E, and eIF-5), have been shown to associate with microtubules (Jansen, 2001; López de Heredia and Jansen, 2004). Although the role of microtubules in mRNA transport and/or anchoring is well established, especially in neurons (Palacios and St Johnston, 2001; López de Heredia and Jansen, 2004), their involvement in protein translation remains unclear. Interestingly, we showed that two non-HIF–related miRNAs were also recruited to Ago2 after disruption of microtubule dynamics (Fig. 7 C), suggesting that additional messages may be susceptible to MTD treatment. Identification of proteins that mediate the association of HIF-1α mRNA with microtubules can provide new therapeutic opportunities to specifically target HIF translation for cancer therapy.
In summary, our work identifies an important, as of yet unrecognized, role of the microtubule cytoskeleton in repressing HIF-1α translation and provides compelling evidence that microtubules are actively involved in regulating protein translation. Understanding the role of microtubules in both the facilitation and repression of HIF-1α translation will have important implications for both cell biology and clinical oncology given the importance of the microtubule cytoskeleton and HIF-1α in tumor biology.
Materials and methods
Chemicals and antibodies
Vinblastine and MG-132 were obtained from EMD, and paclitaxel was obtained from Sigma-Aldrich. 2ME2 was provided by EntreMed Inc. Primary antibodies used were rat anti–α-tubulin (YL1/2; EMD), mouse anti–HIF-1α (BD), rabbit anti-actin (Sigma-Aldrich), mouse anti–GE-1 (sc-8418; Santa Cruz Biotechnology, Inc.), mouse anti-Ago2 (Abcam), sheep anti-p53 (Ab7; Oncogene Science), and mouse anti-GAPDH (6C5; Santa Cruz Biotechnology, Inc.). The following secondary antibodies were used for immunofluorescence: Alexa Fluor 488, 568, and 647, and rhodamine phalloidin (Invitrogen). The following secondary antibodies were used for immunoblotting: Alexa Fluor 680 (Invitrogen) and IRDye 800CW–conjugated antibodies (Rockland Immunochemicals, Inc.).
Cell lines
The cell lines used in this study were MCF7 human breast cancer cells stably expressing EGFP-tubulin (provided by M. Jordan, University of California, Santa Barbara, Santa Barbara, CA), MCF7 cells stably expressing GFP-nucleolin (provided by M. Kastan, St. Jude Children’s Research Hospital, Memphis, TN), and HeLa human cervical carcinoma cells stably expressing EGFP-Ago2 (provided by P. Sharp, Massachusetts Institute of Technology, Cambridge, MA), all maintained in DME (Cellgro; Mediatech, Inc.). 1A9 human ovarian carcinoma, 1A9/PTX10 (Giannakakou et al., 1997), and 1A9/2MRC (Escuin et al., 2009) cells were cultured in RPMI1640 (Cellgro). For hypoxic exposure, cells were sealed in a modular incubator chamber (Billups-Rothenberg, Inc.) and flushed with 1% O2, 5% CO2, and 94% N2 for 4 h.
Western blotting
Proteins (70 µg/lane) were resolved by SDS-PAGE, electrotransferred to PVDF transfer membranes (Immobilon; Millipore), and incubated with the indicated primary antibodies followed by fluorescence-conjugated secondary antibodies. Immunoreactivity was visualized using an infrared imaging system (Odyssey; LI-COR Biosciences). Protein quantification was performed using Odyssey (version 2.1) and ImageJ (National Institutes of Health) software.
Immunofluorescence and confocal microscopy
Exponentially growing cells were plated on 12-mm glass coverslips (Thermo Fisher Scientific) into 24-well plates and treated with the indicated drugs. Cells were fixed with PHEMO buffer (0.068 M Pipes, 0.025 M Hepes, 0.015 M EGTA, 0.003 M MgCl2, and 10% DMSO, pH 6.8) containing 3.7% formaldehyde, 0.05% glutaraldehyde, and 0.5% Triton X-100 for 10 min at room temperature. Coverslips were blocked in 10% goat serum in PBS for 10 min and processed for double-labeling immunofluorescence with rat anti–α-tubulin, mouse anti–HIF-1α, and mouse anti–GE-1 primary antibodies followed by Alexa Fluor 488, 586, and 647 secondary antibodies. Coverslips were then mounted onto glass slides and imaged with a confocal microscope (5 LIVE; Carl Zeiss, Inc.) using AIM software (version 4.2; Carl Zeiss, Inc.) and a 100× 1.4 NA Plan Apochromat objective.
MB design and live cell imaging
HIF-1α MB, 5′-Texas red–TCACCAGCATCCAGAAGTGGTGA–Blackhole quencher 2 (BHQ2)-3′; scrambled MB, 5′-Texas red–TCACCACTTCTGGATGCTGGTGA-BHQ2-3′; and GAPDH MB, 5′-Texas Red–CGACGGAGTCCTTCCACGATACCACGTCG-BHQ2-3′ (Eurofins MWG Operon). Fluorescence intensity was read with a microplate spectrofluorometer (Gemini EM; MDS Analytical Technologies). For live cell imaging, MBs were transfected (Oligofectamine; Invitrogen) for 24 h. Images were acquired by continuous scanning with a confocal (5 LIVE; AIM 4.2 software) or spinning-disk confocal microscope (Ultraview ERS software; PerkinElmer) using a 100× 1.4 NA Plan Apochromat objective and a camera (Orca-ER; Hamamatsu Photonics). A Tokai Hit heating stage (Shizuoka-ken; 418–0074 Japan) was used to incubate cells at 37°C with 5% CO2 in DME without l-glutamine and phenol red (Cellgro). Stack arithmetic and 3D reconstructions were performed using MetaMorph software (MDS Analytical Technologies).
Linear sucrose gradient fractionation
The polysome profile analyses were performed after linear sucrose gradient (LSG) fractionation as previously described (Feng et al., 1997). In brief, cells were washed with PBS containing 100 µg/ml cycloheximide and lysed with 500 µl LSG lysis buffer (100 mM KCl, 20 mM Tris, pH 7.5, 5 mM MgCl2, 0.4% NP-40, 100 µg/ml cycloheximide, 0.1 U RNasin, and complete mini-EDTA–free protease inhibitors [Roche]). Cell lysates were precleared by centrifugation at 14,000 rpm for 15 min at 4°C. The supernatant was loaded onto 15–45% sucrose gradients (in a buffer containing 5 mM MgCl2, 80 mM NaCl, and 20 mM Tris-HCl, pH 7.5). Ultracentrifugation (36,000 rpm for 1.5 h at 4°C) was performed in an SW41 rotor. 12 equal-volume (1 ml) fractions were collected with continuous OD254 measurements (Vis Detector UA-6; Isco UV). Total RNA was isolated from each fraction using RNeasy kit (QIAGEN). cDNA was synthesized with the ProtoScript First Strand cDNA Synthesis kit (New England BioLabs, Inc.). qRT-PCR was performed using iQ SYBR green supermix (Bio-Rad Laboratories) and the following specific primers: HIF-1α (forward, 5′-TGGTGACATGATTTACATTTCTGA-3′; reverse, 5′-AAGGCCATTTCTGTGTGTAAGC-3′), GAPDH (forward, 5′-GGAGTCAACGGATTTGGTCG-3′; reverse, 5′-CTTGATTTTGGAGGGATCTCG-3′), or p53 (forward, 5′-TCACAGCACATGACGGAGGTT-3′; reverse, 5′-TCGGATAAGATGCTGAGGAGG-3′).
shNA and siRNA transfection
SureSilencing shNA plasmids for human EIF2C2 (Ago2) used for Ago2 KD were obtained from SABiosciences. Cells were transfected with 0.40 µg shNA using FuGENE 6 transfection reagent (Roche) for 48 h followed by puromycin selection to generate stable transfections. Silencer select predesigned siRNAs were obtained from Applied Biosystems (siRNA ID nos. 25935, s25930, and s25931). Cells were transfected with 10 nM of each siRNA for 48 h using the AMAXA nucleofector system (Lonza).
Immunoprecipitation of mRNPs
RNA isolation after GFP immunoprecipitation was performed as described previously (Tenenbaum et al., 2002). In brief, cells were lysed in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, 0.5% Nonidet P-40, 1 mM DTT, 100 U/ml RNase OUT, 0.2 mM PMSF, and complete mini-EDTA–free protease inhibitors) and subjected to immunoprecipitation using rabbit anti-GFP antibody (Novus Biologicals), or ChomPure rabbit IgG control (Jackson ImmunoResearch Laboratories, Inc.) bound to protein A agarose beads (EMD) in NT2 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40) for 2 h. Beads were washed five times in NT2 buffer, resuspended in 100 µl NT2 buffer supplemented with 0.1% SDS and 30 µg proteinase K, and incubated at 55°C for 30 min. Bound and unbound RNA was extracted, converted to cDNA, and subjected to qRT-PCR as described above. The percentage of HIF-1α mRNA in P-bodies was calculated as follows: percentage of Ago2-bound HIF-1α mRNA = (Ago2-bound mRNA/[bound + unbound mRNA]) × 100.
GFP-bound miRNA isolation and miRNA validation
miRNAs were isolated from total cell lysates or Ago2-bound fractions using the mirVana miRNA Isolation kit (Ambion). cDNA was synthesized using the MicroRNA Reverse Transcription kit (TaqMan), and qRT-PCR was performed using miRNA (TaqMan). Primers specific for each miRNA were obtained from Applied Biosystems. All values were normalized to expression of the small nucleolar RNA U19. Pre-miR miRNA precursor molecules and anti-miR miRNA inhibitors were obtained from Applied Biosystems and transfected into cells at 10 nmol/liter using FuGENE 6 (Roche) for 48 h.
Luciferase assay with HIF-1α 3′ UTR
A pHIF1α-luc-3-UTR construct (provided by G. Semenza, Johns Hopkins University, Baltimore, MD) was cotransfected into cells with the pre-miR miRNA precursors or anti-miRs for 24 h. The ratio of firefly to renilla luciferase was monitored after each condition using the Dual-Luciferase Reporter Assay System (Promega) and a Sirius Tube Luminometer (Berthold Detection Systems).
Statistical analyses
The p-values for altered translation status (Fig. 1 B) and increased P-body numbers (Fig. 4 C) were calculated using one-way analysis of variance followed by multiple comparisons with Bonferroni adjustments. P-values for Ago2-enriched transcripts (Fig. 5, A and B) were obtained using the Wilcoxon sum-rank test, and the Kruskal-Wallis test was used to calculate the p-value for HIF-MB/Ago2 colocalization. A t test was used to determine the p-value for increased P-body number after glucose starvation (Fig. 6 C) and for Ago2-enriched miRNAs (Fig. 7 C). A t test was also used to determine p-values for fold change in luciferase activity (Fig. 8 B) and Ago2 expression (Fig. 9 A). All statistical analyses were performed using Stata for Windows (release 10).
Online supplemental material
Fig. S1 shows the state of microtubules in the 2ME2- and TX-resistant cell lines after each drug treatment. Fig. S2 shows MB design and controls confirming that GAPDH or antisense MBs do not associate with microtubules. Fig. S3 shows a parallel Western blot and qRT-PCR (Fig. 5), confirming that 2ME2 and TX treatment inhibit HIF-1α protein without affecting total RNA levels. Fig. S4 shows time-lapse confocal microscopy of a single cell after Noc treatment and washout. Videos 1–4 show microtubule-dependent trafficking of HIF-1α mRNA in untreated (Videos 1 and 2), 2ME2 (Video 3)-, or TX (Video 4)-treated cells. Video 5 shows lack of microtubule-dependent trafficking of GAPDH mRNA. Videos 6 and 7 show 3D models of HIF-1α accumulation into Ago2 foci after 2ME2 and TX treatment, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004145/DC1.
Acknowledgments
We thank Drs. Yue Feng (Emory University, Atlanta, GA) and Scott Blanchard (Weill Cornell Medical College [WCMC], New York, NY) for help with sucrose gradient fractionation and Drs. Lilly Yang and Erik Kline (Emory University, Atlanta, GA) for help with MB design. We also thank Drs. Geri Kreitzer, Tim McGraw, and Samie Jaffrey (WCMC) for critical review of our manuscript.
This work was supported by the National Institutes of Health (grants CA114335 and CA100202), Entremed, Inc. (grant 659342 to P. Giannakakou), and the WCMC Clinical and Translational Science Center (grant TL1RR024998 to M. Carbonaro).
Footnotes
Abbreviations used in this paper:
- 2ME2
- 2-methoxyestradiol
- Ago2
- Argonaute 2
- HIF
- hypoxia inducible factor
- KD
- knockdown
- MB
- molecular beacon
- miRNA
- microRNA
- MTD
- microtubule-targeting drug
- Noc
- nocodazole
- ODN
- oligonucleotide
- qRT-PCR
- quantitative RT-PCR
- TX
- taxol
References
- Aizer A., Brody Y., Ler L.W., Sonenberg N., Singer R.H., Shav-Tal Y. 2008. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell. 19:4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. 2002. Stressful initiations. J. Cell Sci. 115:3227–3234 [DOI] [PubMed] [Google Scholar]
- Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Lührmann R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 11:717–727 10.1261/rna.2340405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 11:833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 71:513–521 10.1101/sqb.2006.71.038 [DOI] [PubMed] [Google Scholar]
- Bratu D.P., Cha B.J., Mhlanga M.M., Kramer F.R., Tyagi S. 2003. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA. 100:13308–13313 10.1073/pnas.2233244100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310:486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Séraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escuin D., Kline E.R., Giannakakou P. 2005. Both microtubule-stabilizing and microtubule-destabilizing drugs inhibit hypoxia-inducible factor-1alpha accumulation and activity by disrupting microtubule function. Cancer Res. 65:9021–9028 10.1158/0008-5472.CAN-04-4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escuin D., Burke P.A., McMahon-Tobin G., Hembrough T., Wang Y., Alcaraz A.A., Leandro-García L.J., Rodríguez-Antona C., Snyder J.P., Lavallee T.M., Giannakakou P. 2009. The hematopoietic-specific beta1-tubulin is naturally resistant to 2-methoxyestradiol and protects patients from drug-induced myelosuppression. Cell Cycle. 8:3914–3924 [DOI] [PubMed] [Google Scholar]
- Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1:109–118 10.1016/S1097-2765(00)80012-X [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Taylor S.S. 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 14:111–122 10.1016/j.ccr.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Giannakakou P., Sackett D.L., Kang Y.K., Zhan Z., Buters J.T., Fojo T., Poruchynsky M.S. 1997. Paclitaxel-resistant human ovarian cancer cells have mutant beta-tubulins that exhibit impaired paclitaxel-driven polymerization. J. Biol. Chem. 272:17118–17125 10.1074/jbc.272.27.17118 [DOI] [PubMed] [Google Scholar]
- Giannakakou P., Sackett D.L., Ward Y., Webster K.R., Blagosklonny M.V., Fojo T. 2000. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2:709–717 10.1038/35036335 [DOI] [PubMed] [Google Scholar]
- Giannakakou P., Nakano M., Nicolaou K.C., O’Brate A., Yu J., Blagosklonny M.V., Greber U.F., Fojo T. 2002. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. USA. 99:10855–10860 10.1073/pnas.132275599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guppy M., Brunner S., Buchanan M. 2005. Metabolic depression: a response of cancer cells to hypoxia? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 140:233–239 10.1016/j.cbpc.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., Kriegsheim Av H.F., Hebestreit H.F., Mukherji M., Schofield C.J., et al. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 292:468–472 10.1126/science.1059796 [DOI] [PubMed] [Google Scholar]
- Jansen R.P. 2001. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2:247–256 10.1038/35067016 [DOI] [PubMed] [Google Scholar]
- Jordan M.A., Wilson L. 2004. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 4:253–265 10.1038/nrc1317 [DOI] [PubMed] [Google Scholar]
- Kavallaris M. 2010. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer. 10:194–204 10.1038/nrc2803 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 10.1042/BST0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K.J., Kappel A., Goodall G.J. 2002. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 13:1792–1801 10.1091/mbc.02-02-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 103:18125–18130 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J. 2005a. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. 2005b. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723 10.1038/ncb1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Heredia M., Jansen R.P. 2004. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 16:80–85 10.1016/j.ceb.2003.11.002 [DOI] [PubMed] [Google Scholar]
- Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O. 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 20:709–722 10.1016/j.molcel.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Luke B., Azzalin C.M., Hug N., Deplazes A., Peter M., Lingner J. 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 35:7688–7697 10.1093/nar/gkm912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabjeesh N.J., Escuin D., LaVallee T.M., Pribluda V.S., Swartz G.M., Johnson M.S., Willard M.T., Zhong H., Simons J.W., Giannakakou P. 2003. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 3:363–375 10.1016/S1535-6108(03)00077-1 [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Lührmann R., Tuschl T. 2005. Identification of novel argonaute-associated proteins. Curr. Biol. 15:2149–2155 10.1016/j.cub.2005.10.048 [DOI] [PubMed] [Google Scholar]
- Palacios I.M., St Johnston D. 2001. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17:569–614 10.1146/annurev.cellbio.17.1.569 [DOI] [PubMed] [Google Scholar]
- Pillai R.S. 2005. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 11:1753–1761 10.1261/rna.2248605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309:1573–1576 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Pouysségur J., Dayan F., Mazure N.M. 2006. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 441:437–443 10.1038/nature04871 [DOI] [PubMed] [Google Scholar]
- Schepens B., Tinton S.A., Bruynooghe Y., Beyaert R., Cornelis S. 2005. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 33:6884–6894 10.1093/nar/gki1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 3:721–732 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- Semenza G.L., Wang G.L. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633–636 10.1038/ncb1265 [DOI] [PubMed] [Google Scholar]
- Sivan G., Kedersha N., Elroy-Stein O. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27:6639–6646 10.1128/MCB.00798-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper H.E. 1971. Kinetics of mammary tumor cell growth and implications for therapy. Cancer. 28:1479–1499 [DOI] [PubMed] [Google Scholar]
- Sweet T.J., Boyer B., Hu W., Baker K.E., Coller J. 2007. Microtubule disruption stimulates P-body formation. RNA. 13:493–502 10.1261/rna.355807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum S.A., Lager P.J., Carson C.C., Keene J.D. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 26:191–198 10.1016/S1046-2023(02)00022-1 [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D. 2005. Perspective: machines for RNAi. Genes Dev. 19:517–529 10.1101/gad.1284105 [DOI] [PubMed] [Google Scholar]
- Tyagi S., Kramer F.R. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308 10.1038/nbt0396-303 [DOI] [PubMed] [Google Scholar]
- Yu J.H., Yang W.H., Gulick T., Bloch K.D., Bloch D.B. 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 11:1795–1802 10.1261/rna.2142405 [DOI] [PMC free article] [PubMed] [Google Scholar]