p62 is recruited to the ER at an early-stage autophagosome formation independently of most Atg proteins.
Abstract
Autophagy is an intracellular degradation process by which cytoplasmic contents are degraded in the lysosome. In addition to nonselective engulfment of cytoplasmic materials, the autophagosomal membrane can selectively recognize specific proteins and organelles. It is generally believed that the major selective substrate (or cargo receptor) p62 is recruited to the autophagosomal membrane through interaction with LC3. In this study, we analyzed loading of p62 and its related protein NBR1 and found that they localize to the endoplasmic reticulum (ER)–associated autophagosome formation site independently of LC3 localization to membranes. p62 colocalizes with upstream autophagy factors such as ULK1 and VMP1 even when autophagosome formation is blocked by wortmannin or FIP200 knockout. Self-oligomerization of p62 is essential for its localization to the autophagosome formation site. These results suggest that p62 localizes to the autophagosome formation site on the ER, where autophagosomes are nucleated. This process is similar to the yeast cytoplasm to vacuole targeting pathway.
Introduction
Macroautophagy (which we will simply refer to as autophagy) is an intracellular degradation system mediated by the autophagosome. A small portion of the cytoplasm is engulfed by an isolation membrane/phagophore, which results in the formation of an autophagosome. The outer membrane of autophagosome then fuses with the lysosome membrane, and materials inside the autophagosome are degraded by the lysosomal hydrolases together with the inner autophagosome membrane. Autophagy is involved in various physiological processes such as maintenance of the amino acid pool during starvation and preimplantation embryo development, intracellular quality control, antigen presentation, and killing of intracellular microorganisms (Rubinsztein, 2006; Mizushima, 2007; Cecconi and Levine, 2008; Mizushima et al., 2008; Deretic, 2009; Virgin and Levine, 2009). To date, >30 autophagy-related (ATG) genes have been identified in yeast, many of which are conserved in higher eukaryotes (Suzuki and Ohsumi, 2007; Xie and Klionsky, 2007; Longatti and Tooze, 2009).
Although autophagy has been considered to be a nonselective, bulk process (Kominami et al., 1983; Kopitz et al., 1990), recent studies have revealed that the autophagosome membrane can selectively recognize certain specific proteins and organelles (Yu et al., 2008; Kirkin et al., 2009b; Kraft et al., 2010). The prototype for selective autophagy is the yeast cytoplasm to vacuole targeting (Cvt) pathway (Wang and Klionsky, 2003; Lynch-Day and Klionsky, 2010). Two vacuolar enzymes, aminopeptidase 1 (Ape1) and α-mannnosidase (Ams1), lack typical signal sequences and are synthesized as precursor enzymes in the cytosol. prApe1 dodecamers form an Ape1 complex, which are assembled into a Cvt complex with Ams1 and Atg19, and are then sequestered into Cvt vesicles. The Cvt vesicles are reminiscent of autophagosomes but are smaller in size. The Cvt vesicles finally fuse with the vacuole, releasing the two proenzymes into the vacuolar lumen. Mitochondria can also be selectively degraded by autophagy, and a yeast cargo receptor on the mitochondrial membrane, Atg32, was recently identified (Kanki et al., 2009; Okamoto et al., 2009).
The best-known mammalian autophagy-specific substrate is p62/SQSTM1 (Bjørkøy et al., 2005; Pankiv et al., 2007). p62 has multiple functions in bone metabolism, obesity, caspase activation, inclusion body formation, and tumorigenesis (Wooten et al., 2006; Seibenhener et al., 2007; Moscat and Diaz-Meco, 2009). Selective degradation of p62 by autophagy is physiologically important because accumulation of p62 is cytotoxic, at least in the liver (but not in the brain; Komatsu et al., 2007). It is also hypothesized that p62 functions as an adaptor or cargo receptor in degradation of ubiquitinated proteins, organelles such as peroxisomes and mitochondria (Kim et al., 2008; Kirkin et al., 2009b), and intracellular bacteria (Dupont et al., 2009; Yoshikawa et al., 2009; Zheng et al., 2009). Another structurally related protein, NBR1 (neighbor of BRCA1 gene 1), is also selectively incorporated into autophagosomes and degraded by autophagy (Kirkin et al., 2009a).
These selective substrates/adaptors such as Atg19, Atg32, p62, and NBR1 have an Atg8/LC3-interacting motif (WXXL/I) called the LC3 recognition sequence (LRS) or the LC3-interacting region (Noda et al., 2010). Atg8 and its mammalian homologue LC3 are present on both outer and inner membranes of the autophagosome. Direct binding to Atg8/LC3 allows the cargos to be selectively enclosed by autophagosomes. In yeast, Ape1, Ams1, and Atg19 are present on the preautophagosomal structure (PAS), where most Atg proteins gather and autophagosome are generated (Suzuki et al., 2001; Kim et al., 2002; Chang and Huang, 2007). Importantly, localization of the Cvt complex to the PAS requires Atg19 but does not absolutely depend on other Atg proteins including Atg8, suggesting that cargo recruitment can be achieved independently of the Atg8–cargo interaction (Cao et al., 2008). In contrast, how mammalian selective substrates or cargo receptors are loaded into autophagosomes remains to be determined. It is generally thought that the cargo proteins are recruited to the elongating isolation membrane in an LC3-dependent manner (Kirkin et al., 2009b; Lamark et al., 2009).
In this study, we examined how p62 and NBR1 are loaded onto autophagosomes and found that they are recruited to the autophagosome formation site, which may be equivalent to the yeast PAS. Localization of p62 to this site is dependent on self-oligomerization but independent of most Atg proteins, including LC3. LC3 seems to be critical for later enclosing steps. These results suggest that p62 and NBR1 are already present at autophagosome formation sites. Localization of these proteins may determine where autophagosomes are nucleated.
Results and discussion
p62 can localize to early autophagic structures in an LC3-PE–independent manner
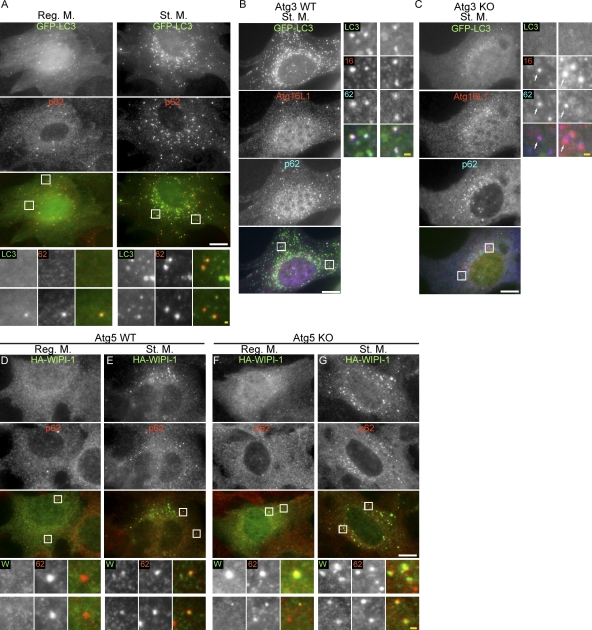
To study p62 loading into the autophagosome, we first determined the requirement of LC3 in p62 association with the autophagic membrane using mouse embryonic fibroblasts (MEFs). When wild-type cells were starved, LC3 punctate structures representing isolation membranes and autophagosomes increased in number (Fig. 1 A). p62 also formed punctate structures, even under nonstarved conditions, although the number increased after starvation (Fig. 1 A). Most of the p62 puncta that were generated under starvation conditions would represent autophagosomes because p62 is incorporated into autophagosomes (Komatsu et al., 2007). The p62 puncta colocalized well with LC3 and Atg16L1 (Fig. 1, A and B), which represented isolation membrane (Mizushima et al., 2001).
Figure 1.
Localization of p62 to the early autophagic structures is independent of LC3. (A) MEFs stably expressing GFP-LC3 were cultured in regular medium or starvation medium for 1 h. Cells were stained with anti-p62 antibodies and analyzed by immunofluorescence microscopy. (B and C) Localization of p62 to the autophagic structures is independent of LC3 lipidation. Wild-type (B) and Atg3 KO MEFs (C) stably expressing GFP-LC3 were cultured in starvation medium for 2 h. Cells were stained with anti-Atg16L1 and anti-p62 antibodies. Structures positive for Atg16L1 and p62 but not for GFP-LC3 are indicated by arrows. (D–G) Localization of p62 to the autophagic structures is independent of Atg5. Wild-type (D and E) and Atg5 KO MEFs (F and G) stably expressing HA–WIPI-1 were cultured in regular (D and F) or starvation medium (E and G) for 2 h. Cells were stained with anti-HA and anti-p62 antibodies. Signal color is indicated by color of typeface. Reg. M., regular medium; St. M., starvation medium; WT, wild type. Bars: (white) 10 µm; (yellow) 1 µm.
Localization of LC3 to the autophagosome depends on conjugation of LC3 with phosphatidylethanolamine, or LC3–PE (LC3-II) formation, which requires Atg3 as a specific E2-like conjugation enzyme. Accordingly, in Atg3 knockout (KO) cells, both LC3–PE formation and LC3 membrane association were completely inhibited (Fig. 1 C; Sou et al., 2008). In these cells, however, Atg16L1-positive puncta were generated, which would represent intermediate structures lacking LC3 (Fig. 1 C; Sou et al., 2008). Unexpectedly, most p62 puncta still colocalized with Atg16L1, even in these Atg3 KO cells. This suggests that LC3 is not essential for the p62 recruitment to the autophagic structures, which is apparently inconsistent with the general idea that p62 recruitment is LC3 dependent (Fig. 1 C). However, the colocalization rate was not as high as in wild-type cells (Fig. 1 B). Approximately 87% and 35% of the Atg16L1 puncta were positive for p62 in wild-type and Atg3 KO cells, respectively. Thus, these results suggest that, although partially important, LC3 is not essential for p62 targeting to the autophagosome intermediates.
In yeast, the hierarchical relationship among the Atg proteins has recently been determined by a systematic analysis (Suzuki et al., 2007). We performed a similar comprehensive analysis using mammalian cells including higher eukaryote-specific factors (Itakura and Mizushima, 2010). The results indicate that the complex containing ULK1 (unc-51–like kinase 1), Atg13, FIP200 (focal adhesion kinase family interacting protein of 200 kD) and Atg101, the ULK1–Atg13–FIP200–Atg101 complex, functions at the most upstream position in the process. This is followed in turn by the Atg14–Beclin 1–Vps34–p150 phosphatidylinositol 3-kinase (PI3-kinase) complex, its putative effecters WIPI-1 (WD-repeat protein interacting with phosphoinosides 1) and DFCP1 (double FYVE domain-containing protein 1), the Atg12–Atg5–Atg16L1 complex, and LC3–PE. LC3 is the most downstream factor among these mammalian Atg proteins. Given this hierarchy, we sought to determine at which step p62 is recruited to the autophagy structures.
Atg5 is covalently attached to the ubiquitin-like protein, Atg12, and further associates with Atg16L1. Atg5 is also essential for LC3–PE formation and LC3 puncta formation (Mizushima et al., 2001). We tested the requirement of Atg5 for the recruitment of p62 to the autophagic membrane. In this assay, we used an upstream factor, WIPI-1, which is present on isolation membranes in a PI3-kinase activity dependent manner (Proikas-Cezanne et al., 2007; Itakura and Mizushima, 2010). Starvation treatment induced WIPI-1 punctate structures, which colocalized with p62 (Fig. 1, D and E). This colocalization was still observed in Atg5-deficient MEFs (Fig. 1, F and G), suggesting that Atg5 is also dispensable for p62 recruitment.
p62 localizes to the ER-associated autophagosome formation site
Autophagosome formation requires PI3-kinase activity; a PI3-kinase inhibitor, wortmannin, suppresses autophagic degradation (Blommaart et al., 1997) and isolation membrane formation (Kovács et al., 2000). Although wortmannin treatment indeed suppresses puncta formation of downstream Atg proteins, such as WIPI-1, DFCP1, Atg5, Atg16L1, and LC3, it does not affect that of upstream Atg proteins such as ULK1 and Atg14 (Itakura and Mizushima, 2010). We proposed that the ULK1- and Atg14-positive structures observed in wortmannin-treated cells represent autophagosome formation sites. We therefore determined whether p62 could localize to these sites.
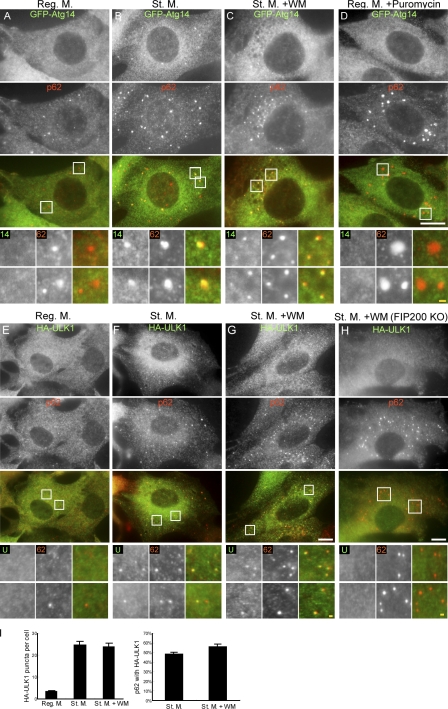
Under nutrient rich conditions, there were quite a few Atg14 puncta, and p62 dots were mostly negative for Atg14 (Fig. 2 A). After starvation treatment, Atg14 punctate structures were induced, most of which colocalized with p62 (Fig. 2 B). Notably, the wortmannin treatment, which should inhibit isolation membrane formation, did not disrupt the Atg14–p62 colocalization (Fig. 2 C). One concern was that the Atg14–p62 colocalization might simply represented cytoplasmic protein aggregates in which both Atg14 and p62 were incorporated. To address this possibility, we treated cells with puromycin, which is commonly used to increase the production of truncated and misfolded proteins, and thereby induce formation of cytoplasmic inclusions containing p62 (Szeto et al., 2006; Pankiv et al., 2007; Kirkin et al., 2009b). However, GFP-Atg14 was not incorporated into the puromycin-induced p62 aggregates (Fig. 2 D), suggesting that the Atg14–p62 colocalization is specific for autophagy induction, and does not simply represent protein aggregates.
Figure 2.
p62 localizes to the autophagosome formation site. (A–D) NIH3T3 cells stably expressing GFP-Atg14 were cultured in regular medium (A), starvation medium with (C) or without (B) 0.2 µM wortmannin for 1 h, or in regular medium containing 50 µg/ml puromycin for 2 h (D). Cells were analyzed by immunofluorescence microscopy using anti-GFP and anti-p62 antibodies. (E–H) Wild-type (E–G) and FIP200 KO MEFs (H) stably expressing HA-ULK1 were cultured in the indicated medium for 1 h. Cells were stained as described in Fig. 1. Signal color is indicated by color of typeface. (I) Quantification of HA-ULK1 puncta per cell (left) and HA-ULK1 positivity (%) of the p62 puncta (right) are shown. Data represent mean ± SEM of 30 images. Reg. M., regular medium; St. M., starvation medium; WM, wortmannin; WT, wild type. Bars: (white) 10 µm; (yellow) 1 µm.
We reported that Atg14 punctate structures are not formed in FIP200 KO cells and proposed that the ULK1-FIP200 complex is the most upstream unit among the autophagy core units (Itakura and Mizushima, 2010). We thus examined ULK1 and confirmed that the starvation-induced puncta of ULK1 colocalized with p62, even in the presence of wortmannin (Fig. 2, E–G and I). This colocalization was regulated by FIP200 because the ULK1 puncta disappeared in FIP200 KO cells, even though a large number of p62 structures were observed in these cells (Fig. 2 H). These data again argue that the ULK1–p62 colocalization does not represent nonspecific incorporation of ULK1 into p62 aggregates. The colocalization of p62 with ULK1 and Atg14 in wortmannin-treated cells suggests that p62 localizes to the autophagosome formation site independently of downstream factors.
p62 localizes to autophagy-related structures even in FIP200 KO cells
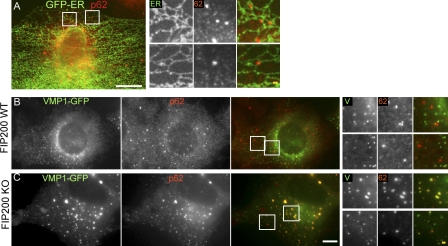
We have found that these ULK1 puncta tightly associate with the ER membrane (Itakura and Mizushima, 2010). Consistent with this observation, some of the p62 dots attached the ER networks and were moving together with the ER (Fig. 3 A and Video 1). The p62–ER association was also observed in the presence of wortmannin (Video 2). Collectively, these data suggest that p62 can accumulate at the ER-associated autophagosome formation site, where Atg proteins are recruited.
Figure 3.
p62 puncta associate with the ER independently of FIP200. (A) MEFs stably expressing GFP-ER (GFP fused to cytochrome b5 residues 95–134) were cultured in starvation medium for 1 h. Cells were fixed, permeabilized, and subjected to immunofluorescence microscopy using anti-GFP and anti-p62 antibodies. (B and C) p62 puncta colocalize with VMP1 in FIP200 KO cells. Wild-type (B) and FIP200 KO MEFs (C) stably expressing VMP1-GFP were cultured in starvation medium for 1 h. Cells were stained with anti-p62 antibodies. Signal color is indicated by color of typeface. Bars: (white) 10 µm; (yellow) 1 µm.
We further tested whether classical Atg proteins are required for generation of the ER-associated p62 punctate structures. We found in a separate study that most Atg proteins including ULK1, Atg14, Atg16L1, WIPI-1, DFCP1 and LC3 do not form puncta in FIP200 KO cells, whereas VMP1 puncta are constitutively generated in FIP200 KO cells (Itakura and Mizushima, 2010). VMP1 is an ER-associated protein required for autophagosome formation (Ropolo et al., 2007). VMP1 transiently associates with the ULK1 puncta and dissociates from them during autophagosome formation (Itakura and Mizushima, 2010). We therefore determined the requirement of FIP200 in targeting of p62 to the autophagosome formation site using VMP1 as a marker. In wild-type cells, only a few p62 puncta colocalized with the VMP1 puncta (Fig. 3 B). In contrast, almost all p62 puncta colocalized with the VMP1 puncta in FIP200 KO cells (Fig. 3 C). These results suggest that targeting of p62 to the autophagosome formation site on or close to the ER membrane is independent of the ULK1–FIP200 complex, and p62 seems to be trapped at the VMP1 structures when autophagosome formation is inhibited.
These results raised another possibility that VMP1 may be required for targeting of p62 to the autophagosome formation site. To this end, we performed siRNA-mediated silencing of VMP1 and analyzed p62 localization. As we previously reported (Itakura and Mizushima, 2010), ULK1 and DFCP1 puncta accumulated in VMP1 knockdown cells. Knockdown of VMP1 also led to accumulation of large p62 puncta, which colocalized with ULK1 and DFCP1, an ER-associated protein (Fig. S1). These results suggest that VMP1 is not necessary for the p62 recruitment to the ER-associated autophagosome formation site.
NBR1 localizes to the autophagosome formation site in a p62-independent manner
We also tested whether another autophagy selective substrate (or cargo receptor), NBR1, is also targeted to the autophagosome formation site in an LC3-independent manner. NBR1 is structurally similar to p62; it has a PB1 domain at the N terminus and a ubiquitin-associated (UBA) domain at the C terminus (Kirkin et al., 2009a). Moreover, NBR1 has an LRS-like sequence (YIII) which interacts with LC3, and is consequently selectively degraded by autophagy. Under nutrient rich conditions, NBR1 formed punctate structures, which colocalized with p62, but were mostly negative for ULK1 (Fig. S2 A). However, starvation treatment induced the ULK1 punctate structures, which colocalized with both p62 and NBR1 (Fig. S2 B). This colocalization was still maintained when cells were treated with wortmannin (Fig. S2 C), suggesting that NBR1 also targets to the autophagosome formation site independently of Atg factors downstream of the PI3-kinase complex, including LC3. This targeting is also independent of p62 because similar results were obtained when we used p62 KO MEFs (Fig. S2, D–F). These results suggest that localization to the autophagosome formation sites is not specific to p62, but may be a general feature of autophagy-selective substrates and cargo receptors.
The PB1 domain but not LRS of p62 is critical for its localization to the autophagosome formation site
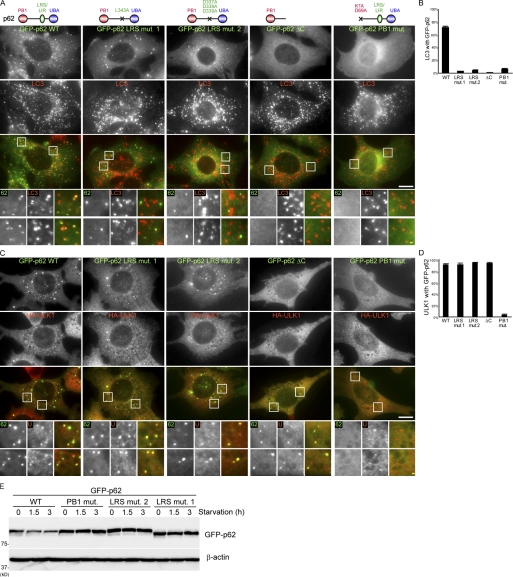
p62 interacts with LC3 via the LRS, which is critical for p62 degradation (Pankiv et al., 2007; Ichimura et al., 2008; Noda et al., 2008). Interaction of LC3 with p62 was found to be severely reduced in the p62 LRS mutant 1 (L343A) and was virtually abolished in the p62 LRS mutant 2 (D337/D338/D339A; Ichimura et al., 2008). We analyzed colocalization of LC3 and mutant p62 proteins in p62-deficient cells to eliminate the effect of endogenous p62. To avoid aggregation caused by overexpression, we used stably transformants expressing GFP-tagged p62 mutants at approximately less than fivefold of the endogenous levels. Consistent with previous observations, colocalization of LC3, with the p62 LRS mutant 1, LRS mutant 2 and p62ΔC (lacking both LRS and the UBA domain) was profoundly inhibited compared with wild-type p62 under starvation conditions (Fig. 4, A and B).
Figure 4.
The PB1 domain but not LRS of p62 is essential for localization to the autophagosome formation site. (A and B) p62 KO MEFs stably expressing GFP-p62 wild type, GFP-p62 LRS mutant 1 (L343A), GFP-p62 LRS mutant 2 (D337A, D338A, and D339A), GFP-p62ΔC (1–265 amino acids), and PB1 mutant (K7A and D69A) were cultured in starvation medium for 1 h. Cells were analyzed by immunofluorescence microscopy using anti-LC3 antibodies. GFP-p62 positivity (%) of the LC3 puncta is shown in B. Data represent mean ± SEM of 30 images. (C and D) p62 KO MEFs stably coexpressing HA-ULK1 and one of the GFP-p62 described in A were cultured in starvation medium containing 0.2 µM wortmannin for 1 h. Cells were analyzed by immunofluorescence microscopy using anti-HA antibodies. GFP-p62 positivity (%) of the LC3 puncta is shown in D. Data represent mean ± SEM of 30 images. (E) p62 KO MEFs stably expressing the indicated GFP-p62 were cultured in regular or starvation medium for 1.5 and 3 h. Cell lysates were analyzed by immunoblot analysis with the anti-p62 and β-actin antibodies. Signal color is indicated by color of typeface. St. M., starvation medium; WM, wortmannin; WT, wild type. Bars: (white) 10 µm; (yellow) 1 µm.
We next determined whether these p62 mutants could localize to the autophagosome formation site by testing colocalization between ULK1 and p62 mutants in the presence of wortmannin to inhibit isolation membrane formation. GFP-p62 (wild type) colocalized with ULK1 after starvation in the presence of wortmannin, as did endogenous p62 (91% of HA-ULK1 puncta were positive for GFP-p62; Fig. 4, C and D). Although p62 LRS mutant 1 and LRS mutant 2 are defective in LC3 binding, almost all ULK1 puncta generated after starvation and wortmannin treatment colocalized with the p62 LRS mutants (92% and 95% of HA-ULK1 puncta were positive for GFP-p62 LRS mutant 1 and mutant 2, respectively; Fig. 4, C and D). Furthermore, GFP-p62ΔC could form puncta and colocalized with ULK1 (Fig. 4, C and D), although it seldom forms puncta without wortmannin (Fig. 4, A and B). These data suggest that the LRS domain can be dispensable for targeting to the autophagosome formation site, although it may be important for incorporation into LC3-positive mature autophagosomes.
We therefore determined whether the other domains in p62 are required for targeting to the autophagosome formation site. p62 has a Phox and Bem1 (PB1) and UBA domains. We tested the effect of a PB1 mutation (K7A/D69A) in p62, which compromises the interaction surface of the PB1 domain and causes loss of p62 self-oligomerization activity (Ichimura et al., 2008). The PB1 mutant of GFP-p62 did not form any punctate structures even in the presence of wortmannin, although LC3- and ULK1-positive structures were generated normally in these cells (Fig. 4, A–D). Thus, the PB1 domain plays a critical role in p62 recruitment to the autophagosome formation site. We also confirmed that starvation-induced p62 degradation was suppressed in the PB1 domain mutant and in the LRS mutants (Fig. 4 E).
Collectively, these results suggest that the interaction with LC3 is not necessary for p62 association with early autophagic structures, but appears to be important to incorporate p62 inside these structures. In contrast, the PB1 domain is important for targeting of p62 to the early autophagic structure.
Self-oligomerization is required for localization of p62 to the autophagosome formation site
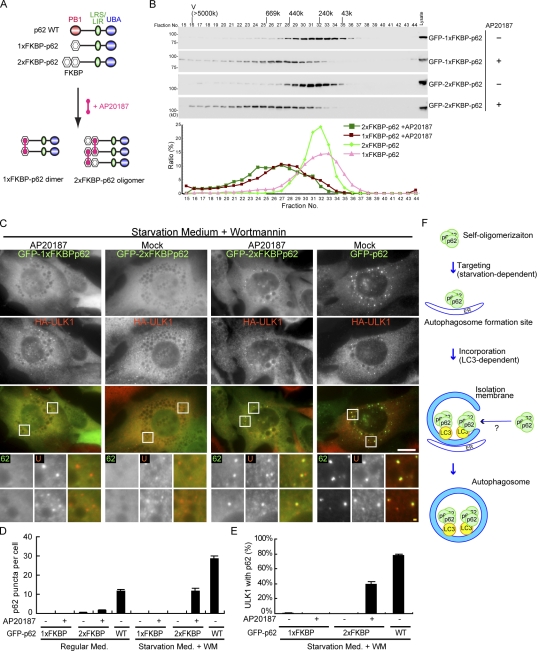
p62 is known to be self-oligomerized through the PB1 domain (Lamark et al., 2003; Wilson et al., 2003). We confirmed that p62 exists as oligomers irrespective of nutritional conditions (Fig. S3). To investigate the effect of p62 oligomerization through the PB1 domain, we developed an artificial self-oligomerization system using FK506-binding protein (FKBP) domain and a small ligand AP20187 (Amara et al., 1997; Clackson et al., 1998). This system is based on the binding between the FKBP domain and the small ligand AP20187, which can chemically link two FKBP domains. We replaced the PB1 domain of p62 with either one or two tandem FKBP domains, and generated p62 KO cells stably expressing GFP-1xFKBP-p62 or GFP-2xFKBP-p62 with HA-ULK1 (Fig. 5 A). After treatment of AP20187, two 1xFKBP-p62 molecules are linked to form a dimmer, whereas multiple 2xFKBP-p62 molecules are linked to form an oligomer. Gel filtration analysis confirmed that 1xFKBP-p62 moved to approximately twofold higher molecular weight fractions by treatment with AP20187 (Fig. 5 B). Treatment of cells expressing 2xFKBP-p62 causes broader shift to much higher molecular weight fractions, suggesting that 2xFKBP-p62 indeed formed oligomers (Fig. 5 B).
Figure 5.
Self-oligomerization of p62 is critical for its localization to the autophagosome formation site. (A) Schematic representation of the artificial p62 oligomerization system using a FKBP domain. The PB1 domain of p62 was replaced by one FKBP or two tandem FKBP domains. The small ligand AP20187 chemically links two FKBP domains. (B) p62 KO MEFs stably expressing GFP-1xFKBP-p62 or GFP-2xFKBP-p62 were treated with ethanol (vehicle) or 0.1 µM AP20187 for 24 h. After homogenization and centrifugation, the resulting supernatant fractions were applied to a Superose 6 column. Each fraction was analyzed by immunoblotting using anti-p62 antibodies. Positions of the molecular mass standards (kD) are shown. (C–E) p62 KO MEFs stably expressing HA-ULK1 and either GFP-1xFKBP-p62 or GFP-2xFKBP-p62 were treated with ethanol (vehicle) or 0.1 µM AP20187 for 24 h and cultured in regular or starvation medium containing 0.2 µM wortmannin with or without 0.1 µM AP20187 for 1 h. Cells were analyzed by immunofluorescence microscopy using anti-HA antibodies. The number of p62 puncta per cell (D) and the ULK1 positivity (%) of the p62 puncta (E) are shown. Data represent mean ± SEM of 30 images. Signal color is indicated by color of typeface. AP, AP20187; WM, wortmannin. (F) Proposed model of p62 localization to autophagic structures. Bars: (white) 10 µm; (yellow) 1 µm.
Using this system, we investigated the localization of these FKBP-p62 oligomers. Treatment of AP20187 did not induce p62 puncta formation in either 1xFKBP-p62– or 2xFKBP-p62–expressing cells cultured in regular medium (Fig. 5 D), suggesting that dimerization/oligomerization is not sufficient for puncta formation of p62. However, 2xFKBP-p62 formed puncta after treatment of AP20187 when cells were cultured in a starvation medium containing wortmannin (Fig. 5, C and D). Although starvation-induced puncta formation of 2xFKBP-p62 was less efficient than that of p62 (wild type), it was more efficient than that of 1xFKBP-p62 (Fig. 5, C and D). We also confirmed that these starvation-induced 2xFKBP-p62 puncta colocalized with ULK1 (Fig. 5, C and E). These results suggest that self-oligomerization of p62 through its PB1 domain is prerequisite to its localization to autophagosome formation sites.
Conclusion
It has been speculated that p62 is recruited to preexisting isolation membranes through interaction with LC3. However, our data suggest a completely different model of the p62 loading (Fig. 5 F). At the first step, oligomerized p62 targets to the autophagosome formation site. This step is clearly accelerated by starvation, but appears to be independent of LC3 and other classical Atg proteins. How p62 targets to the autophagosome formation site is currently unknown. One potential mechanism could be an interaction between p62 and ULK1 (Zhou et al., 2007), but we could not detect this interaction in our system (unpublished data). At the second step, the p62 oligomers are incorporated into autophagosomes probably in an LC3-dependent manner. Additional p62 molecules may be further recruited during autophagosome formation. This model is consistent with previous studies showing that the interaction with LC3 is not sufficient for efficient p62 degradation, and the PB1 domain is also important for its degradation (Ichimura et al., 2008; Lamark et al., 2009). We do not know the exact nature of p62 puncta that become enclosed by the autophagosome. They might be small protein aggregates or soluble oligomeric p62 associated with the autophagosome formation sites.
Because p62 is present on the autophagosome formation site, one might expect that p62 could serve in the machinery for autophagosome formation. However, ULK1 and Atg14 puncta formation are not affected in p62 KO cells (unpublished data), and starvation-induced autophagosome formation and degradation of long-lived proteins are normal in p62 KO cells (Komatsu et al., 2007). Nonetheless, it is still possible that p62 has the ability to recruit Atg proteins and to determine where autophagosome should be generated, although p62 is dispensable for autophagy induction. As p62 is also important for pexophagy (Kim et al., 2008) and elimination of bacteria (Yoshikawa et al., 2009), p62 may play a general role in recruiting Atg proteins to initiate autophagosome formation. It is also possible that other p62-like proteins such as NBR1 may function redundantly with p62 (Kirkin et al., 2009a).
Although the Cvt-specific genes are not conserved in mammals, p62 recruitment to the autophagosome formation site is reminiscent of the yeast Cvt pathway (Wang and Klionsky, 2003). After synthesis in the cytosol, precursors of Ape1 (prApe1) forms a homo-dodecamer and assembles into a Cvt complex. Atg19 serves as a receptor for prApe1 and recruits the Cvt complex to the PAS because Atg19 can also bind Atg8 and Atg11 (Scott et al., 2001; Shintani et al., 2002; Suzuki et al., 2002; Chang and Huang, 2007). An in vivo reconstitution experiment showed that prApe1, Atg19, and Atg11 are sufficient for cargo packaging and, at least in part, for cargo delivery to the PAS, which is the autophagosome/Cvt vesicle formation site in yeast (Cao et al., 2008). These processes may be similar to those of p62 targeting to the autophagosome formation site in mammalian cells. p62 could target to the site in an LC3-independent manner, and self-oligomerization of p62 through the PB1 domain is essential for the targeting (Figs. 4 and 5). The delivery of cargos and cargo receptors to the autophagosome/Cvt vesicle formation site would facilitate efficient incorporation of these cargos into autophagosomes and the Cvt vesicles.
Materials and methods
Cell culture and transfection
MEFs were cultured in DME supplemented with 10% FBS and 50 µg/ml penicillin and streptomycin (regular medium) in a 5% CO2 incubator. NIH3T3 cells were maintained in DME containing 10% bovine calf serum and the antibiotics (regular medium). Atg5 KO (Kuma et al., 2004), Atg3 KO (Sou et al., 2008), FIP200 KO (Gan et al., 2006), and p62 KO (Ichimura et al., 2008) MEFs were generated as described previously. For starvation, cells were washed with PBS and incubated in amino acid–free DME without serum (starvation medium). FuGENE 6 reagent (Roche) was used for transfection. Wortmannin and puromycin were purchased from Sigma-Aldrich.
Plasmids
Mouse p62 cDNAs (WT and mutants) were provided by M. Komatsu (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). An FLJ clone (GenBank accession no. D30756) encoding human NBR1 was obtained from Promega. To generate the pMXs-puro-GFP-p62 and pMXsIP-GFP-NBR1 plasmids, cDNAs were cloned into pMXs-puro or pMXs-IP (provided by T. Kitamura, University of Tokyo, Tokyo, Japan; Kitamura et al., 2003) together with EGFP (Takara Bio Inc.). pMXs-puro-HA-WIPI-1, pMXs-puro-VMP1-GFP, pMXs-IP-GFP-LC3, and pMXs-IP-GFP-Atg14 were described previously (Hara et al., 2008; Itakura et al., 2008; Itakura and Mizushima, 2010). To construct GFP-ER and monomeric RFP (mRFP)–ER, a cDNA of rat cytochrome b5 encoding residues 95–134 (provided by K. Mihara, Kyushu University, Fukuoka, Japan) was subcloned into pMXs-puro-GFP and pMXs-puro-mRFP (provided by R.Y. Tsien, University of California, San Diego, La Jolla, CA; Campbell et al., 2002). To generate the pMRX-IP-GFP-1xFKBP-p62 and pMRX-IP-GFP-2xFKBP-p62 plasmids, 1xFKBP and 2xFKBP amplified from pC4-FV1E or pC4M-FV2E (provided by ARIAD Pharmaceuticals; see http://www.ariad.com/wt/page/regulation_kits) were inserted into pMRX-IP (provided by S. Yamaoka, Tokyo Medical and Dental University, Tokyo, Japan) together with EGFP and p62.
Retroviral infections and generation of stable cell lines
Stable cell lines were generated using a retroviral expression system. pMXs and pMRX vectors were used to transfect Plat E cells to generate recombinant retroviruses. MEF and NIH3T3 cells were infected with recombinant retroviruses and selected in medium containing 1 µg/ml puromycin as previously described (Hara et al., 2008).
Antibodies
Mouse monoclonal anti-HA (16B12) antibodies were purchased from Covance. Rat monoclonal anti-GFP antibodies (GF090R) were purchased from Nacalai Tesque. Guinea pig polyclonal anti-p62 antibodies (for immunocytochemistry) were purchased from Progen. Rabbit polyclonal anti-p62 antibodies (for immunoblotting) were purchased from MBL International. The rabbit polyclonal antibodies against LC3 (SK2-6; Kabeya et al., 2000) and Atg16L1 (Mizushima et al., 2003) have been described previously. Mouse monoclonal anti–β-actin antibodies were purchased from Sigma-Aldrich.
Immunocytochemistry
Cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde in PBS for 10 min at 4°C. Fixed cells were permeabilized with 50 µg/ml digitonin in PBS for 5 min, blocked with 3% BSA in PBS for 30 min, and incubated with primary antibodies for 1 h. After washing, cells were incubated with Alexa Fluor 488–conjugated goat anti–rat, anti–rabbit, or anti–mouse IgG, Alexa Fluor 564–conjugated goat anti–rabbit, anti–mouse, or anti–guinea pig IgG or Alexa Fluor 660–conjugated goat anti–guinea pig IgG secondary antibodies (Invitrogen) for 30 min and examined under a fluorescence microscope (IX81; Olympus) equipped with a charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). A 60× 1.42 NA Plan Apo oil immersion lens (Olympus) was used. Images were acquired using MetaMorph image analysis software (version 7.0; MDS Analytical Technologies).
Time-lapse imaging
Cells were plated onto a glass-bottom dish and mounted in a chamber for live recording, and conditions were maintained with 5% CO2 at 37°C. Data were analyzed using MetaMorph image analysis software.
Immunoprecipitation and immunoblotting
Cell lysates were prepared in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethanesulfonyl fluoride, 1 mM Na3VO4, and protease inhibitor cocktail [Complete EDTA free; Roche]). The lysates were subjected to immunoprecipitation using specific antibodies in combination with protein G–Sepharose (GE Healthcare). Immunoblot analysis was performed with the indicated antibodies and visualized with SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific; Itakura et al., 2008; Itakura and Mizushima, 2010).
Gel filtration analysis
Gel filtration analysis was performed as previously described (Hosokawa et al., 2009). Cells were scrapped from the dish with a rubber policeman and homogenized in hypotonic buffer (40 mM Tris-HCl, pH 7.5, and protease inhibitors cocktail) by repeated passage (30 times) through a 1-ml syringe with a 27-G needle. The 100,000 g supernatant fraction (1.6 mg protein in 800 µl) was applied to a Superose 6 column (GE Healthcare) and eluted at a flow rate of 0.5 ml/min with 40 mM Tris-HCl, pH 7.5, and 150 mM NaCl. 0.5-ml fractions were examined by immunoblotting. The column was calibrated with 669-kD thyrogloblin, 440-kD ferritin, 232-kD catalase, and 43-kD ovalbumin.
RNAi
Stealth RNAi oligonucleotides (Invitrogen) were used for siRNA experiments. The following sequences were used for VMP1 siRNA: human VMP1 siRNA antisense, 5′-UAUACGUUGCACAUACUGUUGAUGC-3′; and sense, 5′-GCAUCAACAGUAUGUGCAACGUAUA-3′ (Itakura et al., 2008; Itakura and Mizushima, 2010). For a negative control, a medium GC duplex of Stealth RNAi negative control duplexes (Invitrogen) was used. The Stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocols. After 2 d, the cells were again transfected with the same siRNA and cultured for an additional 3 d before analysis unless otherwise specified.
Online supplemental material
Fig. S1 shows that VMP1 knockdown does not affect p62 targeting. Fig. S2 shows that NBR1 localizes to the early autophagic structures independently of PI3K and p62. Fig. S3 shows that oligomerization of p62 is not promoted by starvation. Video 1 shows that p62 puncta are closely associated with the ER during starvation. Video 2 shows that p62 puncta are closely associated with the ER during starvation with wortmannin treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201009067/DC1.
Acknowledgments
We thank Dr. Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science) for providing Atg3 KO MEFs, p62 KO MEFs, and p62 cDNA constructs, Dr. Tetsuro Ishii (University of Tsukuba) for p62 KO MEFs, Dr. Roger Y. Tsien (University of California at San Diego) for mRFP cDNA, Dr. Katsuyoshi Mihara (Kyushu University) for rat cytochrome b5 cDNA, Dr. Toshio Kitamura (University of Tokyo) for the retroviral vectors and Plat-E cells, Dr. Shoji Yamaoka (Tokyo Medical and Dental University) for pMRX-IP plasmid, Dr. Jun-Lin Guan (University of Michigan) for FIP200 KO cells, and ARIAD Pharmaceuticals for providing AP20187 chemical dimerizer and the pC4-FV1E and pC4M-FV2E plasmids. We also thank Drs. Naotada Ishihara and Chieko Kishi for helpful discussions.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N. Mizushima) and by grants for research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to E. Itakura), the Toray Science Foundation, and the Takeda Science Foundation (to N. Mizushima).
Footnotes
Abbreviations used in this paper:
- Cvt
- cytoplasm to vacuole targeting
- KO
- knockout
- LRS
- LC3 recognition sequence
- MEF
- mouse embryonic fibroblast
- mRFP
- monomeric RFP
- PAS
- preautophagosomal structure
- PI3-kinase
- phosphatidylinositol 3-kinase
- UBA
- ubiquitin associated
References
- Amara J.F., Clackson T., Rivera V.M., Guo T., Keenan T., Natesan S., Pollock R., Yang W., Courage N.L., Holt D.A., Gilman M. 1997. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 94:10618–10623 10.1073/pnas.94.20.10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart E.F.C., Krause U., Schellens J.P.M., Vreeling-Sindelárová H., Meijer A.J. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882 10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Cheong H., Song H., Klionsky D.J. 2008. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J. Cell Biol. 182:703–713 10.1083/jcb.200801035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F., Levine B. 2008. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 15:344–357 10.1016/j.devcel.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.Y., Huang W.P. 2007. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol. Biol. Cell. 18:919–929 10.1091/mbc.E06-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T., Yang W., Rozamus L.W., Hatada M., Amara J.F., Rollins C.T., Stevenson L.F., Magari S.R., Wood S.A., Courage N.L., et al. 1998. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. USA. 95:10437–10442 10.1073/pnas.95.18.10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. 2009. Multiple regulatory and effector roles of autophagy in immunity. Curr. Opin. Immunol. 21:53–62 10.1016/j.coi.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N., Lacas-Gervais S., Bertout J., Paz I., Freche B., Van Nhieu G.T., van der Goot F.G., Sansonetti P.J., Lafont F. 2009. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 6:137–149 10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J.L. 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 175:121–133 10.1083/jcb.200604129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Itakura E.M., Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. 2009. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 17:98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P.E., Klionsky D.J. 2002. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277:763–773 10.1074/jbc.M109134200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. 2008. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 105:20567–20574 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009a. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kirkin V., McEwan D.G., Novak I., Dikic I. 2009b. A role for ubiquitin in selective autophagy. Mol. Cell. 34:259–269 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J.I., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Kominami E., Hashida S., Khairallah E.A., Katunuma N. 1983. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J. Biol. Chem. 258:6093–6100 [PubMed] [Google Scholar]
- Kopitz J., Kisen G.O., Gordon P.B., Bohley P., Seglen P.O. 1990. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J. Cell Biol. 111:941–953 10.1083/jcb.111.3.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács A.L., Réz G., Pálfia Z., Kovács J. 2000. Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-ethyladenine. Cell Tissue Res. 302:253–261 10.1007/s004410000275 [DOI] [PubMed] [Google Scholar]
- Kraft C., Peter M., Hofmann K. 2010. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12:836–841 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568–34581 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Lamark T., Kirkin V., Dikic I., Johansen T. 2009. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 8:1986–1990 [DOI] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., Klionsky D.J. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584:1359–1366 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116:1679–1688 10.1242/jcs.00381 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T. 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 137:1001–1004 10.1016/j.cell.2009.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. 2008. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 13:1211–1218 10.1111/j.1365-2443.2008.01238.x [DOI] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584:1379–1385 10.1016/j.febslet.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 17:87–97 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T., Ruckerbauer S., Stierhof Y.D., Berg C., Nordheim A. 2007. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 581:3396–3404 10.1016/j.febslet.2007.06.040 [DOI] [PubMed] [Google Scholar]
- Ropolo A., Grasso D., Pardo R., Sacchetti M.L., Archange C., Lo Re A., Seux M., Nowak J., Gonzalez C.D., Iovanna J.L., Vaccaro M.I. 2007. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282:37124–37133 10.1074/jbc.M706956200 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 443:780–786 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Scott S.V., Guan J., Hutchins M.U., Kim J., Klionsky D.J. 2001. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 7:1131–1141 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Geetha T., Wooten M.W. 2007. Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581:175–179 10.1016/j.febslet.2006.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.P., Stromhaug P.E., Klionsky D.J. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581:2156–2161 10.1016/j.febslet.2007.01.096 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kamada Y., Ohsumi Y. 2002. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell. 3:815–824 10.1016/S1534-5807(02)00359-3 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Szeto J., Kaniuk N.A., Canadien V., Nisman R., Mizushima N., Yoshimori T., Bazett-Jones D.P., Brumell J.H. 2006. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2:183–193 [DOI] [PubMed] [Google Scholar]
- Virgin H.W., Levine B. 2009. Autophagy genes in immunity. Nat. Immunol. 10:461–470 10.1038/ni.1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Klionsky D.J. 2003. The molecular mechanism of autophagy. Mol. Med. 9:65–76 [PMC free article] [PubMed] [Google Scholar]
- Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L. 2003. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell. 12:39–50 10.1016/S1097-2765(03)00246-6 [DOI] [PubMed] [Google Scholar]
- Wooten M.W., Hu X., Babu J.R., Seibenhener M.L., Geetha T., Paine M.G., Wooten M.C. 2006. Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62’s role in neurodegenerative disease. J. Biomed. Biotechnol. 2006:62079 10.1155/JBB/2006/62079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T., et al. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11:1233–1240 10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- Yu L., Strandberg L., Lenardo M.J. 2008. The selectivity of autophagy and its role in cell death and survival. Autophagy. 4:567–573 [DOI] [PubMed] [Google Scholar]
- Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J.H. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183:5909–5916 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- Zhou X., Babu J.R., da Silva S., Shu Q., Graef I.A., Oliver T., Tomoda T., Tani T., Wooten M.W., Wang F. 2007. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. USA. 104:5842–5847 10.1073/pnas.0701402104 [DOI] [PMC free article] [PubMed] [Google Scholar]