Individual or joint knockdown of the transcriptional coactivators SRC-2 and SRC-3 inhibits lipid accumulation in adipocytes by decreasing PPARγ activity.
Abstract
The related coactivators SRC-2 and SRC-3 interact with peroxisome proliferator activated receptor γ (PPARγ) to coordinate transcriptional circuits to promote adipogenesis. To identify potential coactivator redundancy during human adipogenesis at single cell resolution, we used high content analysis to quantify links between PPARγ, SRC-2, SRC-3, and lipogenesis. Because we detected robust increases and significant cell–cell heterogeneity in PPARγ and lipogenesis, without changes in SRC-2 or SRC-3, we hypothesized that permissive coregulator levels comprise a necessary adipogenic equilibrium. We probed this equilibrium by down-regulating SRC-2 and SRC-3 while simultaneously quantifying PPARγ. Individual or joint knockdown equally inhibits lipid accumulation by preventing lipogenic gene engagement, without affecting PPARγ protein levels. Supporting dominant, pro-adipogenic roles for SRC-2 and SRC-3, SRC-1 knockdown does not affect adipogenesis. SRC-2 and SRC-3 knockdown increases the proportion of cells in a PPARγhi/lipidlo state while increasing phospho-PPARγ–S114, an inhibitor of PPARγ transcriptional activity and adipogenesis. Together, we demonstrate that SRC-2 and SRC-3 concomitantly promote human adipocyte differentiation by attenuating phospho-PPARγ–S114 and modulating PPARγ cellular heterogeneity.
Introduction
The dominant cellular basis of obesity is increased fat cell size during the adipocyte differentiation process. The process is marked by accretion of triglycerides within intracellular lipid droplets (Farmer, 2006). Adipogenesis is tightly regulated by peroxisome proliferator activated receptor γ (PPARγ), a member of the ligand-activated nuclear receptor superfamily of transcription factors. Mechanistically, exogenous (thiazolidinediones) or endogenous (eicosanoids) ligands activate PPARγ by stabilizing the active conformation of the ligand-binding domain (Nolte et al., 1998) to induce or repress a wide array of differentiation-dependent and adipose-specific genes. PPARγ mRNA and protein expression are robustly induced in a feed-forward loop with CAAT/enhancer binding protein-α (C/EBPα) during adipogenesis (Wu et al., 1999; Rosen et al., 2002). The process is initially stimulated by several up-stream transcription factors: C/EBPβ, C/EBPδ (Yeh et al., 1995; Wu et al., 1996; Zuo et al., 2006), and coregulators, including the p160 class of steroid receptor coactivators (SRCs; Louet and O’Malley, 2007).
A critical step required for adipogenesis is the down-regulation of kinase signaling pathways targeting PPARγ to permit its full transcriptional activity (Hu et al., 1996; Adams et al., 1997; Camp and Tafuri, 1997). Specifically, the pro-adipogenic function of PPARγ is decreased by mitogen-activated protein kinase (MAPK) phosphorylation in the N-terminal A/B region (mouse S112/human S114), which concomitantly reduces thiazolidinedione affinity for PPARγ (Shao et al., 1998). Overexpression of a nonphosphorylatable form of PPARγ promotes insulin sensitization and elevated adipogenesis in 3T3L1 (Hu et al., 1996; Shao et al., 1998). Additionally, mouse embryonic fibroblasts expressing a serine-to-alanine substitution at codon 112 (Rangwala et al., 2003) exhibit a similar effect. PPARγ phosphorylation at S112/S114 also decreases interactions with SRCs (Shao et al., 1998), resulting in a potential negative cooperative effect on PPARγ-regulated, adipocentric genes.
The p160 family of SRCs (SRC-1, SRC-2, and SRC-3) defined the first class of coregulators (CoR) that enhance nuclear receptor transactivation in a ligand-dependent manner, bridging NRs to other components of the basal transcriptional machinery and integrating both genomic and nongenomic signals (Oñate et al., 1995; Anzick et al., 1997; Hong et al., 1997). Depending on the ligand context, biochemical assays have shown that each of the SRCs potentiate the transcriptional activity of PPARγ through direct interactions (McInerney et al., 1998; Kodera et al., 2000; Rocchi et al., 2001; Louet et al., 2006). However, SRC-2 and SRC-3 share the highest degree of sequence homology and promote adipogenesis in knockout mouse models. SRC-2−/− mice are protected from obesity because of enhanced energy expenditure, decreased white adipocyte differentiation, and increased thermogenic activity of brown adipose (Picard et al., 2002). SRC-3 ablation leads to lean mice with increased energy expenditure and decreased adipogenesis (Louet et al., 2006; Coste et al., 2008). However, upon high-fat diet feeding, SRC-1−/− knockout mice are slightly prone to obesity because of both a reduced capacity for fatty acid oxidation and decreased energy expenditure (Picard et al., 2002). Moreover, double knockout of SRC-1 and SRC-3 results in a lean phenotype and increased metabolic rate (Wang et al., 2006), similar to SRC-3−/− mice, which provides evidence of a dominant pro-adipogenic role for SRC-3.
Although the similarities observed among SRC-2 and SRC-3 knockout mice indicate a functional overlap and dominant pro-adipogenic roles, no data exists on the contributions of cell–cell variability between SRC-2, SRC-3, and PPARγ that collectively and/or redundantly promote human adipogenesis. Accordingly, the purpose of this study was to focus upon SRC-2 and SRC-3 and dissect the early interplay between these coactivators and PPARγ that converts a human preadipocyte into a mature fat cell. Here, we developed and used a high-throughput microscopy-based, high-content analysis (HCA) approach to quantify the effects of SRC loss of function on the cell-to-cell population dynamics of PPARγ. Our results emphasize the novel regulatory role of steady-state levels of SRC-2 and SRC-3 in human adipogenesis, specifically by promoting lipid accumulation under both high and low PPARγ phenotypes marked by attenuation of PPARγ phosphorylation at S114.
Results
Human adipocyte differentiation occurs independently of static SRC mRNA profiles
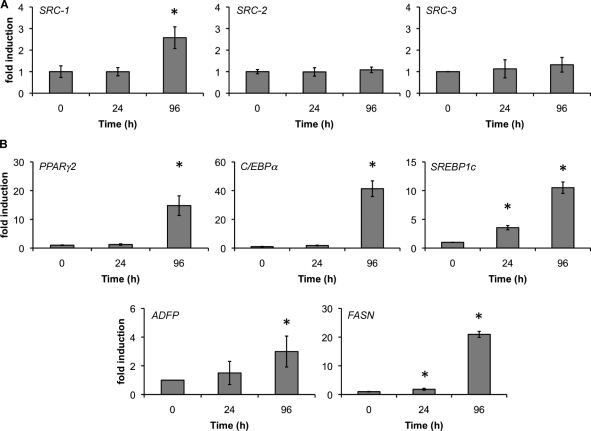
SRCs have been shown to be critical elements of the murine adipogenic gene program (Picard et al., 2002; Louet et al., 2006; Coste et al., 2008). Given these results, we used quantitative RT-PCR (qPCR) to measure mRNA expression during the first 4 d of human adipocyte differentiation with rosiglitazone, dexamethasone, 3-isobutyl-1-methylxanthine (IBMX), and insulin (MIX). Surprisingly, we found that although SRC-1 mRNA levels were increased twofold, SRC-2 and SRC-3 were not changed during the first 4 d of differentiation (Fig. 1 A). We also measured mRNA levels of several transcription factors that stimulate differentiation and markers of lipogenesis (Fig. 1 B). The mRNA levels of C/EBPα (40-fold), PPARγ (10-fold), SREBP1c (10-fold), FASN (20-fold), and ADFP (threefold) were up-regulated in response to 96 h of treatment with MIX. These initial experiments suggested that human adipocytes maintain constant SRC-2 and SRC-3 mRNA levels while up-regulating SRC-1 transcripts during early differentiation.
Figure 1.
Expression of SRC-2, SRC-3, PPARγ, and adipocentric genes during human adipocyte differentiation. Subcutaneous human preadipocytes were differentiated for 4 d, and total mRNA was isolated at each indicated interval. The mRNA levels of SRC-1, SRC-2, and SRC-3 (A); and PPARγ2, C/EBPα, SREBP1c, ADFP, and FASN (B) were determined by qPCR. β-actin served as an internal control. RNA was collected from at least two independent isolations and error bars represent SEM. Asterisks indicate levels of induction statistically different from 0 h (P < 0.05).
HCA of human adipocyte cell populations validates mRNA results for SRCs and PPARγ
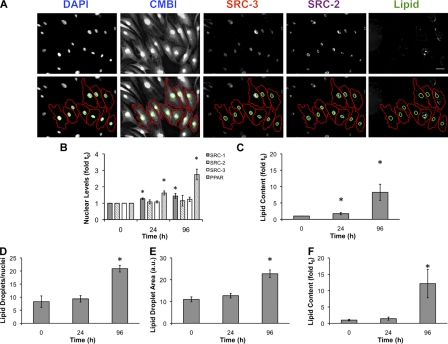
To further understand the dynamics of SRC-1, SRC-2, SRC-3, and PPARγ during differentiation, we validated the mRNA profiling (Fig. 1) with protein and lipid measurements at the single cell level using HCA. Subcutaneous preadipocytes were differentiated for 4 d, then fixed and labeled for DNA (DAPI), SRC-1, SRC-2, SRC-3, or PPARγ. Lipid droplets were labeled with a fluorescent neutral lipid dye to mark differentiating cells. We next quantified changes in these properties for each cell (≥1,000 cells/condition/experiment), by automated cell and nucleus identification (Fig. 2 A) with both in-house algorithms and commercially available software. As indicated in Fig. 2 B, mean PPARγ protein levels increased monotonically (2.8-fold) over the first 4 d, which correlates with an approximately eightfold induction of lipid accumulation (Fig. 2 C). After 7 d, 88.1% of cells were positive for lipids, which suggests that the preadipocyte cell population responds robustly to chemical induction of adipogenesis in a unidirectional manner (Fig. S1 A). Consistent with the qPCR results (Fig. 1 A), SRC-1 was significantly increased (50%), whereas modest, statistically insignificant changes in SRC-2 (15%) and SRC-3 (22%) were detected. Using additional software with built-in lipid droplet analysis tools (McDonough et al., 2009), we detected increases in the number of lipid droplets/nuclei (+250%; Fig. 2 D), total lipid droplet area (+120%; Fig. 2 E), and lipid content/cell (12-fold induction; Fig. 2 F). Dose response experiments using rosiglitazone (BRL49653) or a natural PPARγ ligand (15-deoxy-Δ12,14-prostaglandin J2; 15dPGJ2) showed that both ligands increase lipogenesis (Fig. S1 B), PPARγ (Fig. S1 C), and SRC-1 levels (Fig. S1 D). Increasing concentrations of 15dPGJ2 and BRL49653 did not significantly alter the protein expression of SRC-2 (Fig. S1 E) and SRC-3 (Fig. S1 F).
Figure 2.
Development of an image-based analysis platform to study PPARγ and coactivator expression in human adipocytes. (A) Shown are representative grayscale images of adipocytes differentiated for 96 h, immunolabeled with antibodies to SRC-2 or SRC-3, and then stained with DAPI, CMBl, and LipidTOX (Lipid). Binary nuclear and cellular masks were generated by a combination of watershed and threshold image transformations (Pipeline Pilot; Accelrys). Nuclear masks are indicated in green; whole cell masks are shown in red. (B–F) Nuclear and cellular masks were used to extract pixel-based measurements that describe nuclear PPARγ, SRC-1, SRC-2, SRC-3 levels (B) and lipid accumulation (C) during a 96-h differentiation period. Additional software (CyteSeer; Vala Sciences) was used to calculate the number of lipid droplets/nuclei (D), lipid droplet area (E), and fold induction of lipid (F). Experiments shown are the mean of 11 independent experiments. Values are the mean fold induction. Error bars indicate SEM (*, P < 0.05 compared with 0 h). Bar, 50 µm.
Cell-to-cell measurements of lipids as a function of PPARγ represent population heterogeneity during human adipogenesis
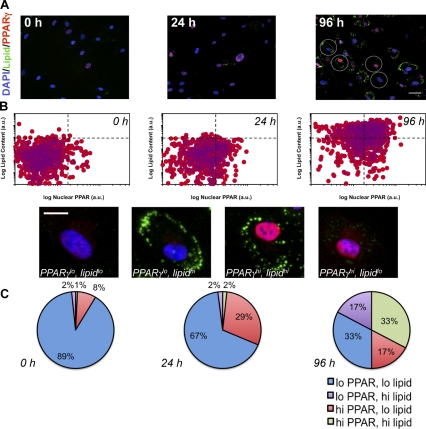
At the population level, the time-dependent induction of PPARγ was positively correlated with the accumulation of lipids, which is consistent with previously established deterministic paradigms of PPARγ-mediated adipogenesis (Rosen et al., 1999). However, when the differentiating fraction of preadipocytes was quantitatively examined at the single cell level, a large degree of population heterogeneity was observed, which is in agreement with recent studies on the 3T3L1 differentiation program (Loo et al., 2009). These subpopulation changes were apparent as early as 24 h after MIX treatment, and more visible at 96 h (Fig. 3 A). This heterogeneity is illustrated in Fig. 3 B by plotting, on a cell-by-cell basis, cellular lipid content as a function of nuclear PPARγ protein levels. At 96 h after induction by MIX, 100-fold variation in PPARγ and a range of three orders of magnitude in lipid content were observed. This was an undetectable response at the median protein (immunofluorescence; Fig. 2) and mRNA levels (qPCR; Fig. 1). To explore this heterogeneity across experiments, we subdivided the cell populations. For each experiment and time point, individual cell measurements of PPARγ and lipid were normalized to the median intensity at 96 h, which set the threshold for quadrant subdivision for both properties arbitrarily equal to 1. This threshold was then applied to each time point. Subsequently, this created four subpopulation quadrants (Fig. 3 C): high PPARγ/high lipid (PPARγhi/lipidhi), high PPARγ/low lipid (PPARγhi/lipidlo), low PPARγ/low lipid (PPARγlo/lipidlo), and low PPARγ/high lipid (PPARγlo/lipidhi). Temporal analysis of these quadrants showed that the PPARγhi/lipidhi population increased from 2% after 24 h of differentiation to 33% at 96 h. More interestingly, however, was the up-regulation of the PPARγhi/lipidlo population at 24 h (21% change) followed by a decrease (−12%) at 96 h. Based on these results, the nonessential role of SRC-1 (Fig. S2), and the central functions of SRC-2 and SRC-3 in adipogenesis (Picard et al., 2002; Louet et al., 2006; Wang et al., 2006; Coste et al., 2008), we proposed that variation in PPARγ (Fig. 3 C) might be dictated by factors, specifically SRC-2 and SRC-3, whose overall expression level was not regulated by differentiation but nonetheless were important for the early human adipogenic phenotype.
Figure 3.
Population dynamics of PPARγ protein expression as a function of lipid content during the early phases of human adipocyte differentiation. (A) Representative images of adipocytes during differentiation were immunolabeled for PPARγ, and lipids were labeled and imaged by high throughput microscopy (HTM). Bar, 50 µm. (B) Cell-to-cell variation in PPARγ and lipids during the first 96 h of differentiation was monitored. One representative experiment is shown. Individual cell measurements of PPARγ and lipid were normalized to the median intensity at 96 h, which set a threshold (dotted lines) that was applied to each time point to create the PPARγx/lipidy populations. An example cell is shown from each quadrant. Bar, 20 µm. (C) Pie charts are shown that indicate the change in population distribution over this time period after median threshold application (n = 5 independent experiments).
SRC-2 and SRC-3 levels are correlated with PPARγ-dependent lipogenesis
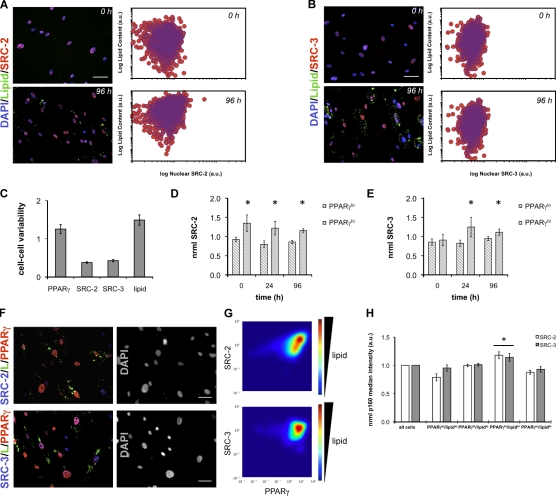
Biochemical studies and mouse models have indicated that pro-adipogenic transcriptional activity of PPARγ is maintained by functional interactions with the coactivators SRC-2 and SRC-3 (McInerney et al., 1998; Rocchi et al., 2001; Louet et al., 2006). Therefore, we sought to understand the single cell relationship between these SRCs and PPARγ. First, we analyzed the level of heterogeneity that existed for both SRC-2 and SRC-3 in individual cells during the first 96 h of human adipocyte differentiation. Analysis of the cell–cell heterogeneity in both SRC-2 (Fig. 4 A) and SRC-3 (Fig. 4 B) levels indicated a <10-fold lower range of expression levels compared with PPARγ (>100-fold). This tighter range of SRC-2 or SRC-3 levels was consistent with (96 h) and without (0 h) differentiation, whereas lipid content increased independent of single cell SRC-2 and SRC-3 expression. We further represented this contrast in variability by calculating the coefficient of variation (CV = σ/μ) after 96 h of differentiation, where a higher CV indicates wider system heterogeneity. In agreement with scatter plot representations of PPARγ (Fig. 3 B), SRC-2 (Fig. 4 A), and SRC-3 (Fig. 4 B), CVs for PPARγ and lipid were significantly greater than those calculated for SRC-2 and SRC-3.
Figure 4.
Cell-to-cell relationships between SRC-2, SRC-3, and PPARγ. (A and B) SRC-2 (A) and SRC-3 (B) were monitored along with lipids at the single cell level at the indicated time points. (C) Cell–cell variability, measured as the CV (σ/µ), of the indicated properties at 96 h. (D and E) SRC-2 (D) and SRC-3 (E) normalized median intensities were monitored as a function of time in PPARγlo and PPARγhi cell populations by immunofluorescence. For each experiment and time point, individual cell measurements of PPARγ and SRC were normalized to the median intensity at 96 h. PPARγ normalization set the threshold for binary subdivision into PPARγlo and PPARγhi populations arbitrarily equal to 1. This threshold was then applied to each time point, and normalized SRC levels were calculated (*, P < 0.05 for PPARγlo vs. PPARγhi). (F) Shown are immunofluorescence images of SRC/PPARγ/lipid (L) after 96 h of adipocyte differentiation from one representative experiment with (G) contour mapping of SRC/PPARγ/lipid relationships. Density plots show normalized lipid expression as a function of normalized SRC and PPARγ levels (n ≥ 1,900 cells). (H) Cells were divided into four quadrants based on their median PPARγ and lipid levels, followed by calculation of the normalized median SRC-2 or SRC-3 intensity inside each population. In all experiments, SRC levels were normalized to the median intensity at 96 h (n = 6 independent experiments; *, P < 0.05). Error bars indicate SEM. Bars, 50 µm.
Next, we simultaneously detected SRC/PPARγ (Fig. 4 F) to determine if there were cells with SRC-2 or SRC-3 levels that correlated with PPARγ and/or lipids. Individual cell measurements of PPARγ and SRC were normalized to their respective median intensities at 96 h. PPARγ normalization set the threshold for binary subdivision into PPARγlo and PPARγhi populations arbitrarily equal to 1. This threshold was then applied to each time point, and normalized SRC levels were calculated. As shown in Fig. 4 D, SRC-2 levels were higher in the PPARγhi population across all time points. SRC-3 (Fig. 4 E) showed a similar pattern at 24 h and 96 h only. The SRC-2 and SRC-3 levels in the PPARγlo and PPARγhi were not affected by differentiation, which suggested that these populations might represent, in terms of SRC and PPARγ, similar biological states. Single cell analysis of the correlation (Pearson’s r [Pr]) between SRC and PPARγ levels at 96 h showed significant, positive correlation between both SRC-2 and PPARγ (Pr = 0.39, n = 6) and SRC-3 and PPARγ (Pr = 0.33, n = 6). Contour mapping (Fig. 4 G) of lipid intensity, as a function of SRC and PPARγ, showed that high SRC-2 and PPARγ correlated with increased lipids. SRC-3, PPARγ, and lipids exhibited a less pronounced relationship. Specifically, SRC-2 and SRC-3 intensities within cell subpopulations defined by PPARγ and lipid levels in Fig. 3 showed that the PPARγhi/lipidhi cells had significantly higher levels of SRC-2 or SRC-3 than the other quadrants (Fig. 4 H). Interestingly, this population also had significantly higher correlation coefficients between SRC-2/PPARγ and SRC-3/PPARγ. In contrast, PPARγhi/lipidlo cells exhibited the lowest SRC/PPARγ correlation (Table I).
Table I.
Pearson’s product moment correlation coefficients were calculated for PPARγ/SRC-2 and PPARγ/SRC-3 inside of PPARγx/lipidy subpopulations
| Phenotype | Lipidlo | Lipidhi |
| PPARγ/SRC-2 | ||
| PPARγlo | 0.21 ± 0.05 | 0.25 ± 0.10 |
| PPARγhi | 0.16 ± 0.06 | 0.43 ± 0.10 |
| PPARγ/SRC-3 | ||
| PPARγlo | 0.17 ± 0.10 | 0.13 ± 0.06 |
| PPARγhi | 0.09 ± 0.08 | 0.33 ± 0.13 |
Data are presented as the mean ± SEM. n = 6.
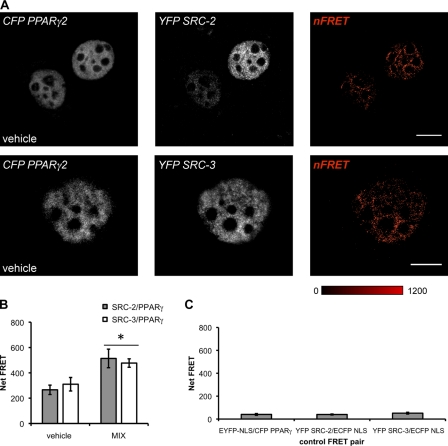
The correlations between SRCs and PPARγ established a quantitative relationship between lipid, PPARγ, and SRC levels in human adipocytes, but did not indicate the nature of the interaction between these proteins. To visualize the interactions between PPARγ/SRC-2 and PPARγ/SRC-3 occurring in response to differentiation cues, we used sensitized emission fluorescence resonance energy transfer (seFRET). CFP PPARγ2/YFP SRCs were cotransfected for 48 h and subsequently treated with MIX or vehicle (DMSO) for 2 h. A strong FRET signal, chiefly localized in a heterotypic pattern within the nucleus, was observed when CFP-PPARγ/YFP–SRC-2 or CFP-PPARγ2/YFP–SRC-3 were coexpressed in HeLa cells (Fig. 5 A). FRET was measured both treatment conditions implying differentiation-independent interactions between SRCs and PPARγ. Although vehicle treatment exhibited a high basal level of FRET, statistically significant increases (>1.5-fold change) in FRET were detected in the presence of MIX (Fig. 5 B). Cells coexpressing YFP-SRC/ECFP-NLS or CFP-PPARγ2/EYFP-NLS were used as negative controls and exhibited significantly less FRET (Fig. 5 C) than PPARγ–SRC fusion pairs.
Figure 5.
SRC-2 and SRC-3 interact with PPARγ in a differentiation-independent manner. (A) seFRET was used to evaluate the interactions between CFP-PPARγ2/YFP SRC-2 or CFP-PPARγ2/YFP SRC-3 after exposure to either vehicle (DMSO) or MIX for 2 h in wild-type HeLa cells. Representative images are shown from one experiment for a single channel (CFP or YFP) with the calculated FRET image. Bars, 10 µm. (B) For each cell, the net FRET between CFP-PPARγ2 and YFP-SRC was determined using the softWoRx user interface. FRET was measured within nucleoplasmic regions of interest only. (C) Control plasmids, ECFP or EYFP fused to a NLS sequence, were coexpressed, and FRET was determined. On average, CFP-PPARγ2/YFP-SRC FRET signals were 8–20× greater than those measured in vector control experiments (n ≥ 22 cells measured over three independent experiments; *, P < 0.05 compared with vehicle treatment). FRET signals were scaled between minimum and maximum signals (0–1,200 pixels), and intensity was colored as shown. Error bars indicate SEM.
SRC-2 and SRC-3 are essential for the adipocentric phenotype
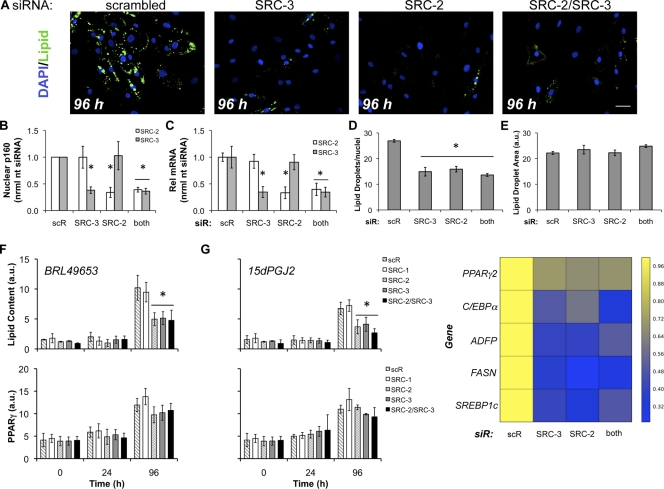
The combination of qPCR, HCA, and FRET results led us to hypothesize that static, permissive SRC-2 and SRC-3 levels, occurring in the entire population contribute to an equilibrium condition that controls human adipogenesis. To perturb the postulated SRC-2 and SRC-3 equilibrium, we performed siRNA-based knockdowns (individually or in tandem) while simultaneously using antibodies to detect cell-to-cell changes in individual SRC protein levels. This approach uniquely allows cell-by-cell monitoring of target knockdown and any effect on differentiation. After a 48-h siRNA knockdown, preadipocytes were induced to differentiate for up to 96 h, and the extent of lipid accumulation and SRC levels was quantified by HCA. Shown in Fig. 6 A, single knockdown of SRC-2 or SRC-3 resulted in decreased lipid accumulation. In line with previous observations (Louet et al., 2006), SRC-1 siRNA had no effect on lipogenesis or adipocentric gene expression (Fig. S2). Moreover, there was no apparent synergistic or additive inhibition of adipogenesis with dual SRC-2/SRC-3 siRNA knockdown. Additionally, siRNA targeting SRC-2 or SRC-3 did not alter the expression of the other CoR detected at the immunofluorescence level (Fig. S3 A) and quantified by HCA (Fig. 6 B). qPCR analyses of SRC levels in the median population levels for single or dual siRNA (Figs. 6 C and S3, B–F) showed consistent knockdown for both messages, validating our protein (HCA) measurements 6 d after transfection.
Figure 6.
Single or double siRNA knockdown of SRC-2 and SRC-3 disrupts adipocyte differentiation without affecting PPARγ protein induction. (A) Preadipocytes were reverse-transfected with scrambled (scR) or siRNA to SRC-2, SRC-3, or both SRC-2/SRC-3 for 48 h followed by induction of differentiation, and imaging. Bar, 50 µm. (B) HCA detection of p160 levels after siRNA transfection. (C) qPCR was used to validate measurements of SRC-2 or SRC-3 knockdown by HCA. (D and E) Lipid droplet count (D) and lipid droplet area (E) were determined after siRNA knockdown and 4 d of differentiation (n = 7 independent experiments). (F and G) The effects on the rate of lipogenesis and PPARγ induction were determined by differentiation of preadipocytes for the indicated time points in the presence of either 3 µM BRL49653 (rosiglitazone; F) or 30 µM 15dPGJ2 (G) after SRC siRNA transfection (n = 3 independent experiments). (H) Heat map summary of the downstream effects of siRNA to SRC-2/SRC-3 on lipid accumulation markers as measured by qPCR: PPARγ2, C/EBPα, ADFP, FASN, and SREBP1c. RNA was isolated from two independent experiments. Asterisks indicate measured variables statistically different from the nontargeting siRNA control at the 95% confidence level (*, P < 0.05). Error bars indicate SEM.
Quantification over a large span of experiments (n = 7 independent replications, >1,000 cells/condition) showed that a >60% reduction in either or both SRC-2 and SRC-3 led to a 40% decrease (Fig. 6 D) in the number of lipid droplets/nuclei without altering lipid droplet size (Fig. 6 E). We next determined the effect of SRC-2/SRC-3 on the rate of lipogenesis in the presence of a synthetic (BRL49653/rosiglitazone) or the natural PPARγ agonist, 15dPGJ2 (Forman et al., 1995; Kliewer et al., 1995). In these loss-of-function experiments, SRC-2/SRC-3 single or joint knockdown slowed lipogenesis at 96 h by at least 50% (compared with scrambled siRNA control) without significantly affecting the induction of PPARγ. Additionally, the effect was observed under differentiation with rosiglitazone (Fig. 6 F) or 15dPGJ2 (Fig. 6 G). This result suggests that SRC-2 and SRC-3 are critical components of the basal adipogenic machinery, in agreement with in vivo data (Picard et al., 2002; Louet et al., 2006; Coste et al., 2008). By qPCR, down-regulation of SRC-2, SRC-3, or SRC-2/SRC-3 resulted in decreases in central adipogenic transcription factors (Fig. 6 H): PPARγ (>29%) and C/EBPα (>40%). Further, reductions in lipogenic gene expression were also measured (Fig. 6 H): ADFP (>57%), SREBP1c (>59%), and FASN (>64%). To probe the action of SRCs in gain-of-function experiments, SRC-1, SRC-2, and SRC-3 were overexpressed by lentiviral infection. Importantly, we ignored expression level artifacts that present themselves as protein aggregates (Stenoien et al., 2000, 2002; Feige et al., 2005). Although single or double SRC-2/SRC-3 siRNA decreased lipogenesis, moderate (1.5–2.5 times endogenous protein) overexpression of SRC-1, SRC-2, and SRC-3 showed modest increases in lipogenesis (<1.6-fold compared with FLAG control) after 96 h of differentiation (Fig. S5). Each of these results suggested that equilibrium levels of only SRC-2 and SRC-3 are needed for human adipocyte differentiation and lipogenic gene regulation.
SRC-2 and SRC-3 promote adipocyte heterogeneity and attenuate PPARγ phosphorylation
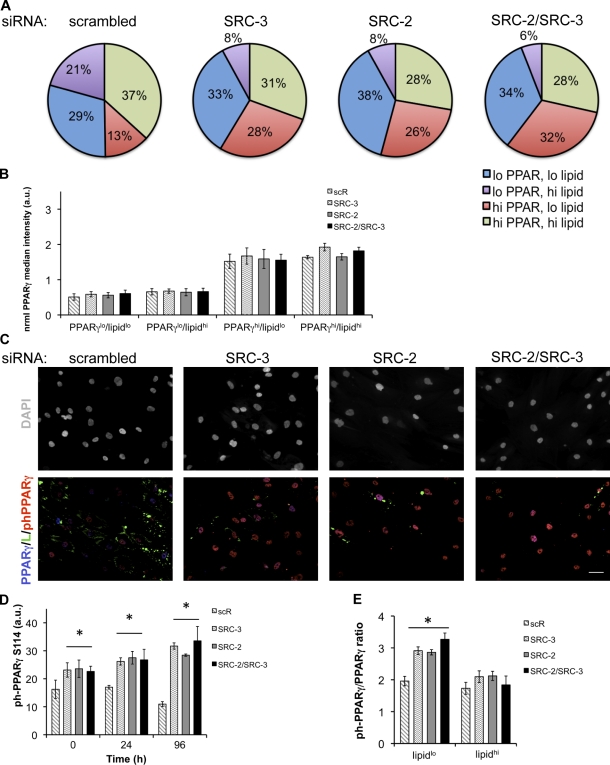
Although our experiments revealed a central function of SRC-2 and SRC-3 in promoting human adipogenesis both phenotypically (HCA) and transcriptionally (qPCR), follow-up experiments showed that when PPARγ mRNA was reduced by 29% (Fig. 6 H), PPARγ protein levels and induction were not significantly altered (Fig. 6, F and G). We then analyzed the cell–cell variability of PPARγ and lipids after SRC siRNA knockdown (Fig. 7 A). For each experiment and transfection, individual cell measurements of PPARγ and lipid were normalized to the median intensity of the scrambled control, which set the threshold for quadrant subdivision for both properties arbitrarily equal to 1. This threshold was then applied to each siRNA transfection to create the PPARγx/lipidy populations. As validation of the PPARγ/lipid gating, PPARγ siRNA inhibits lipid accumulation (Fig. S4 C) and shifts the cells to a predominantly (73%) PPARγlo/lipidlo state by decreasing PPARγhi fractions (Fig. S4 D). Although the total PPARγ was largely unchanged at the whole population (Fig. 6 F) and subpopulation level (Fig. 7 B), siRNA to SRC-2 and/or SRC-3 caused shifts in each PPARγ/lipid subpopulation. SRC siRNA increased the proportion of cells in a PPARγlo/lipidlo state by >4% while decreasing the PPARγhi/lipidhi percentage >6%. Concurrent with these changes in subpopulation distributions, more significant effects were detected on the PPARγlo/lipidhi (−13% to −15%) and the PPARγhi/lipidlo lipid populations (+15% to +19%). The changes in population variation indicate a role for SRC-2 and SRC-3 in controlling cell heterogeneity that promotes lipogenesis over a wide continuum of PPARγ expression. Additionally, up-regulation of the PPARγhi/lipidlo population, along with decreases in downstream PPARγ-dependent (ADFP) and adipocentric/lipogenic genes (Fig. 6 H), led us to hypothesize that the loss of the coactivators resulted in higher levels of PPARγ phosphorylation at S114, leading to delayed/reduced adipogenesis.
Figure 7.
SRC-2/SRC-3 single or double knockdown alters PPARγ heterogeneity and phosphorylation status. (A) PPARγ was immunolabeled and imaged by HTM with DAPI/CMBl and lipid counterstains under conditions of SRC-2, SRC-3, or SRC-2/SRC-3 siRNA. The effects of SRC-2/SRC-3 siRNA on subpopulation distributions (*, P < 0.05, n = 3) were tabulated. For each experiment and transfection, individual cell measurements of PPARγ and lipid were normalized to the median intensity of the scrambled control, setting the threshold for quadrant subdivision for both properties arbitrarily equal to 1. This threshold was then applied to each siRNA transfection to create the PPARγx/lipidy populations. (B) The normalized median PPARγ level was determined in each subpopulation for scrambled (scR) and SRC siRNA conditions. (C) Human preadipocytes were reverse-transfected with siRNA to SRC-2, SRC-3, or both coactivators and treated with MIX for up to 96 h. Subsequent to the perturbations, cells were immunolabeled with antibodies to phosphoPPARγ-S114 and total PPARγ, followed by HTM imaging. Bar, 50 µm. (D) After imaging, phosphoPPARγ-S114 was quantified for 0, 24, and 96 h of differentiation (*, P < 0.05; n = 3). (E) The single cell intensity ratios of phosphoPPARγ-S114 to PPARγ were determined for the lipidlo and lipidhi populations at 96 h (*, P < 0.05; n = 3). Error bars indicate SEM.
MAPK–ERK phosphorylation of PPARγ at S112/S114 diminishes its ligand affinity, transcriptional activity, adipogenic capacity, and interactions with SRCs (Hu et al., 1996; Adams et al., 1997; Shao et al., 1998; Rangwala et al., 2003). We postulated that our PPARγhi/lipidlo populations might represent higher levels of phospho-PPARγ S114 and that the presence of SRC-2 and/or SRC-3 minimizes this proportion of cells to promote adipocyte differentiation. To test this hypothesis, we knocked down SRC-2 and/or SRC-3 with siRNA and evaluated the levels of phospho-PPARγ S114 at 0, 24, and 96 h after differentiation. Upon immunolabeling with a specific antibody to phospho-PPARγ S114 and total PPARγ (Fig. 7 C), higher levels of phospho-PPARγ were present when SRC-2 and/or SRC-3 levels were reduced by siRNA. Further quantitative analysis indicated increases in phospho-PPARγ (Fig. 7 D) at each time point for individual knockdowns of SRC-2 (2.89-fold, 96 h) or SRC-3 (2.6-fold, 96 h), or when SRC-2 and SRC-3 (2.75-fold, 96 h) are both knocked down, respectively. Contrasting the effect of SRC siRNA, scrambled siRNA (scR) reduces phospho-PPARγ S114 by 32% over the 96-h differentiation period. As shown in Fig. 7 A and Fig. 7 D, several correlated findings are suggested: (a) up-regulation of phospho-PPARγ levels coincided with (b) reduction of lipid accumulation and (c) enrichment of cells in a PPARγhi/lipidlo state. Simultaneous immunofluorescence detection of phospho-PPARγ and total PPARγ in our subpopulation analyses also showed that, with respect to the nontargeting control, the mean single cell intensity ratio of phospho-PPARγ to PPARγ (ph-PPARγ/PPARγ) was highest in the lipidlo populations (Fig. 7 E). This finding was consistent with our hypothesis that the observed defect in lipogenesis was caused by a specific subpopulation up-regulation of phospho-PPARγ. Collectively, data from Figs. 6 and 7 suggest that permissive levels of the coactivators SRC-2 and SRC-3 attenuate phospho-PPARγ to promote a full adipogenic response.
Discussion
The contribution of transcription factors and the associated regulatory machinery to the development of functional heterogeneity among white fat depots remains largely undiscovered, especially in the context of human adipogenesis. Recent cell culture studies (Le and Cheng, 2009; Loo et al., 2009) have indicated that concurrent physiological and molecular states may exist in differentiating 3T3L1 preadipocytes largely as a response to systemic or growth factor stimulation. Additionally, PARγ titrated knock-in transgenic mice (Tsai et al., 2009) demonstrated that selective reduction of PPARγ only affected the accumulation of perigonadal fat, without decreasing retroperitoneal, inguinal, mesenteric, or subcapsular adipose mass, an indication that PPARγ functional heterogeneity at the whole animal level can exist without compromising generalized adipogenesis. Our single cell–oriented data are consistent and expand upon these findings, showing a wide cell-to-cell variability (Fig. 3) in the early human adipocyte differentiation cascade. During this period, cells exhibit and support continuous PPARγ states with and without lipids, even while robust activation of pro-adipogenic genes occurs at the population level (Fig. 1).
Synthetic and natural ligands bind PPARγ in a relatively large, promiscuous ligand-binding pocket that alters receptor conformation to assemble active transcriptional machinery (Nolte et al., 1998). Distinct from the hormonal up-regulation of PPARγ and its effects upon its downstream targets, bulk mRNA (Fig. 1) and protein (Fig. 2) levels of pro-adipogenic coregulators SRC-2 and SRC-3 remain quite constant during the first 96 h of differentiation independent of natural (15dPGJ2) and synthetic (rosiglitazone) ligands (Fig. S2). Further examination of the cell–cell correlations between SRC-2, SRC-3, and PPARγ revealed that (a) PPARγhi/lipidhi cells also exhibited the highest SRC levels and a correlation between PPARγ/SRC, whereas (b) all other PPARγ/lipid populations showed little or no correlation between PPARγ and SRC (Table I). In addition to correlations between SRCs, PPARγ, and lipids, we have also shown a ligand-independent direct (FRET) interaction between SRC-2/PPARγ and SRC-3/PPARγ. Recent data suggests that the N-terminal A/B domain of PPARγ can act as a docking site for coregulators, in the absence of a ligand, to maintain a basal level of constitutive transcriptional activity, but it can also direct and enhance target gene specificity of the receptor (Gelman et al., 1999; Feige et al., 2005; Molnár et al., 2005; Tudor et al., 2007).
In contrast to the SRC-1/SRC-3 double knockout mouse (Wang et al., 2006), the SRC-2/SRC-3 and SRC-1/SRC-2 mutant mice are lethal (Mark et al., 2004; Xu et al., 2009), making our experiments the first to analyze compensation between SRC-2 and SRC-3 during human adipogenesis. Although overexpression suggested that SRC-1, SRC-2, and SRC-3 modestly increase adipogenesis, siRNA established that the endogenous levels of SRC-2 and SRC-3 are essential for differentiation. To determine if a functional overlap exists specifically between SRC-2 and SRC-3 during differentiation of cultured human adipocytes, we used a dual siRNA approach. Strikingly, we found that single or double knockdown of SRC-2 and SRC-3 inhibited adipogenesis to the same extent for both natural (15dPGJ2) and synthetic PPARγ ligands with comparable PPARγ induction. Consistent with pro-adipogenic SRCs being dominant in adipocyte development (Wang et al., 2006), SRC-1 siRNA had no effect on differentiation. This result suggests that SRC-2 and SRC-3 are fundamental components of the basal, preligand adipogenic machinery driven by PPARγ. The data at both the phenotypic (Fig. 6, A–G) and gene regulatory level (Fig. 6 H) did not exhibit any apparent compensation by the nontargeted SRCs in single siRNA treatments. In support of this finding, it has been proposed that SRC-2 and SRC-3 preferentially pair and interact to promote gene transcription (Zhang et al., 2004).
Upon single or double SRC-2/SRC-3 siRNA treatment, PPARγ protein levels were unchanged; interestingly, there was an enrichment of cells in a PPARγhi/lipidlo state, with decreases in the PPARγhi/lipidhi and PPARγlo/lipidhi proportions (Fig. 7 A). Coincident with decreased expression of PPARγ downstream genes, the increased PPARγhi/lipidlo population reflected an increase in the amount of phospho-PPARγ S114 (Fig. 7 D). When the levels of coactivator are reduced, the signal transduction environment elevates phospho-PPARγ S112/S114 status, lowers ligand (thiazolidinedione/eicosanoid) affinity, and increases interactions with the corepressors SMRT (Shao et al., 1998) or PER2 (Grimaldi et al., 2010) to reduce transcriptional activity (Lavinsky et al., 1998). Collectively, these results imply that SRC-2 and SRC-3, together, collaborate to promote adipocyte differentiation through potential multimerization (McKenna et al., 1998) and/or dimerization via protein dimerization/interaction domains (Lodrini et al., 2008). To add to this model, we combined quantitative analysis of SRC/PPARγ/lipid kinetics, correlations, FRET data, and loss-of-function experiments at single cell resolution. These important single cell characteristics suggest a novel mechanism of action. Specifically, permissive, homeostatic levels of SRC-2 and SRC-3 can interact with the PPARγ in the absence of MIX to regulate PPARγ heterogeneity, reduce inhibitory PPARγ phosphorylation, and promote adipogenesis.
Our quantitative, cell-by-cell approach has identified a unique interplay between SRC-2, SRC-3, and PPARγ that promotes adipogenesis. Small molecule inhibitors that block SRC recruitment or disrupt the predifferentiation complex between SRCs and PPARγ might maintain the positive (insulin sensitization) while reducing negative (weight gain) effects of thiazolidinediones (Reginato et al., 1998; Rocchi et al., 2001; Michalik et al., 2006). Collectively, the data presented here have implications for targeting the PPARγ–SRC interaction surface as strategies for new therapeutics to prevent the onset of obesity associated with the treatment of type 2 diabetes.
Materials and methods
Primary cell culture and differentiation
Cryopreserved, subcutaneous primary human preadipocytes from normal female donors with a mean body mass index of 27.51 were provided by Zen-Bio Inc. Cells were maintained at 5% CO2/37°C in DME/F12 (Mediatech, Inc.) with 10% FBS (Gemini Bio-Products), 100 U/ml penicillin, and 100 µg/ml streptomycin (growth media). Medium was replaced during routine maintenance every 2 d. Cells were received at passage 2, and experiments were performed before cells reached passage 10. Experiments were performed using pooled human preadipocytes from five individual female donors (Lot SL0033).
Unless otherwise indicated, all components were purchased from Sigma-Aldrich. After seeding to the appropriate experimental format (coverslips, 96- or 384-well plate format), cells were differentiated using growth media supplemented with 100 nM human insulin, 0.250 mM IBMX, 500 nM dexamethasone, and either rosiglitazone (BRL49653; Cayman Chemical Company) or 15dPGJ2 (Cayman Chemical Company). Unless otherwise indicated, differentiation was performed with IBMX, dexamethasone, human insulin, and 3 µM rosiglitazone.
Antibodies
The following antibodies were purchased from commercial sources and used for immunofluorescence: mouse monoclonal AIB1/SRC-3 (BD), mouse monoclonal TIF2/SRC-2 (BD), rabbit polyclonal phospho-PPAR S112/S114 (Abcam), rabbit monoclonal PPARγ (Cell Signaling Technology), mouse monoclonal PPARγ (clone E-8; Santa Cruz Biotechnology, Inc.), SRC-1 (BD), and mouse monoclonal FLAG-M2 (Sigma-Aldrich). Rabbit polyclonal antibody to SRC-3 was provided by B. O’Malley (Baylor College of Medicine, Houston, TX).
Immunofluorescence
For fluorescence detection of antibodies and neutral lipid content in multiwell plates, the following protocol was performed on the BioMek NX (Beckman Coulter). The well plate systems used were: 96-well and 384-well (Sensoplate Plus; Greiner). Aspirations and plate washes were performed with an ELx405 (BioTek). After differentiation, media was aspirated, and 4% paraformaldehyde (ultrapure; Electron Microscopy Sciences) in PBS was immediately added for 30 min at room temperature. Plates were then quenched with 100 mM ammonium chloride. After quenching, plates were washed three times with TBS. Fixed adipocytes were permeabilized with 0.1% Triton X-100 in TBS for 10 min and washed three times with TBS. A 2% BSA in TBS/0.01% saponin (antibody diluent) blocking solution was added for 30 min at room temperature followed by three TBS washes. Antibodies were then diluted at a 1:200 concentration in antibody diluent and incubated overnight at 4°C. Subsequently, plates were washed with TBS and incubated with secondary antibodies for 1 h at room temperature. Alexa Fluor 647–-conjugated anti–mouse and Alexa Fluor 568–conjugated anti–rabbit secondary antibodies (Invitrogen) were used. Cells were again washed three times and incubated with 1 µg/ml CellMask blue (CMBl; Invitrogen), 1:1,000 LipidTOX green (Invitrogen), and 10 µg/ml DAPI in PBS for 45 min at room temperature. Dyes were then aspirated and PBS/0.01% azide was added. Plates were then sealed and imaged immediately.
seFRET imaging
CFP/YFP FRET experiments were performed using PPARγ2-ECFP (provided by F. Schaufele, University of California, San Francisco, CA), EYFP-SRC-2 (R. Michalides, Netherlands Cancer Institute, Amsterdam, Netherlands) and EYFP-SRC-3 (Amazit et al., 2007) expressed in HeLa cells grown on standard 12-mm glass coverslips. Constructs were cotransfected using Lipofectamine 2000 (Invitrogen). Media was removed and replaced with fresh DME/F12 with 5% FBS 24 h after transfection. After a further 24 h, cells were treated for 2 h with either DMSO or differentiation cocktail (IBMX, human insulin, dexamethasone, and rosiglitazone), both prepared in growth media (DME/F12, 5% FBS). Treatment was followed with these steps: fixation 4% PFA (30 min), quench 100 mM NH4Cl (10 min), and mount with SlowFade Gold (Invitrogen). After fixation, cells were washed with PBS++ three times, while all other wash steps were performed with Pipes/Hepes/EGTA/MgCl2 (PEM) buffer, prepared at a final pH of 6.8.
FRET imaging was performed as described previously (Trinkle-Mulcahy et al., 2001; Chusainow et al., 2006) with the DeltaVision Core Image Restoration Microscope (Applied Precision). Z stacks were imaged at 0.2 µm separation and a frame size of 1,024 × 1,024 pixels at 1 × 1 binning with a microscope (IX71; Olympus) using a 60×, 1.42 NA Plan Achromat objective (Olympus) and a charge-coupled device camera (CoolSnap HQ2; Photometrics). Filter sets were as follows, with a dichroic to split CFP and YFP: excitation 430 nm/emission 470 nm (CFP), excitation 500 nm/emission 535 nm (YFP), and excitation 430 nm/emission 535 nm (FRET). Z series stacks were deconvolved with the DeltaVision constrained iterative algorithm. After deconvolution (softWoRx; Applied Precision), FRET calculations were performed using the Applied Precision FRET user interface. FRET measurements on individual nuclei were acquired on maximum intensity projections of the derived FRET image. Spectral bleed-through was corrected for by acquiring specimens containing only CFP-PPARγ2 and YFP-p160. Standard values for α and β coefficients were 0.6 (CFP) and 0.12 (YFP) acquired from single donor/acceptor plasmid expression experiments.
siRNA transfection
siRNA to SRC-2 and SRC-3 oligomers was provided by E. Lader (QIAGEN, Germantown, MD). Before transfection, optical quality 384-well plates (SensoPlate Plus; Greiner) were coated with 20 µl of FBS (Gemini Bio-Products) overnight at 37°C. Cells were reverse-transfected with siRNA or mismatch control at a final concentration of 10 nM using HiPerFect transfection reagent (QIAGEN). siRNA and transfection reagents were mixed in OptiMEM I reduced serum media (Invitrogen) and allowed to complex at room temperature for 20 min. Preadipocytes, diluted to a final density of 5,000 cells/well, were then added to the HiPerFect–siRNA complexes followed by immediate seeding to plates. The total volume of cells and transfection reagent was 30 µl/well. Final volume of each condition upon seeding to the 20 µl FBS coat was 50 µl/well. After reverse transfection, cells were incubated for 48 h at 5% CO2/37°C before induction of differentiation for up to 96 h.
Production of lentiviral particles
SRC-1, SRC-2, and SRC-3 cDNAs were cloned into the lentiviral expression vector pCDH–CMV-MCS-EF1-Puro (System Biosciences) by XbaI–NheI digestion. Pseudolentiviruses were produced in 293TN cells by cotransfecting lentiviral expression constructs and the pPACK packaging plasmid mix (System Biosciences). Pseudoviral particles were harvested 48 h after transfection and were concentrated using a PEG-it virus precipitation solution kit (System Biosciences).
RNA extraction and qPCR analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To measure the relative mRNA levels of SRC-2, SRC-3, PPARγ, and adipocentric genes (ADFP, SREBP1c, FASN, and C/EBPα), quantitative real-time RT-PCR was performed using the Taqman RT-PCR one-step master mix in conjunction with an ABI 7500 real-time PCR system (Applied Biosystems). Each sample was tested in duplicate in three independent experiments. β-actin was used as the invariant control. For C/EBPα, relative mRNA was evaluated using the TaqMan Gene Expression Assay (Assay ID Hs00269972_s1). The following primer and probe sets were used to detect SREBP1c, FASN, SRC-2, SRC-3, PPARγ2, and ADFP. SREBP1c: 5′-TGCGCAAGACAGCAGATTTA-3′ (forward), 5′-CGCTCCTCCATCAATGACA-3′ (reverse), and Roche Universal ProbeLibrary probe No. 77 (probe); FASN: 5′-CGGAGTGAATCTGGGTTGAT-3′ (forward), 5′-CAGGCACACACGATGGAC-3′ (reverse), and Roche Universal Probe Library probe No. 11 (probe); SRC-2: 5′-GGACCTGGTAAGAAGGTGTATTCAG-3′ (forward), 5′-TGCCTCTTAGCATAGGACACAGA-3′ (reverse), and 5′-TCCATGCGCAGCATGAAGGAGA-3′ (probe); SRC-3: 5′-TTCAGGAAAGGTTGTCAATATAGATACA-3′ (forward), 5′-AATACACCTTCGGATTATATCTTCAAA-3′ (reverse), and 5′-TTCACTGAGATCCTCCATGAGGCCTG-3′ (probe); ADFP: 5′-GTGACTGGCAGTGTGGAGAAG-3′ (forward), 5′-TCCGACTCCCCAAGACTGT-3′ (reverse), and 5′-CCAAGTCTGTGGTCAGTGGCAGCA-3′ (probe); PPARγ2: 5′-ATGCTGGCCTCCTTGATGA-3′ (forward), 5′-GCTTTCGCAGGCTCTTTAGAA-3′ (reverse), and 5′-TCTCATATCCGAGGGCCAAGGCTTC-3′ (probe); SRC-1: 5′-AACCCAGCAGGTGCAACAG-3′ (forward), 5′-GTCCCCGCCTACCAGATTC-3′ (reverse); and 5′-CAGGTGTTTGCTGACGTCCAGTGTACA-3′ (probe).
Imaging and microscopy
For high-speed image acquisition and subsequent analysis, cells were imaged using the Cell Laboratory IC-100 Image Cytometer (IC-100; Beckman Coulter) controlled by CytoShop 2.0 (Beckman Coulter). The IC-100 platform was equipped with an inverted microscope (Nikon; Eclipse TE2000-U) and a triple band filter set (Chroma 82000; Chroma Technology Corp.). A progressive scan camera (COHU) functioned as the focusing camera. The imaging camera (Hamamatsu Photonics) was set to capture 8-bit images at 2 × 2 binning (672 × 512 pixels, 0.684 × 0.684 µm2/pixel) with five images captured per field (DAPI, CMBl, LipidTOX, Alexa Fluor 568, and Alexa Fluor 647 secondary antibodies). All high-throughput microscopy experiments were performed with an S Fluor 20×/0.75 NA objective lens (Nikon). In general, 12–16 images were captured per well for image analysis. All imaging was performed at room temperature.
Image analysis
Images were analyzed using custom algorithms developed with the Pipeline Pilot (v7.5) software platform (Accelrys) in a similar workflow as described previously (Szafran et al., 2008, 2009) and summarized in the following steps. After background subtraction, nuclear and cell masks were generated using a combination of nonlinear least squares and watershed-from-markers image manipulations of the DAPI images. Specifically, a nonlinear least squares threshold was applied to a DAPI image to create a binary image. This image was subsequently eroded and distance transformed to generate a marker image identifying the approximate center for each nuclei. This marker image in combination with the original DAPI image was used in a watershed-from-markers operation to define the full nuclear mask for each nucleus. A final morphological open operation was used to generate the final nuclear masks. Then, cellular masks were created by applying watershed segmentation on the CellMask images using nuclei regions as seeds. To prevent cell body oversegmentation, cell regions were trimmed so their boundaries did not exceed an empirically determined maximal distance from the nucleus. All events with whole cell masks bordering the edge of the image were additionally eliminated from analysis. Both whole cell and nucleus segmentation generate regions under which single cell intensity features were extracted. Cell populations were filtered to discard events with cell aggregates, mitotic cells, apoptotic cells, cellular debris, or poor segmentation. Applied gates were based upon nuclear area, nuclear circularity, and cell size/nucleus ratio. In general, these filters removed 10% of the population of segmented cells. An additional image analysis platform, CyteSeer (Vala Sciences), was also used to support the Pipeline Pilot-driven algorithms. Measurements extracted using lipid droplet analysis intrinsic to CyteSeer software (McDonough et al., 2009) included lipid droplet count, total integrated intensity of the lipid mask on the lipid image, and lipid droplet area. Post-analysis measurements were exported to spreadsheet software (Excel; Microsoft) for further analysis.
Statistical analyses
Data presented were acquired from a minimum of two (qPCR) or three (HCA) independent experiments performed on multiple days, unless otherwise indicated. Analysis of variance (ANOVA) was first used to compare the effects of time or siRNA treatment. If significant differences were identified, then data were compared with Tukey’s HSD post-hoc tests. All tests were performed at the 95% confidence interval using JMP-IN 7 (SAS).
Online supplemental material
Fig. S1 shows the extension of the human adipogenesis assay to later time points and dose response experiments for BRL49653 and 15dPGJ2 with detection of SRC-1, SRC-2, SRC-3, PPARγ, and lipid content. Fig. S2 shows the effect of SRC-1 siRNA on human adipogenesis. Fig. S3 displays immunofluorescent detection of SRC-2 and SRC-3 under single or double knockdown conditions. Fig. S3 also redisplays mRNA data represented in Fig. 6 H. Fig. S4 shows the validation of subpopulation analyses using PPARγ siRNA. Fig. S5 describes SRC gain-of-function experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004026/DC1.
Acknowledgments
The authors thank Drs. N. McKenna and Z.D. Sharp for critically reviewing the manuscript; I.P. Uray for qPCR assay design; J.Y. Newberg, A.T. Szafran, M.G. Mancini, L. Vergara, and J. Broughman for technical resource support; and P. McDonough and J.H. Price (Vala Sciences) for longstanding support in automated cytometry. Benjamin Buehrer is an employee of Zen-Bio, Inc.
This work was funded by National Institutes of Health (NIH) grant 5R01DK055622, the Hankamer Foundation, and pilot grant and equipment support from the John S. Dunn Gulf Coast Consortium for Chemical Genomics (to M.A. Mancini). Additional funding was provided by NIH 1F32DK85979 (to S.M. Hartig), 5T32HD007165 (to B.W. O’Malley), 5K01DK081446 (to B. He), and 5R01CA090464 (to E. Chang). Imaging resources were supported by Specialized Cooperative Centers Program in Reproduction U54 HD-007495 (to B.W. O’Malley), P30 DK-56338 (to M.K. Estes), P30 CA-125123 (to C.K. Osborne), and the Dan L. Duncan Cancer Center of Baylor College of Medicine.
Footnotes
Abbreviations used in this paper:
- C/EBPα
- CAAT/enhancer binding protein-α
- CMBl
- CellMask blue
- CV
- coefficient of variation
- HCA
- high-content analysis
- HTM
- high throughput microscopy
- IBMX
- 3-isobutyl-1-methylxanthine
- PPARγ
- peroxisome proliferator activated receptor γ
- qPCR
- quantitative RT-PCR
- seFRET
- sensitized emission fluorescence resonance energy transfer
- SRC
- steroid receptor coactivator
References
- Adams M., Reginato M.J., Shao D.L., Lazar M.A., Chatterjee V.K. 1997. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J. Biol. Chem. 272:5128–5132 10.1074/jbc.272.43.27295 [DOI] [PubMed] [Google Scholar]
- Amazit L., Pasini L., Szafran A.T., Berno V., Wu R.C., Mielke M., Jones E.D., Mancini M.G., Hinojos C.A., O’Malley B.W., Mancini M.A. 2007. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol. Cell. Biol. 27:6913–6932 10.1128/MCB.01695-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick S.L., Kononen J., Walker R.L., Azorsa D.O., Tanner M.M., Guan X.Y., Sauter G., Kallioniemi O.P., Trent J.M., Meltzer P.S. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 277:965–968 10.1126/science.277.5328.965 [DOI] [PubMed] [Google Scholar]
- Camp H.S., Tafuri S.R. 1997. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J. Biol. Chem. 272:10811–10816 10.1074/jbc.272.16.10811 [DOI] [PubMed] [Google Scholar]
- Chusainow J., Ajuh P.M., Trinkle-Mulcahy L., Sleeman J.E., Ellenberg J., Lamond A.I. 2006. FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA. 11:1201–1214 10.1261/rna.7277705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A., Louet J.F., Lagouge M., Lerin C., Antal M.C., Meziane H., Schoonjans K., Puigserver P., O’Malley B.W., Auwerx J. 2008. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 105:17187–17192 10.1073/pnas.0808207105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S.R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4:263–273 10.1016/j.cmet.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Gelman L., Tudor C., Engelborghs Y., Wahli W., Desvergne B. 2005. Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J. Biol. Chem. 280:17880–17890 10.1074/jbc.M500786200 [DOI] [PubMed] [Google Scholar]
- Forman B.M., Tontonoz P., Chen J., Brun R.P., Spiegelman B.M., Evans R.M. 1995. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 83:803–812 10.1016/0092-8674(95)90193-0 [DOI] [PubMed] [Google Scholar]
- Gelman L., Zhou G., Fajas L., Raspé E., Fruchart J.C., Auwerx J. 1999. p300 interacts with the N- and C-terminal of PPARgamma2 in a ligand-independent and -dependent manner. J. Biol. Chem. 274:7681–7688 10.1074/jbc.274.12.7681 [DOI] [PubMed] [Google Scholar]
- Grimaldi B., Bellet M.M., Katada S., Astarita G., Hirayama J., Amin R.H., Granneman J.G., Piomelli D., Leff T., Sassone-Corsi P. 2010. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 12:509–520 10.1016/j.cmet.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Kohli K., Garabedian M.J., Stallcup M.R. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17:2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E.D., Kim J.B., Sarraf P., Spiegelman B.M. 1996. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 274:2100–2103 10.1126/science.274.5295.2100 [DOI] [PubMed] [Google Scholar]
- Kliewer S.A., Lenhard J.M., Willson T.M., Patel I., Morris D.C., Lehmann J.M. 1995. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell. 83:813–819 10.1016/0092-8674(95)90194-9 [DOI] [PubMed] [Google Scholar]
- Kodera Y., Takeyama K., Murayama A., Suzawa M., Masuhiro Y., Kato S. 2000. Ligand type-specific interactions of peroxisome proliferator-activated receptor gamma with transcriptional coactivators. J. Biol. Chem. 275:33201–33204 10.1074/jbc.C000517200 [DOI] [PubMed] [Google Scholar]
- Lavinsky R.M., Jepsen K., Heinzel T., Torchia J., Mullen T.M., Schiff R., Del-Rio A.L., Ricote M., Ngo S., Gemsch J., et al. 1998. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. USA. 95:2920–2925 10.1073/pnas.95.6.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.T., Cheng J.X. 2009. Single-cell profiling reveals the origin of phenotypic variability in adipogenesis. PLoS One. 4:e5189 10.1371/journal.pone.0005189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodrini M., Münz T., Coudevylle N., Griesinger C., Becker S., Pfitzner E. 2008. P160/SRC/NCoA coactivators form complexes via specific interaction of their PAS-B domain with the CID/AD1 domain. Nucleic Acids Res. 36:1847–1860 10.1093/nar/gkn029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L.H., Lin H.J., Singh D.K., Lyons K.M., Altschuler S.J., Wu L.F. 2009. Heterogeneity in the physiological states and pharmacological responses of differentiating 3T3-L1 preadipocytes. J. Cell Biol. 187:375–384 10.1083/jcb.200904140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet J.F., O’Malley B.W. 2007. Coregulators in adipogenesis: what could we learn from the SRC (p160) coactivator family? Cell Cycle. 6:2448–2452 10.4161/cc.6.20.4777 [DOI] [PubMed] [Google Scholar]
- Louet J.F., Coste A., Amazit L., Tannour-Louet M., Wu R.C., Tsai S.Y., Tsai M.J., Auwerx J., O’Malley B.W. 2006. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc. Natl. Acad. Sci. USA. 103:17868–17873 10.1073/pnas.0608711103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M., Yoshida-Komiya H., Gehin M., Liao L., Tsai M.J., O’Malley B.W., Chambon P., Xu J.M. 2004. Partially redundant functions of SRC-1 and TIF2 in postnatal survival and male reproduction. Proc. Natl. Acad. Sci. USA. 101:4453–4458 10.1073/pnas.0400234101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough P.M., Agustin R.M., Ingermanson R.S., Loy P.A., Buehrer B.M., Nicoll J.B., Prigozhina N.L., Mikic I., Price J.H. 2009. Quantification of lipid droplets and associated proteins in cellular models of obesity via high-content/high-throughput microscopy and automated image analysis. Assay Drug Dev. Technol. 7:440–460 10.1089/adt.2009.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney E.M., Rose D.W., Flynn S.E., Westin S., Mullen T.M., Krones A., Inostroza J., Torchia J., Nolte R.T., Assa-Munt N., et al. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357–3368 10.1101/gad.12.21.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N.J., Nawaz Z., Tsai S.Y., Tsai M.J., O’Malley B.W. 1998. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA. 95:11697–11702 10.1073/pnas.95.20.11697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik L., Auwerx J., Berger J.P., Chatterjee V.K., Glass C.K., Gonzalez F.J., Grimaldi P.A., Kadowaki T., Lazar M.A., O’Rahilly S., et al. 2006. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol. Rev. 58:726–741 10.1124/pr.58.4.5 [DOI] [PubMed] [Google Scholar]
- Molnár F., Matilainen M., Carlberg C. 2005. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. J. Biol. Chem. 280:26543–26556 10.1074/jbc.M502463200 [DOI] [PubMed] [Google Scholar]
- Nolte R.T., Wisely G.B., Westin S., Cobb J.E., Lambert M.H., Kurokawa R., Rosenfeld M.G., Willson T.M., Glass C.K., Milburn M.V. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 395:137–143 10.1038/25931 [DOI] [PubMed] [Google Scholar]
- Oñate S.A., Tsai S.Y., Tsai M.J., O’Malley B.W. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 270:1354–1357 10.1126/science.270.5240.1354 [DOI] [PubMed] [Google Scholar]
- Picard F., Géhin M., Annicotte J.S., Rocchi S., Champy M.F., O’Malley B.W., Chambon P., Auwerx J. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 111:931–941 10.1016/S0092-8674(02)01169-8 [DOI] [PubMed] [Google Scholar]
- Rangwala S.M., Rhoades B., Shapiro J.S., Rich A.S., Kim J.K., Shulman G.I., Kaestner K.H., Lazar M.A. 2003. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev. Cell. 5:657–663 10.1016/S1534-5807(03)00274-0 [DOI] [PubMed] [Google Scholar]
- Reginato M.J., Bailey S.T., Krakow S.L., Minami C., Ishii S., Tanaka H., Lazar M.A. 1998. A potent antidiabetic thiazolidinedione with unique peroxisome proliferator-activated receptor gamma-activating properties. J. Biol. Chem. 273:32679–32684 10.1074/jbc.273.49.32679 [DOI] [PubMed] [Google Scholar]
- Rocchi S., Picard F., Vamecq J., Gelman L., Potier N., Zeyer D., Dubuquoy L., Bac P., Champy M.F., Plunket K.D., et al. 2001. A unique PPARgamma ligand with potent insulin-sensitizing yet weak adipogenic activity. Mol. Cell. 8:737–747 10.1016/S1097-2765(01)00353-7 [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M. 1999. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 4:611–617 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Hsu C.H., Wang X.Z., Sakai S., Freeman M.W., Gonzalez F.J., Spiegelman B.M. 2002. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 16:22–26 10.1101/gad.948702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D.L., Rangwala S.M., Bailey S.T., Krakow S.L., Reginato M.J., Lazar M.A. 1998. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 396:377–380 10.1038/24634 [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Mancini M.G., Patel K., Allegretto E.A., Smith C.L., Mancini M.A. 2000. Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol. Endocrinol. 14:518–534 10.1210/me.14.4.518 [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Mielke M., Mancini M.A. 2002. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 4:806–810 10.1038/ncb859 [DOI] [PubMed] [Google Scholar]
- Szafran A.T., Szwarc M., Marcelli M., Mancini M.A. 2008. Androgen receptor functional analyses by high throughput imaging: determination of ligand, cell cycle, and mutation-specific effects. PLoS One. 3:e3605 10.1371/journal.pone.0003605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran A.T., Hartig S.M., Sun H., Uray I.P., Szwarc M., Shen Y., Mediwala S.N., Bell J., McPhaul M.J., Mancini M.A., Marcelli M. 2009. Androgen receptor mutations associated with androgen insensitivity syndrome: a high content analysis approach leading to personalized medicine. PLoS One. 4:e8179 10.1371/journal.pone.0008179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Sleeman J.E., Lamond A.I. 2001. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114:4219–4228 [DOI] [PubMed] [Google Scholar]
- Tsai Y.S., Tsai P.J., Jiang M.J., Chou T.Y., Pendse A., Kim H.S., Maeda N. 2009. Decreased PPAR gamma expression compromises perigonadal-specific fat deposition and insulin sensitivity. Mol. Endocrinol. 23:1787–1798 10.1210/me.2009-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor C., Feige J.N., Pingali H., Lohray V.B., Wahli W., Desvergne B., Engelborghs Y., Gelman L. 2007. Association with coregulators is the major determinant governing peroxisome proliferator-activated receptor mobility in living cells. J. Biol. Chem. 282:4417–4426 10.1074/jbc.M608172200 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Qi C., Krones A., Woodring P., Zhu X.Y., Reddy J.K., Evans R.M., Rosenfeld M.G., Hunter T. 2006. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 3:111–122 10.1016/j.cmet.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Wu Z.D., Bucher N.L.R., Farmer S.R. 1996. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol. Cell. Biol. 16:4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.D., Rosen E.D., Brun R., Hauser S., Adelmant G., Troy A.E., McKeon C., Darlington G.J., Spiegelman B.M. 1999. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 3:151–158 10.1016/S1097-2765(00)80306-8 [DOI] [PubMed] [Google Scholar]
- Xu J., Wu R.C., O’Malley B.W. 2009. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer. 9:615–630 10.1038/nrc2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh W.C., Cao Z.D., Classon M., McKnight S.L. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168–181 10.1101/gad.9.2.168 [DOI] [PubMed] [Google Scholar]
- Zhang H., Yi X., Sun X.J., Yin N., Shi B., Wu H.J., Wang D., Wu G., Shang Y.F. 2004. Differential gene regulation by the SRC family of coactivators. Genes Dev. 18:1753–1765 10.1101/gad.1194704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Qiang L., Farmer S.R. 2006. Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J. Biol. Chem. 281:7960–7967 10.1074/jbc.M510682200 [DOI] [PubMed] [Google Scholar]