ORP5 works together with Niemann Pick C-1 to facilitate exit of cholesterol from endosomes and lysosomes.
Abstract
Oxysterol-binding protein (OSBP) and its related proteins (ORPs) constitute a large and evolutionarily conserved family of lipid-binding proteins that target organelle membranes to mediate sterol signaling and/or transport. Here we characterize ORP5, a tail-anchored ORP protein that localizes to the endoplasmic reticulum. Knocking down ORP5 causes cholesterol accumulation in late endosomes and lysosomes, which is reminiscent of the cholesterol trafficking defect in Niemann Pick C (NPC) fibroblasts. Cholesterol appears to accumulate in the limiting membranes of endosomal compartments in ORP5-depleted cells, whereas depletion of NPC1 or both ORP5 and NPC1 results in luminal accumulation of cholesterol. Moreover, trans-Golgi resident proteins mislocalize to endosomal compartments upon ORP5 depletion, which depends on a functional NPC1. Our results establish the first link between NPC1 and a cytoplasmic sterol carrier, and suggest that ORP5 may cooperate with NPC1 to mediate the exit of cholesterol from endosomes/lysosomes.
Introduction
Sterols are indispensable eukaryotic membrane components, and serve to modulate membrane rigidity, fluidity, and permeability (Maxfield and Tabas, 2005; Chang et al., 2006). Membrane sterols play key roles in many important cellular processes ranging from membrane trafficking to signal transduction. Abnormal distribution and/or metabolism of cholesterol can have serious cellular consequences that may lead to devastating human diseases such as atherosclerosis (Maxfield and Tabas, 2005). Therefore, mammalian cells have developed complex yet elegant mechanisms to maintain a constant level and proper distribution of cholesterol (Goldstein et al., 2006; Mesmin and Maxfield, 2009).
An important means for cells to acquire cholesterol is the receptor-mediated endocytosis of low-density lipoproteins (LDLs). The endocytic pathway sorts and delivers LDL from early endosomes to late endosomes/lysosomes (LEs/LYs) for the hydrolysis of cholesteryl esters, and the released free cholesterol exits LE/LY efficiently to reach the plasma membrane (PM) and/or the ER for structural and regulatory functions, respectively (Chang et al., 2006; Kristiana et al., 2008). The exit of LDL-derived cholesterol (LDL-C) from LE/LY has been under intensive investigation in recent years because of the Niemann Pick Type C (NPC) disease, an autosomal recessive and neurodegenerative disorder that is characterized by the accumulation of LDL-C in LE/LY of cultured NPC fibroblasts (Liscum et al., 1989). Approximately 95% of NPC cases are caused by mutations in the NPC1 gene (Carstea et al., 1997), which encodes an LE/LY membrane protein with 13 transmembrane domains (TMDs) and three large lumenal loops (Davies and Ioannou, 2000). Mutations in NPC2 are responsible for the rest of NPC cases, and the NPC2 protein is a soluble, cholesterol-binding protein that resides in the lysosomal lumen (Storch and Xu, 2009). Recently, the N-terminal lumenal domain of NPC1 has been shown to also bind cholesterol, but in an orientation that is opposite to NPC2 (Infante et al., 2008; Kwon et al., 2009). It has been proposed that NPC2 likely accepts and delivers LDL-C to the N-terminal domain of NPC1, which then inserts LDL-C directly into the lysosomal membrane for export (Kwon et al., 2009). Putative cytoplasmic cholesterol-binding proteins may be required to transport LDL-C from the LE/LY membranes to other membrane destinations for regulatory and structural functions (Kwon et al., 2009).
The endocytic pathway plays a critical role in cholesterol trafficking. Conversely, the level of cholesterol within endosomal compartments can also have a major impact on the sorting and transport of endosomal proteins at multiple steps (Gruenberg, 2003). In yeast, sterols have been demonstrated to regulate both the internalization step of endocytosis and a postinternalization step (Heese-Peck et al., 2002). In mammalian early endosomes, annexin II interacts with cholesterol to regulate the biogenesis of multivesicular transport intermediates destined for LEs (Mayran et al., 2003). The recycling rate of GPI (glycosylphosphatidylinositol)-anchored proteins through recycling endosomes can be greatly increased by reducing cellular cholesterol (Mayor et al., 1998). The role of cholesterol in the dynamics of LEs has been characterized in more detail because cholesterol can be trapped in LEs by genetic and pharmacological means. The motility of cholesterol-laden LEs is greatly reduced, which may be caused by the increased membrane association of Rab7 (Lebrand et al., 2002). Cholesterol accumulation in NPC cells also interferes with the retrograde transport from endosomes to the TGN, which delivers receptors, enzymes, and some bacterial toxins to the TGN. The cation-independent mannose-6 receptors (CI-MPR) localize to the TGN at steady-state but accumulate in NPC LEs, possibly because of increased membrane sequestration of Rab9 as a result of cholesterol accumulation (Kobayashi et al., 1999; Ganley and Pfeffer, 2006). These observations further highlight the need to understand the trafficking mechanisms of intracellular cholesterol.
Cholesterol transport can be performed by membrane vesicles and also by carrier proteins in a nonvesicular manner, although the identity of these bona fide sterol carriers remains unclear (Yang, 2006; Prinz, 2007; Mesmin and Maxfield, 2009). The oxysterol-binding protein (OSBP) and its related proteins (OSBP-related protein [ORP]), which constitute a large conserved protein family in eukaryotes (Ridgway et al., 1992; Yan and Olkkonen, 2008), have recently been characterized as putative sterol carriers. OSBP was first identified as a high-affinity cytosolic receptor for oxysterols, such as 25-hydroxycholesterol. Homologues to OSBP have subsequently been isolated in most eukaryotes, including 12 members in humans and 7 members in budding yeast (Beh et al., 2001; Lehto et al., 2001). These proteins all share a conserved ∼400 amino acid OSBP-related domain (ORD) found at the C terminus of OSBP, which has been shown to bind cholesterol and oxysterols (Wang et al., 2005b; Suchanek et al., 2007). In addition, OSBP and ORPs possess membrane-targeting domains/motifs, such as the pleckstrin homology (PH) domain and an FFAT motif (diphenylalanine in an acidic tract), allowing OSBP and ORPs to target different membranes (Wyles et al., 2002; Loewen et al., 2003). It has been suggested that sterol and membrane binding may encourage reciprocal changes in ORP conformation, facilitating a sterol transport cycle between donor and acceptor membranes (Im et al., 2005). Indeed, all yeast ORPs and mammalian ORP9 have been found to be able to transfer sterol between liposomes in vitro, and phosphoinositides (PIPs) also play a key role in this transfer (Raychaudhuri et al., 2006; Ngo and Ridgway, 2009). In addition, the sterol transport between PM and ER in yeast was severely compromised when all seven yeast ORPs were missing (Raychaudhuri et al., 2006). Despite these advances, the exact location and mode of action for many mammalian ORPs in intracellular cholesterol trafficking remains to be elucidated.
In this study, we examine the cellular function of a previously uncharacterized ORP, ORP5, and we show that siRNA-mediated knockdown of ORP5 impairs LDL-C transport to the ER and causes mislocalization of TGN proteins. Interestingly, cholesterol appears to accumulate on the limiting membranes of LE/LY, an effect that is dependent on a functional NPC1. Our results suggest that ORP5 may cooperate with NPC1 to deliver cholesterol from LE/LY to the ER.
Results
Interaction between yeast NPC1 and ORPs
Our laboratory previously demonstrated that the NPC1 orthologue in the budding yeast, Ncr1p, localized to the limiting membrane of the yeast vacuole through the vacuolar protein sorting pathway (Zhang et al., 2004). We then attempted to identify interacting protein partners of Ncr1p in the budding yeast Saccharomyces cerevisiae by immunoprecipitation followed by mass-spectrometric analysis. Among other proteins, we identified two members of yeast OSBP homologues, Osh6p and Osh7p (unpublished data). We confirmed the interaction between Ncr1p and Osh6/7p in vivo by using the split-ubiquitin yeast two-hybrid (sp-ubY2H) assay (Fig. S1, A and B; Johnsson and Varshavsky, 1994). Next, we investigated whether changing sterol levels in the vacuolar membrane affects the interaction between Ncr1p and Osh6p or Osh7p. We deleted the ARV1 gene from the wild-type reporter strain used for the sp-ubY2H system. arv1Δ mutants are temperature sensitive (Fig. S1 C) and may have depleted levels of sterol in vacuolar membranes (Kajiwara et al., 2008). We found that the interaction between Ncr1p and either Osh6p or Osh7p was nearly abolished in the arv1Δ background (Fig. S1, D and E), which suggested that Ncr1p loses its affinities for Osh6p and Osh7p when vacuolar membranes are depleted of sterols. We also conducted in vitro pull-down experiments: vacuoles were isolated and incubated with bacterially derived Osh6–GST. Ncr1p was efficiently pulled-down by Osh6p only in the presence of exogenous ergosterol ranging from 0.2 to 5 µM (Fig. 1 F). Ncr1p is highly homologous (∼40% identical, ∼75% similar) to human NPC1 (Malathi et al., 2004; Zhang et al., 2004) and has been shown to be able to complement the NPC1 defect in mammalian cells (Malathi et al., 2004). Among all human ORPs, ORP5 shows highest sequence identity with Osh6/7p (National Center for Biotechnology Information Basic Local Alignment Search Tool [BLAST] E value, 2 × 10−57; Altschul et al., 1990). Led by the interaction between Ncr1p and Osh6/7p in yeast, we next aim to characterize the mammalian ORP5 and investigate its role in NPC1-mediated cholesterol trafficking.
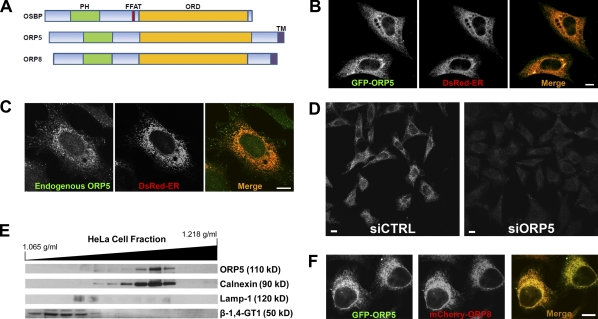
Figure 1.
ORP5 localizes to the endoplasmic reticulum. (A) Schematic diagram of the domain structures of human OSBP, ORP5, and ORP8. FFAT, diphenylalanine in an acidic tract. (B) HeLa cells grown on coverslips in medium A were cotransfected with EGFP-ORP5 and DsRed-ER for 24 h and processed for confocal fluorescence microscopy. Representative confocal images are shown. (C) HeLa cells grown on coverslips in medium A were cotransfected with DsRed-ER for 24 h and processed for immunofluorescence staining with a goat polyclonal antibody to ORP5. Specificity of staining was verified by using normal goat IgG as a negative control in the experiment. Representative confocal images are shown. (D) HeLa cells grown in medium A were transfected with siRNA against ORP5 (siORP5) or a universal control siCTRL for 72 h, followed by processing for immunofluorescence staining as in C. Representative confocal images are shown. (E) Subcellular distribution of ORP5. Subcellular fractionation of membranes from HeLa cells was performed as described in Materials and methods. The migration of ORP5, ER (calnexin), LY (Lamp-1), and Golgi (β-1,4-galactosyltransferase I, β-1,4GT1) markers with the approximate molecular weights is shown. (F) HeLa cells grown on coverslips in medium A were cotransfected with EGFP-ORP5 and mCherry-ORP8 for 24 h followed by processing for confocal fluorescence microscopy. Bars, 10 µm.
ORP5 localizes to the ER through its C-terminal TMD
Human ORP5 is ubiquitously expressed (unpublished data). Like OSBP and ORP8, ORP5 has a well-conserved PH domain near its N terminus and ORD near its C terminus. Similar to ORP8 but different from all other mammalian ORPs, ORP5 has a putative C terminal TMD (Fig. 1 A). To gain insights into subcellular localization of ORP5, we tagged EGFP to the N terminus of ORP5 and transfected HeLa cells with the pEGFP-ORP5 plasmid and pDsRed2-ER (an ER marker). Expression of EGFP-ORP5 showed an ER-like reticular fluorescence pattern (Fig. 1 B), which suggests that ORP5 associates with ER membranes. Endogenous ORP5, which was detected by immunofluorescence staining using a goat polyclonal antibody to N-terminal amino acids 2–14 of human ORP5, also colocalized with the ER marker (Fig. 1 C). The specificity of immunofluorescence staining was assessed in cells treated with siRNA against ORP5. As shown in Fig. 1 D, ORP5 staining was clearly seen in control cells but became much weaker in cells treated with siRNA targeting the endogenous ORP5. The ER localization of ORP5 was further confirmed by subcellular fractionation. In HeLa cells, ORP5 was predominantly detected in ER-enriched, calnexin-positive fractions (Fig. 1 E). ORP8 has been recently localized to the ER via its C-terminal TMD (Yan et al., 2008). We constructed a pmCherry-ORP8 plasmid and cotransfected it with pEGFP-ORP5 into HeLa cells. EGFP-ORP5 and mCherry-ORP8 almost completely colocalized in HeLa cells, as revealed by fluorescence microscopy (Fig. 1 F). We also created an ORP5 truncated construct lacking the predicted C-terminal TMD (ORP5ΔC). The truncated protein lost the reticular localization pattern (not depicted), similar to ORP8ΔC (Yan et al., 2008). These data demonstrate that ORP5, like ORP8, is also anchored in the ER membrane via its C-terminal TMD.
ORP5 transports sterols in vitro
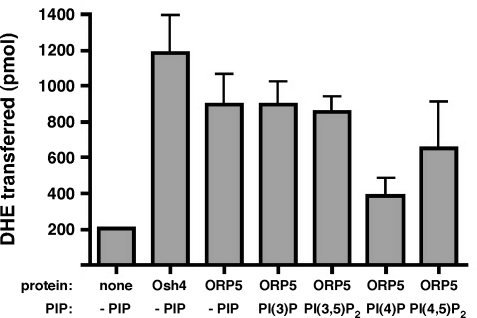
ORP5 has been shown to bind cholesterol and 25-hydroxycholesterol in a photo-cross-linking assay in live cells (Suchanek et al., 2007). However, the ORP5–cholesterol interaction is weak and it is unclear whether ORP5 can transfer sterols from the donor to the acceptor membrane directly. To test whether ORP5 can transport sterols in vitro, we purified the ORDs of ORP5 and yeast Osh4p, and performed a sterol transfer assay as described previously (Schulz et al., 2009). The assay was performed between dehydroergosterol (DHE)-incorporated donor liposomes and dansyl-phosphatidylethanolamine (dansyl-PE)–incorporated acceptor liposomes in the absence or presence of various PIPs with 40 pmol of purified proteins. The energy transfer between DHE and dansyl-PE in the acceptor liposomes reflects DHE transfer from donor to acceptor membranes. We found that purified ORP5ORD can transfer DHE in vitro at a similar rate to that of Osh4p (Fig. 2). The addition of PIPs, such as PI(3)P, PI(3,5)P2, or PI(4,5)P2, to the donor membranes did not seem to affect the transfer rate, except for PI(4)P, which appeared to partially inhibit DHE transfer by ORP5ORD (Fig. 2). These results suggest that ORP5 is able to transfer cholesterol in vitro and that the transfer may be affected by certain lipid compositions (e.g., PI4P) of donor membranes.
Figure 2.
ORP5 transfers sterols in vitro. DHE transfer by 40 pmol of purified ORP5LBD from donor to acceptor liposomes was determined fluorometrically as described in Materials and methods. The amount of DHE transferred in 1 h (30°C) was calculated and the transferred DHE in the absence or presence of different species of PIPs is shown. The results are expressed as means + SD (error bars) and are representative of two independent experiments.
Knockdown of ORP5, but not ORP8, impairs the transport of LDL-C to the ER
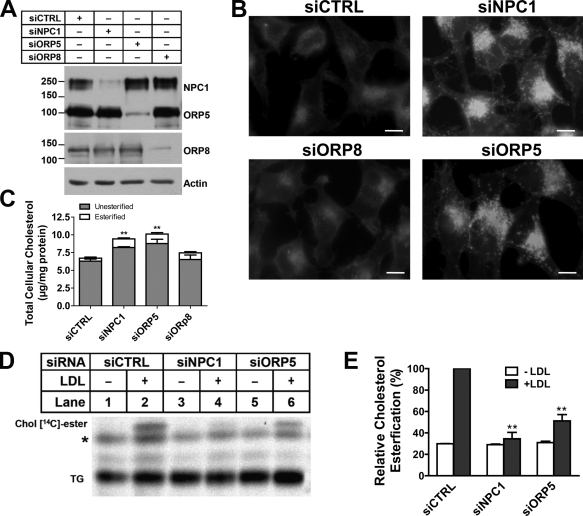
To assess the in vivo role of ORP5 in intracellular cholesterol transport, HeLa cells were used as a model because we were able to efficiently silence ORP5 and NPC1 expression using siRNA in these cells (Fig. 3 A). siRNA-transfected cells were incubated with LDL overnight, and filipin staining experiments were performed to detect LDL-C. NPC1 depletion produced a typical LDL-C staining pattern that is usually seen in NPC1-deficient cells (Fig. 3 B). Interestingly, cholesterol accumulation also occurred in ORP5 knockdown cells, but not in ORP8 knockdown cells (Fig. 3 B). A significant increase of intracellular free cholesterol in ORP5 and NPC1 knockdown cells was detected when cytoplasmic filipin fluorescence was quantified as described previously (Fig. S2 A; Ge et al., 2008). To biochemically determine the cholesterol content in siRNA-transfected cells, we measured the amounts of total cholesterol in cells with the same treatment. Consistent with the filipin staining observation, the total and free cholesterol content was increased in both NPC1 and ORP5 knockdown cells, but not in ORP8 knockdown cells (Fig. 3 C). This, together with the intracellular, endosome/LY-like filipin staining pattern, indicates that ORP5 may be involved in the intracellular transport of LDL-C.
Figure 3.
ORP5 knockdown causes endosomal cholesterol accumulation and inhibits LDL-stimulated cholesterol esterification. (A) HeLa cells grown in medium A were transfected with siNPC1, siORP5, siORP8, or a universal control siCTRL for 48 h, followed by transfection with pcDNA4-ORP5 or pmCherry-ORP8 for 24 h. Efficiency of knockdown was analyzed by immunoblotting using polyclonal anti-NPC1, anti-ORP5, or anti-DsRed antibodies. The molecular mass of the proteins is indicated in kilodaltons. (B) HeLa cells grown in medium A were transfected with the indicated siRNAs for 54 h. Cells then received medium D supplemented with 50 µg/ml LDL for 18 h followed by processing for filipin staining. Fluorescent images are representative of four independent experiments with similar results. Bars, 10 µm. (C) Cells that received the same treatments as in B were lysed by 0.1 M NaOH and processed for measuring cellular free cholesterol as described in Materials and methods. Values were normalized to total cell proteins and are relative to siCTRL-transfected cells. The results are expressed as means + SD (error bars). **, P < 0.01. (D) HeLa cells grown in medium A were transfected with the indicated siRNAs for 48 h, followed by incubation in medium D for 18 h. Cells were then chased with LDL (50 µg/ml) and [14C]-palmitate in medium D for 6 h. Lipids extracted from cell lysates were standardized for total cell proteins and separated by TLC. Cholesteryl [14C]-esters were revealed by phosphorimaging. A representative phosphorimage of three experiments with similar results is shown. TG, triacylglycerols; *, unknown nonspecific lipids. (E) Quantification of cholesteryl [14C]-esters formed in D by densitometry. Values of siCTRL + LDL were arbitrarily set as 100, against which experimental data were normalized. The results are expressed as means + SD (error bars) and are representative of three independent experiments. Significant differences were show between siCTRL and siNPC1 or siORP5 in the presence of LDL. **, P < 0.01.
To further investigate the effect of ORP5 knockdown on LDL-C transport to the ER, we performed a cholesterol esterification assay, which is a standard measurement of cholesterol transport to the ER, where Acyl-CoA:cholesterol acyltransferase (ACAT) is located (Kristiana et al., 2008). Cells were starved of cholesterol and treated with radio-labeled fatty acids together with LDL. Lipids extracted from cells were separated by TLC, and the formation of radio-labeled cholesteryl esters catalyzed by ACAT was visualized by phosphorimaging (Fig. 3 D). We then quantified relative cholesterol esterification by densitometry and normalized the values to those of the cells transfected with control siRNAs (Fig. 3 E). As expected, NPC1 knockdown blocked LDL-C transport to the ER and almost completely abolished cholesterol esterification activity in HeLa cells (Fig. 3 D, lane 4; and Fig. 3 E). Consistent with the filipin staining results, ORP5 knockdown in HeLa cells reduced the rate of cholesterol esterification by nearly 50% (Fig. 3 D, lane 6; and Fig. 3 E).
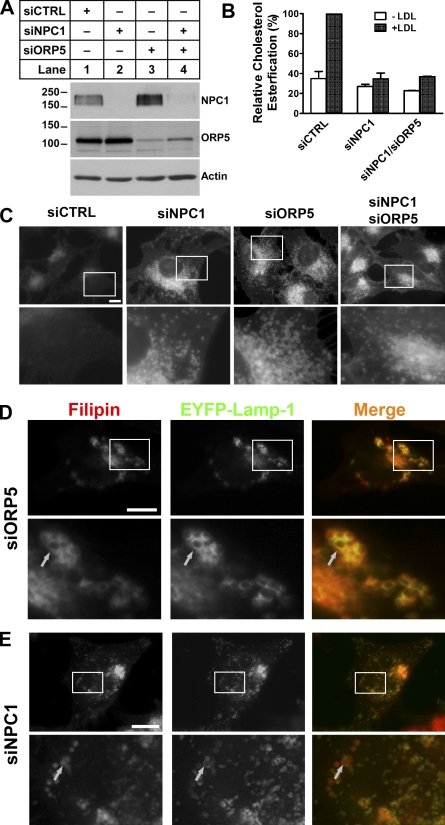
Cholesterol accumulates on the limiting membrane of LE/LY in ORP5 knockdown cells
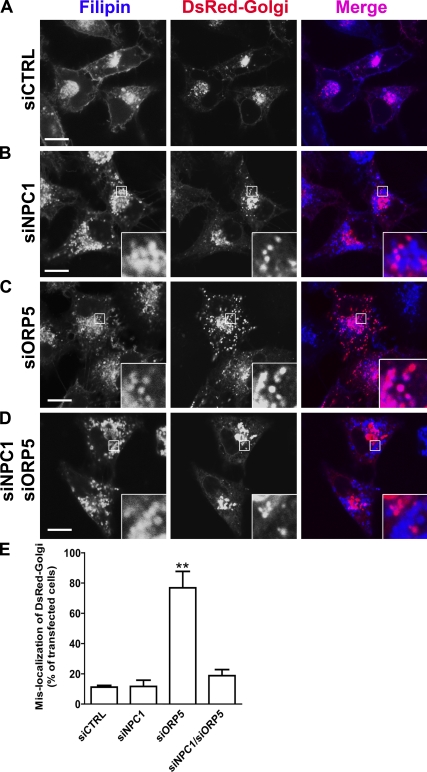
Given the phenotypic similarity between NPC1 and ORP5 knockdown cells, we next investigated the possible functional interactions between ORP5 and NPC1. We were able to knock down ORP5 and NPC1 expression simultaneously (Fig. 4 A, lane 4). The rate of cholesterol esterification was decreased similarly between NPC1 single and NPC1/ORP5 double knockdown cells (Fig. 4 B). A closer examination of the filipin staining pattern of ORP5 knockdown cells revealed that cholesterol appears to accumulate in large, ring-like punctate structures, and that cholesterol is concentrated on the limiting membrane of these compartments (Fig. 4 C, siORP5, bottom; and Fig. S5 B). Colocalization studies showed that most of these compartments were Lamp1-positive LE/LY, and that cholesterol overlapped with Lamp1, a limiting membrane protein of LE/LY (Fig. 4 D). In contrast, cholesterol appeared to fill the whole LE/LY compartment in NPC1 knockdown cells (Fig. 4, C and E). Interestingly, the pattern of cholesterol distribution in NPC1/ORP5 double knockdown cells was identical to that of NPC1, but not ORP5 single knockdown cells (Fig. 4 C). Together, these observations suggest that ORP5 may cooperate with NPC1 in facilitating the egress of LDL-C from LE/LY, and that the ORP5 function depends on a functional NPC1.
Figure 4.
Cholesterol accumulates on the limiting membrane of LE/LY in ORP5 knockdown cells. (A) HeLa cells grown in medium A were transfected with siNPC1, siORP5, siNPC1, and siORP5, or a universal control siCTRL for 48 h, followed by transfection with pcDNA4-ORP5 for 24 h. Efficiency of knockdown was analyzed by immunoblotting using polyclonal anti-NPC1, anti-ORP5 antibodies. The molecular mass of the proteins is indicated in kilodaltons next to the gel blot. (B) HeLa cells grown in medium A were transfected with the indicated siRNAs for 48 h, followed by incubation in medium D for 18 h. Cells were then chased with 50 µg/ml LDL and [14C]-palmitate in medium D for 6 h. A cholesterol esterification assay was performed as described in Fig. 3 D, and relative cholesterol esterification was quantified as described in Fig. 3 E. The results are expressed as means + SD (error bars). (C) HeLa cells grown in medium A were transfected with indicated siRNAs for 54 h. Cells then received medium D supplemented with 50 µg/ml LDL for 18 h followed by processing for filipin staining. Fluorescent images are representative of four independent experiments with similar results. Bars, 10 µm. (D and E) HeLa cells grown in medium A were transfected with siORP5 (D) or siNPC1 (E) for 48 h, followed by transfection with pEYFP-Lamp1 for 6 h. Cells then received medium D supplemented with 50 µg/ml LDL for 18 h followed by processing for filipin staining and fluorescence microscopy. Enlarged views of the boxed regions are shown in the bottom panels. Arrows indicate the overlapping of accumulated cholesterol with EYFP-Lamp-1 in ORP5 knockdown cells but not in NPC1 knockdown cells. Bars, 10 µm.
Knockdown of ORP5 causes mislocalization of trans-Golgi proteins to endosomal compartments, and this effect depends on a functional NPC1
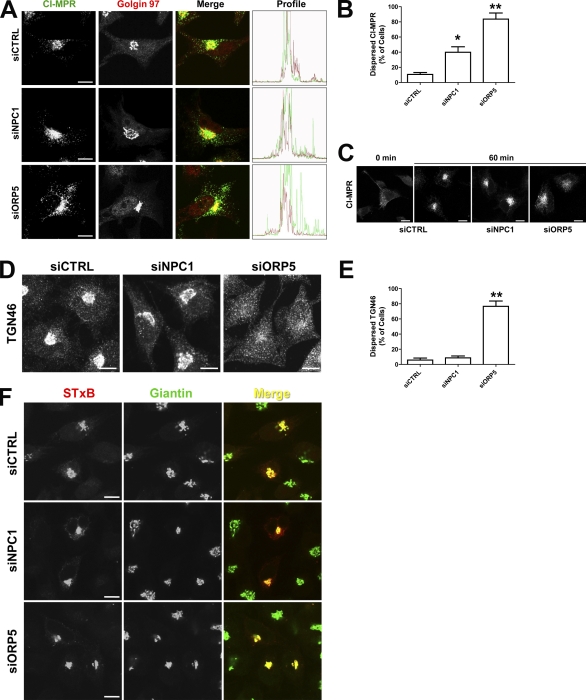
As the knockdown of another ORP, ORP9, disrupted the Golgi, which subsequently caused cholesterol accumulation in the endosomes (Ngo and Ridgway, 2009), the effect of ORP5 knockdown on cholesterol trafficking may be indirect. We therefore examined the morphology of the ER, lipid droplets, and Golgi in ORP5 knockdown cells. Lipid droplet staining was performed with LipidTOX, and fluorescence microscopy revealed that lipid droplet formation remained unaltered in ORP5 knockdown cells, and that accumulated cholesterol in these cells was not associated with lipid droplets (Fig. S2 B, insets). We also transfected cells with the ER marker DsRed2-ER to see if the ER structure was affected. No gross alteration of ER structure was observed at the resolution of light (fluorescence) microscopy in ORP5 knockdown cells compared with NPC1 knockdown or control cells (Fig. S2 C). We next transfected RNAi-treated cells with the fluorescently tagged Golgi marker DsRed-Monomer Golgi (DsRed-Golgi), which expresses Ds-Red fused with the N-terminal 81 amino acids of 1,4-galactosyltransferase (Fig. 5, A–D). Similar to control cells, NPC1 knockdown showed no significant impact on the localization of the DsRed-Golgi (Fig. 5, A and B). LDL-C that accumulated within LE/LY in NPC1 knockdown cells was clearly dissociated from the Golgi marker (Fig. 5 B, merged image inset). In contrast, the DsRed-Golgi displayed a much more dispersed, punctate localization in the majority of ORP5 knockdown cells, and cholesterol colocalized with these structures (Fig. 5 C), which indicates a mislocalization of DsRed-Golgi to endosomal compartments but not fragmentation of the Golgi. Indeed, careful examination of Golgi structure by various markers indicated an intact Golgi (Fig. S2, D and E). Therefore, DsRed-Golgi appeared to be trapped in cholesterol-filled LE/LY upon ORP5 knockdown. Most interestingly, in NPC1/ORP5 double knockdown cells, the localization of DsRed-Golgi appeared to be broadly similar to that observed in control or NPC1 knockdown cells (Fig. 5, D and E), where the accumulated LDL-C was clearly not associated with DsRed-Golgi (Fig. 5 D, merged image inset). These data suggest that the mislocalization of DsRed-Golgi to the LE/LY upon ORP5 knockdown depends on a functional NPC1. These effects were not observed in ORP8 knockdown cells (unpublished data), which further indicates a specific role of ORP5. Collectively, these results provide further evidence that ORP5 may be involved in endosomal LDL-C transport and may functionally interact with NPC1.
Figure 5.
The localization of DsRed-Golgi in cells depleted for ORP5, NPC1, or both. (A–D) HeLa cells grown in medium A were transfected with siCTRL (A), siNPC1 (B), siORP5 (C), or siNPC1 and siORP5 (D) for 48 h, followed by transfection with the Golgi marker DsRed-Golgi for 6 h. Cells were treated with 50 µg/ml LDL in medium D for 18 h followed by processing for filipin staining. Confocal fluorescence images are shown. Data are representative of three independent experiments with similar results. Enlarged views of the boxed regions are shown in the insets. Bars, 10 µm. (E) Quantification of DsRed-Golgi mislocalization in A–D. Values are the percentage of transfected cells with mislocalized DsRed-Golgi marker (means + SD [error bars], n > 80; **, P < 0.01).
The TGN plays a central role in protein sorting and receives proteins by retrograde transport from the endosomes (Bonifacino and Rojas, 2006). Our results indicate that ORP5 knockdown causes cholesterol accumulation in the endosomes, resulting in partial disruption of the retrograde transport pathway (Fig. 5). To investigate this further, we examined the localization of the CI-MPR, which is recycled between LE and TGN but is mostly present in the TGN at steady-state. HeLa cells depleted of NPC1 or ORP5 were double-stained for endogenous CI-MPR or the Golgi marker Golgin 97. CI-MPR, but not Golgin 97, showed a much more dispersed localization in cells depleted for ORP5 than normal cells or cells depleted for NPC1 (Fig. 6, A and B). Consistent with these observations, the green fluorescence profile (CI-MPR) was much broader in ORP5 knockdown cells than that in control or NPC1 knockdown cells (Fig. 6 A, right). We also performed a CI-MPR antibody internalization assay to monitor the endosomal trafficking of endogenous CI-MPR in HeLa cells. As shown in Fig. 6 C, depletion of ORP5 has much more severe effects on retrograde transport of CI-MPR than that of NPC1 depletion. We next examined the effects of ORP5 depletion on the trafficking of TGN46 and Shiga toxin subunit B (STxB), two other model cargos that cycle between the PM and TGN. Both TGN46 and STxB reach the TGN via early but not LEs, whereas STxB may also transit through the recycling endosomes (Lieu and Gleeson, 2010). In contrast to NPC1 depletion, ORP5 knockdown caused dramatic, dispersed distribution of TGN46 (Fig. 6, D and E). Neither NPC1 nor ORP5 knockdown, however, had any apparent effect on the trafficking of STxB to the TGN (Fig. 6 F). These results suggest that ORP5 knockdown affects trafficking events through early and late but probably not recycling endosomes. The trafficking defects could likely result from the accumulation of LDL-C in endosomal compartments in the absence of ORP5.
Figure 6.
Depletion of ORP5 impairs the transport of CI-MPR and TGN46, but not STxB. (A) HeLa cells were transfected with the indicated siRNAs for 72 h, followed by double-labeling with antibodies to CI-MPR and giantin. Confocal images are shown to indicate a more dispersed distribution of CI-MPR in cells treated with siORP5 rather than with siNPC1. Individual fluorescent profiles in merged images were quantified using the Olympus FluoView version 2.0 Viewer software and are shown on the right. (B) Quantification of cells with dispersed CI-MPR in A. Values are the percentage of cells with dispersed localization of CI-MPR (means + SD [error bars], n > 100; *, P < 0.05; **, P < 0.01). (C) HeLa cells were transfected with the indicated siRNA for 72 h, incubated on ice for 10 min, and chased with mouse monoclonal antibodies to CI-MPR for 30 min on ice. Antibody internalization was performed at 37°C for 60 min. Cells were then fixed by 4% paraformaldehyde, and internalized CI-MPR was detected using Alexa Fluor 488–conjugated goat anti–mouse antibodies. (D) HeLa cells were transfected with the indicated siRNAs for 72 h, followed by labeling with antibodies to TGN46. Confocal images are shown to indicate a dispersed distribution of TGN46 in cells treated with siORP5. (E) Quantification of cells with dispersed TGN46 in D. Values are the percentage of cells with dispersed localization of TGN46 (means + SD [error bars], n > 100; **, P < 0.01). Data are representative of three independent experiments with similar results. (F) HeLa cells, transfected with the indicated siRNA for 72 h in medium A, were incubated with Cy3-conjugated STxB in DME, 1% BSA, and 25 mM Hepes, pH 7.4, for 15 min at 37°C. Cells were washed in PBS and incubated for 1 h at 37°C in medium A before fixation by 4% paraformaldehyde, followed by labeling with antibodies to giantin. Bars, 10 µm.
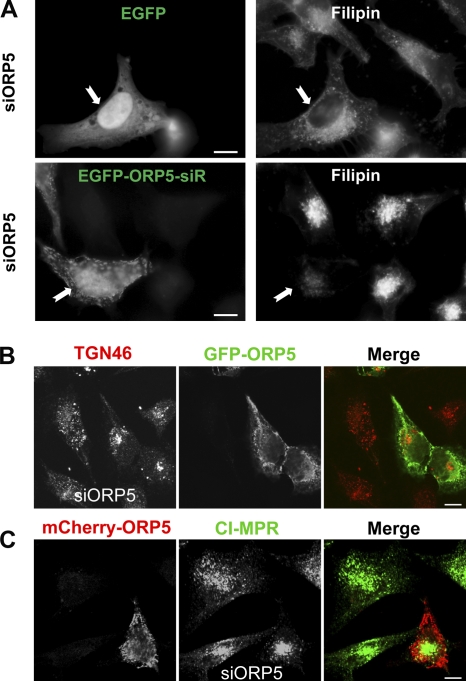
To ascertain the specificity of the effects observed upon depletion of ORP5, we performed rescue experiments in which the RNAi-treated cells were transfected with GFP-ORP5 under a cytomegalovirus promoter. Although GFP-ORP5 was depleted drastically, a significant amount of the protein still remained (Fig. S3 A). The mislocalization of DsRed-Golgi was almost completely rescued in cells expressing GFP-ORP5 (Fig. S3 B). Interestingly, expression of the ORD domain of ORP5, which was not targeted by the siRNA, also corrected the mislocalization of Ds-Red Golgi due to depletion of ORP5 (Fig. S3 B). In a separate approach, we generated RNAi-resistant ORP5 constructs by introducing silent mutations into the wild-type ORP5 sequence targeted by siRNA. Although the expression of EGFP did not affect intracellular cholesterol accumulation in ORP5-depleted cells, the expression of RNAi-resistant EGFP-ORP5 significantly alleviated accumulated cholesterol, as revealed by filipin staining (Fig. 7 A). Similarly, only those cells that expressed fluorescent protein–tagged, RNAi-resistant ORP5 restored normal localization of TGN46 and CI-MPR in cells transfected with siRNA against ORP5 (Fig. 7, B and C), which further confirmed the specificity of ORP5 knockdown effects.
Figure 7.
Rescue effects of ORP5 expression. (A) HeLa cells grown in medium A were transfected with siORP5 for 48 h, followed by transfection with EGFP alone or RNAi-resistant GFP-OPR5 (GFP-ORP5-siR) for 6 h. Cells then received medium D supplemented with 50 µg/ml LDL for 18 h followed by processing for filipin staining and fluorescence microscopy. Images are shown to indicate that cholesterol accumulation was significantly alleviated in GFP-ORP5-siR–expressing cells (71 ± 2%) but not in GFP-expressing cells (5 ± 3%). (B) HeLa cells grown in medium A were transfected with siORP5 for 48 h, followed by transfection with RNAi-resistant GFP-OPR5 for 24 h. Cells were fixed and labeled with antibodies to TGN46. Confocal images are shown to indicate that the dispersed distribution of TGN46 was rescued in cells expressing GFP-ORP5. (C) HeLa cells grown in medium A were transfected with siORP5 for 48 h, followed by transfection with RNAi-resistant mCherry-OPR5 for 24 h. Cells were fixed and labeled with antibodies to CI-MPR. Confocal images are shown to indicate that the dispersed distribution of CI-MPR was rescued in cells expressing mCherry-ORP5. Bars, 10 µm.
ORP5 coimmunoprecipitates with NPC1
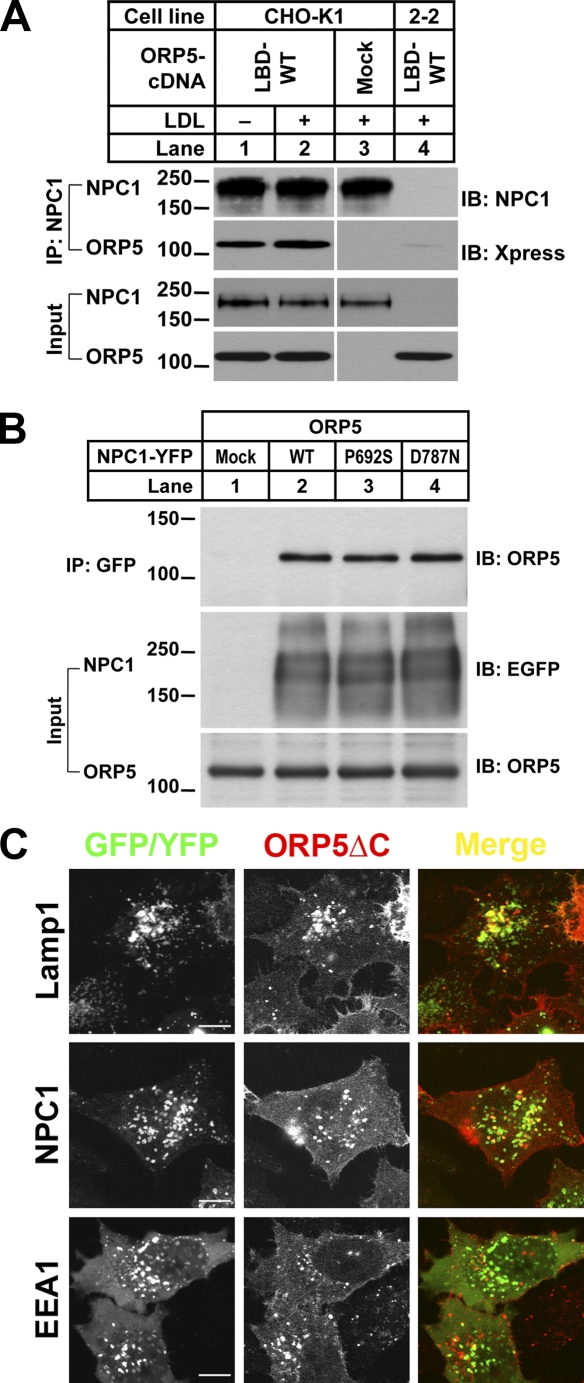
Given the functional connections between NPC1 and ORP5, it is likely that NPC1 and ORP5 may function in the same multiprotein complex that mediates the export of cholesterol from LE/LY. To test this possible physical link, we first performed coimmunoprecipitation (coIP) experiment in HEK293 cells. We cotransfected HEK293 cells with cDNAs encoding the C-terminal ligand-binding domain (amino acids 266–826) of ORP5 (ORP5LBD; Suchanek et al., 2007) and NPC1, or a negative control (GFP). We found that ORP5LBD could pull down YFP-tagged NPC1 but not GFP alone (Fig. S3 C, IP: Xpress, lane 2 vs. 3). Conversely, YFP-tagged NPC1, but not GFP, could pull down ORP5LBD (Fig. S3 C, IP: GFP, lane 2 vs. 3). We then attempted to address whether transiently expressed ORP5 can be immunoprecipitated by endogenous NPC1 in CHO cells. We chose CHO cells because we have in routine use an NPC1-deficient CHO cell line (the 2-2 cell line, a gift from L. Liscum). The mutant cell line lacks a mature NPC1 (Wojtanik and Liscum, 2003) and was therefore used as a negative control in our experiment. We transfected wild-type CHO-K1 and mutant 2-2 cells with cDNAs encoding ORP5LBD or mock vector as a control. Transiently expressed ORP5LBD was coimmunoprecipitated with NPC1 in CHO-K1 cells (Fig. 8 A, lanes 1 and 2) but not in mutant 2-2 cells (Fig. 8 A, lane 4).
Figure 8.
ORP5 coimmunoprecipitates with NPC1 and interacts with LEs. (A) CHO-K1 and 2–2 (Npc1-null) cells grown in medium B were transfected with the indicated cDNAs for 6 h. PBS-washed cells were incubated in medium D for 18 h and then chased with or without 50 µg/ml LDL for 4 h. Cell lysates were processed for immunoprecipitation with rabbit polyclonal anti-NPC1 antibodies. Input and immunoprecipitated complexes were blotted with monoclonal anti-Xpress or anti-NPC1 antibodies. Input represents ∼5% of the whole cell lysates. LBD, ligand binding domain. (B) HEK-293 cells grown in medium A were cotransfected with cDNAs encoding full-length ORP5 and pEGFP (Mock), or different NPC1 (WT, P692S, D787N). Cell lysates were processed for immunoprecipitation with rabbit polyclonal anti-GFP antibodies. Input and the immunoprecipitated complexes were blotted with goat polyclonal anti-ORP5 or anti-GFP antibodies. Input represents ∼2.5% of the whole cell lysates. The molecular mass of the proteins is indicated in kilodaltons next to the gel blots. (C) HeLa cells grown on coverslips in medium A were cotransfected with mCherry-ORP5ΔC and EYFP-Lamp1, EYFP-NPC1, or EGFP-FYVE (EEA1) for 48 h and processed for fluorescent microscopy. Representative confocal images are shown. Bars, 10 µm.
Next, we examined whether mutations in NPC1 could affect the ORP5–NPC1 interaction. A single amino acid change in the sterol-sensing domain of NPC1 (P692S) has been previously shown to abolish cholesterol binding (Ohgami et al., 2004) and represents a loss-of-function mutation of the protein (Millard et al., 2005). Thus, NPC1-P692S mutant was unable to complement the NPC phenotype in NPC1-deficient 2-2 cells, as indicated by filipin staining (not depicted). In contrast, NPC1-D787N, a gain-of-function mutant (Millard et al., 2005), could correct the accumulation of free cholesterol in 2-2 cells (unpublished data). We transfected HEK-293 cells with full-length ORP5 cDNA together with plasmids encoding YFP-tagged wild-type NPC1 or either of the two mutants. ORP5 was immunoprecipitated by a polyclonal anti-GFP antibody in the presence of wild-type NPC1, NPC1-P692S, or NPC1-D787N (Fig. 8 B, lanes 2–4). These data suggest that the two mutations within the sterol-sensing domain of NPC1 have no effects on the ORP5–NPC1 interaction.
How would an ER resident protein associate with an endosomal membrane protein? It is known that ER can form transient junctions with other organelles, such as mitochondria (Kornmann et al., 2009). Recently, it has been established that membrane contacts between endosomes and ER enable interaction between ER and endosomal membrane proteins (Eden et al., 2010). In addition, cholesterol and ORP1L regulate the formation of ER–LE contact sites where the ER protein VAP interacts in trans with the Rab7-RILP receptor to control the intracellular positioning of the LEs (Rocha et al., 2009). It is therefore likely that ORP5 may also transiently associate with LEs. ORP5 predominantly localizes to the ER at steady-state due to the tail anchor (Fig. 1), and we reason that if ORP5 does associate with LEs, ORP5 may localize to LEs upon removal of the tail anchor. Indeed, ORP5 without its tail anchor (ORP5-ΔC) was found in the cytoplasm and also in punctate structures (in ∼30% of transfected cells), which partially colocalized with Lamp1 and NPC1 but not EEA1, an early endosomal marker (Fig. 8 C).
Discussion
NPC1 and NPC2 are key proteins that govern the exit of LDL-C from LE/LY compartments. Recent elegant biochemical and crystallographic studies suggest that NPC2 delivers LDL-C to the N-terminal domain of NPC1, which then inserts the cholesterol into the lysosomal membrane for export (Kwon et al., 2009). How cholesterol moves from the lysosomal membrane to the PM and ER, and whether NPC1 or other proteins play a role in this process, remains unknown. As an attractive model, cytoplasmic sterol-binding proteins such as OSBP/ORPs could extract cholesterol molecules from LE/LY membranes and deliver them directly to the ER or the PM. However, little evidence exists to support such a hypothesis. LDL-C may also take a SNARE-dependent, vesicular pathway to reach the Golgi before arriving at the ER, as suggested by recent studies (Urano et al., 2008; Ngo and Ridgway, 2009). Here we show for the first time that a protein outside of LE/LY, ORP5, is required for efficient cholesterol egress from this compartment. We found that ORP resides in the ER and interacts with NPC1 in the LE/LY membrane, probably at regions of close contact between these two organelles. These findings suggest that ORP5 is required for efficient nonvesicular transfer of LDL-C from LE/LY to the ER.
OSBP and ORPs bind sterols and possess multiple membrane-targeting motifs, features well-suited for a role in moving cholesterol between membranes. It has recently been demonstrated that for efficient sterol transfer, the ORD domain present in all ORPs can simultaneously bind two closely apposed membranes, such as the membrane contact sites (Holthuis and Levine, 2005; Schulz et al., 2009). The ER is known to form contact sites with other organelles, including the mitochondria and LE (Kornmann et al., 2009; Eden et al., 2010). A close apposition between the ER, but not the Golgi, and NPC1 LE/LY has been observed repeatedly (Ko et al., 2001). ORP5 is an ER-localized, tail-anchored protein that has a PH domain near its N terminus. It is therefore likely that ORP5 may operate at the membrane contact sites between ER and LE to transfer LDL-C directly back to the ER. In support of this, we show that ORP5 can transport sterols in vitro, that cholesterol accumulates in the LE/LY of ORP5-silenced cells, and that ORP5 can make contact with Lamp1-positive LEs. Interestingly, ORP5 and NPC1 may exist in the same multiprotein complex through direct or indirect interactions, as ORP5 and NPC1 can be coimmunoprecipitated. This raises the possibility that NPC1 may not only mediate the incorporation of LDL-C to lysosomal membranes through its interaction with NPC2 in the lysosomal lumen, but may also facilitate the extraction of cholesterol by ORP5 in the cytoplasm.
An interesting observation is that cholesterol appears to accumulate on the limiting membranes of LE/LY upon ORP5 knockdown. This differs from NPC1 knockdown cells, as when NPC1 is absent, the carbohydrate glycocalyx lining the lumenal side of the lysosomal membrane would keep NPC2 from interacting with the limiting membrane, preventing cholesterol from transiting the glycocalyx and reaching the limiting membrane (Kwon et al., 2009). Therefore, cholesterol appears to occupy the entire lumenal space of LE/LY in NPC1-depleted cells. With a fully functional NPC1 and NPC2, LDL-C should be able to reach the limiting membrane, but it stays there if subsequent transport machinery is disrupted. ORP5 could be an essential component of such machinery, as cholesterol accumulates in the limiting membrane upon ORP5 depletion. The fact that knocking down both ORP5 and NPC1 exhibits a phenotype that is similar to NPC1 knockdown alone suggests that NPC1 may function upstream of ORP5: ORP5 depends on NPC1 for its function.
The functional relationship between NPC1 and ORP5 has been further supported by examining the localization of Golgi markers, including DsRed-Golgi, CI-MPR, and TGN46. These markers localize to the Golgi at steady-state in normal cells, but are mislocalized to LE/LY upon ORP5 depletion. Interestingly, the mislocalization of DsRed-Golgi is corrected when both ORP5 and NPC1 are knocked down simultaneously (Fig. 5). Why would ORP5 knockdown have a seemingly more severe phenotype than depleting both ORP5 and NPC1? One possibility is that when ORP5 is compromised, cholesterol accumulates on the limiting membrane, which may affect the retrograde transport to the Golgi that involves the recruitment of coat proteins (e.g., retromer) and vesicle budding or tubule formation from specific domains on the limiting membrane of LE (Bonifacino and Rojas, 2006). As is the case when NPC1 is knocked down, lumenal accumulation of cholesterol may not severely disturb the physical properties of the limiting membrane, and therefore is less disruptive to the retrograde transport pathway. Alternatively, though less likely, silencing of ORP5 may impair TGN function and disrupt the vesicular transport pathway between the LEs and the TGN, indirectly causing cholesterol accumulation in the LE/LY. In this regard, the depletion of TGN-specific SNAREs has been shown to reduce the transport of LDL-C by >50% (Urano et al., 2008). ORP5, like the yeast ORP Osh4p, may also exert its effect on the TGN by modulating the pool of PIPs needed for the proper function of Golgi membranes (Fairn et al., 2007; Schaaf et al., 2008).
In summary, our results provide the first characterization of a novel ORP, ORP5, and uncover an intimate relationship between ORP5 and NPC1. Our data support a role for ORP5 in the nonvesicular transport of LDL-C from LE/LY to the ER. We propose a simple model to speculate on how cholesterol can be delivered to the ER from endosomal compartments by ORP5 (Fig. S4). In response to an increased cholesterol level on the limiting membrane, LE/LY may form transient junctions/contact sites with the ER that allow efficient removal of endosomal cholesterol by ORP5 (Holthuis and Levine, 2005). NPC1, ORP5, or yet-to-be identified proteins may play a structural role in establishing such a junction. With direct or indirect assistance from NPC1, ORP5 may move cholesterol from LE/LY directly to the ER, as ORP5 can transport sterols in vitro (Fig. 2). Another interesting protein is the AAA ATPase VPS4/SKD1, which has been shown to regulate endosomal cholesterol trafficking and also interact with both yeast ORPs and NPC1 (Bishop and Woodman, 2000; Wang et al., 2005a; Ohsaki et al., 2006). Given the molecular function of AAA ATPases, it is likely that VPS4/SKD1 may dismantle such a junction once cholesterol delivery is completed (Yang, 2006). In this regard, VPS4/SKD1 interacts with ORPs and NPC1 much more efficiently upon sterol depletion (Wang et al., 2005a; Ohsaki et al., 2006). Although the exact nature of the ORP5–NPC1 interaction remains to be elucidated, this study identifies for the first time a cytoplasmic sterol carrier in NPC1-mediated cholesterol transport. Further characterization of the NPC1–ORP5 pathway should shed new light on the molecular function of NPC1 and also on the mechanisms of intracellular cholesterol transport.
Materials and methods
Materials
DME, DME/Ham’s Nutrient Mixture F-12 (DME/F12), newborn calf serum (NCS), FBS, penicillin–streptomycin, Dulbecco’s PBS, and LipidTOX were obtained from Invitrogen. Cholesterol (5-cholesten-3β-ol) was obtained from Steraloids. Protein A–Sepharose CL-4B, [1α,2α(n)-3H]-Cholesterol (41.0 Ci/mmol), and [1-14C]-Palmitate (specific activity, 51 mCi/mmol) were obtained from GE Healthcare. Lipoprotein-deficient serum was isolated from NCS by ultracentrifugation as described previously (Goldstein et al., 1983). LDL was subfractionated by density gradient ultracentrifugation from the plasma of healthy male volunteers (Brown et al., 1996). Compactin (mevastatin), mevalonate, filipin, saponin, and protease inhibitor cocktail were obtained from Sigma-Aldrich. Yeast extract, peptone, glucose, tryptone, and yeast nitrogen base were obtained from BD. Zymolase 20T was obtained from US Biological. For the in vitro cholesterol transfer assay, all lipids were obtained from Avanti Polar Lipids, Inc., except for DHE (Sigma-Aldrich), and the di-(16:0)-PI(3)P and di-(16:0)-PI(3,5)P2, which were obtained from Echelon Biosciences Inc.
Split-ubiquitin yeast two-hybrid assay
The bait and prey constructs for the split-ubiquitin yeast two-hybrid (sp-ubY2H) system (Dualsystems Biotech AG) was made by subcloning the cDNAs encoding full-length proteins (Ncr1p and Osh proteins, respectively) into the appropriate vectors. The sp-ubY2H screening was performed according to the manufacturer’s instructions. To construct the arv1Δ strain, a PCR-based homologous recombination method was used to delete the entire coding sequence of ARV1, replacing it with a kanamycin cassette. The resultant strain was confirmed by diagnostic PCR and temperature-sensitive phenotype.
Cell culture and transfection
CHO-K1 cells were a gift of M.S. Brown and J.L. Goldstein (University of Texas Southwestern Medical Center at Dallas, Dallas, TX). The 2-2 NPC1-defective cells, which contain an extra nucleotide at position 1335 of NPC1, resulting in a frameshift and early translation termination after the 450th amino acid (Wojtanik and Liscum, 2003), were provided by L. Liscum (Tufts University, Boston, MA). HEK-293 cells were a gift of D. James (Garvan Institute, Sydney, Australia). HeLa cells were purchased from the American Type Culture Collection (ATCC HTB-22). Monolayers of cells were maintained in specified medium supplemented with 10% serum, 100 U/ml penicillin, and 100 µg/ml streptomycin sulfate in 5% CO2 at 37°C. For HeLa and HEK-293 cells, DME and 10% FBS (medium A) were used; for CHO-K1 and Npc1-null 2-2 cells, the 1:1 mixture of DME/F12 and 10% NCS (medium B) was used. For cholesterol starvation treatments, PBS-rinsed cells were cultured overnight in medium containing 5% lipoprotein-deficient serum instead of NCS or FBS (medium C), plus 10 µM mevastatin and 50 µM mevalonate (medium D). Cell transfection was performed using Lipofectamine LTX and Plus Reagent (Invitrogen) according to manufacturer’s instructions. For each transfection, 1–2 µg/well of cDNA was used in 6-well plates, and 5–10 µg/dish of cDNA was used in 100-mm dishes.
Antibodies
Antibodies used were goat polyclonal to ORP5 (Abcam) and β-1,4-galactosyltransferase I (β-1,4-GT1; Santa Cruz Biotechnology, Inc.), rabbit polyclonal to the C-terminal region of human NPC1 and golgin 97 (Abcam), GFP (Invitrogen), syntaxin 5 (Subramaniam et al., 1997), calnexin (Cell Signaling Technology), DsRed (Takara Bio, Inc.), giantin (Covance), and TGN46 (AbD Serotec), mouse monoclonal to the Xpress tag (Invitrogen), actin (Abcam), GST, Lamp-1 and GFP (Santa Cruz Biotechnology, Inc.), and GM130 (BD). For immunoblotting, we obtained horseradish peroxidase–conjugated secondary antibodies from Jackson ImmunoResearch Laboratories, Inc. For immunofluorescence, we obtained Alexa Fluor–conjugated secondary antibodies from Invitrogen.
cDNA constructs
pcDNA4-ORP5LBD encoding the Xpress epitope-tagged C terminus of ORP5 ligand-binding domain was a gift from V. Olkkonen (National Public Health Institute, Helsinki, Finland). The full-length human ORP5 and ORP8 open reading frames were obtained from Thermo Fisher Scientific and cloned into pGEX-4T-1, pcDNA4/HisMaxC (Invitrogen), pEGFP-C1, or pmCherry-C1 vectors (Takara Bio Inc.) by standard subcloning procedures: pGST-ORP5ORD encodes N-terminal GST fusion of human ORP5 ORD (266–826), pcDNA4-ORP5 encodes N-terminal Xpress epitope-tagged ORP5, and pmCherry-ORP5 and pmCherry-ORP8 encode the corresponding proteins with fluorescence protein tagged to the N termini. DsRed2-ER and pDsRed-Monomer-Golgi (N-terminal 81 amino acids of β-1,4-GT1) were obtained from Takara Bio Inc. pEGFP-FYVE and pEYFP-Lamp1 were gifts from R. Teasdale (Institute of Molecular Bioscience, University of Queensland, Queensland, Australia). NPC1-FP constructs encoding mouse NPC1 with YFP fused to the C terminus of the protein were provided by M. Scott (Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA).
Subcellular fractionation
HeLa cells grown in two T175 flasks were harvested and homogenized using 18 strokes of the pestles of a Dounce homogenizer. Homogenates were centrifuged at 750 g for 5 min at 4°C. Postnuclear supernatants were collected, applied to a 10–40% Nycomed OptiPrep (Invitrogen) continuous gradient, and centrifuged at 48,000 g in an SW41 rotor for ∼18 h at 4°C. Fractions (0.8 ml) were collected and subjected to SDS-PAGE and immunoblotting analysis with antibodies to ORP5, calnexin (ER), lamp-1 (LE/LY), or β-1,4-GT1 (Golgi).
RNAi
siRNA transfection was performed in cells grown in medium A at ∼25% confluence, according to standard methods and using Lipofectamine RNAiMAX transfection reagent (Invitrogen). The targeting sequence of siRNAs against NPC1 (QIAGEN) was 5′-ACCAATTGTGATAGCAATATT-3′, and the sequence against ORP5 (Invitrogen) was 5′-CCCTGCCCAGCAGCTACCTGATCTT-3′. A Stealth RNAi universal negative control (Invitrogen) was used as a negative control in all RNAi transfections. 40 pmol of duplexes of siRNA was used for transfection of one well of cells grown in a 6-well plate. Cells transfected for 48–72 h were used for filipin staining, immunofluorescence, immunoblotting, or a cholesterol esterification assay. To generate ORP5 RNAi-resistant constructs, silent mutations in the siRNA targeting sequence, where AGCAGC (-Ser-Ser-) is replaced by TCGTCG (-Ser-Ser-), were produced by two-step QuikChange site-directed mutagenesis (Wang and Malcolm, 1999) using the following oligos: forward, 5′-ACACAGCCCCTGCCCTCGTCGTACCTGATCTTCAGG-3′; and reverse, 5′-CCTGAAGATCAGGTACGACGAGGGCAGGGGCTGTGT-3′.
Filipin staining and fluorescence microscopy
Cells grown on coverslips were fixed with 4% paraformaldehyde for 30 min at RT. Cells were stained with freshly prepared 50 µg/ml filipin in PBS for 1 h at RT. Stained cells were mounted with Prolong Antifade solution (Invitrogen) and imaged at room temperature using a microscope (CTR5500) equipped with an EL6000 fluorescent lamp and a DFC300 FX digital camera (Leica). The filipin signal was visualized with a Cube A filter (a 360/40-nm band-pass excitation filter, a 400-nm dichromatic mirror, and a 470/50-nm band-pass emission filter; Leica). EGFP/YFP signal was visualized with a GFP filter (a 470/40-nm band-pass excitation filter, a 500-nm dichromatic mirror, and a 525/50-nm band-pass emission filter; Leica). Images were processed using Leica LAS AF software and ImageJ (National Institutes of Health, http://rsb.info.nih.gov/ij). Quantification of intracellular free cholesterol was performed as described previously (Ge et al., 2008). In brief, outlines of the whole cell and the nucleus were manually drawn and the filipin fluorescence intensities were measured individually using ImageJ. Intracellular cholesterol content was expressed by subtracting the intensity of the nucleus from that of the whole cell. Relative intracellular cholesterol was presented by normalizing values from transfected cells against those of nontransfected cells, which were arbitrarily defined as 1. For each condition, >50 cells were randomly selected and calculated.
In vitro cholesterol transfer
Recombinant protein expression and purification, preparation of liposomes, and DHE transfer assay were performed essentially as described previously (Schulz et al., 2009). In brief, GST-fused ORP5ORD, expressed in Escherichia coli cells, was purified, and the GST-free recombinant protein was subsequently prepared and concentrated. All liposomes were prepared in standard vesicle buffer (20 mM Hepes, pH 7.3, 100 mM NaCl, and 1 mM EDTA). Donor liposomes without PIPs contained 67:20:4:9 PC/PE/PS/DHE, and donor liposomes with PIPs contained 67:20:3:1:9 PC/PE/PS/PI(x)P/DHE or 67:20:3:0.5:9 PC/PE/PS/PI(x,y)P/DHE. Acceptor liposomes contained 73.5:20:4:2.5 PC/PE/PS/dansyl-PE. The in vitro sterol transfer assay was performed at the same protein and lipid concentrations. The assay was terminated by cooling on ice, and samples were diluted 1:1 immediately before fluorescence measurement. All fluorometric readings were taken using a photomultiplier (detector voltage set to 750 W; model 814; Photon Technology International [PTI]) with a short arc lamp (75W Xenon; Ushio) as a light source. Data were collected and analyzed using the FeliX software (version 32; PTI). All readings were taken as 100 µl samples in a 3-mm quartz cell (Starna Cells).
Cholesterol measurement
HeLa cells plated in 12-well plates were transfected with the indicated siRNA for 54 h, then switched to medium D supplemented with 50 µg/ml for 18 h. Cellular free cholesterol concentrations were measured using an Amplex Red cholesterol assay kit according to manufacturer’s instructions (Invitrogen). In brief, cells were lysed and incubated with cholesterol oxidase, horseradish peroxidase, and Amplex red in the absence or presence of cholesterol esterase. The amount of cholesterol was determined indirectly by measuring resorufin absorbance at 560 nm. The values were normalized to the total cellular protein levels, which were determined with a BCA protein assay kit (Thermo Fisher Scientific).
Cholesterol esterification
Cells were treated with or without LDL and metabolically labeled with [1-14C]-palmitate for 4–6 h. Cell lysis, lipid extraction, and TLC were performed as described previously (Du et al., 2004). Silica Gel 60 F254 TLC plates were then exposed to a BAS-MS imaging plate (Fujifilm) for 72–96 h at RT. The imaging plate was visualized using the FLA-5100 phosphoimager (Fujifilm). The relative intensities of bands corresponding to cholesteryl ester were quantified using Sciencelab ImageGauge 4.0 Software (Fujifilm) or ImageJ.
Immunofluorescence and confocal microscopy
All immunofluorescence steps were performed at room temperature, and cells were extensively rinsed with PBS after each step. Cells grown on glass coverslips were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min. Cell permeabilization was performed using 0.05% Triton X-100/PBS for 5 min, followed by blocking with 1% BSA/0.2 M glycine in PBS for 1 h. For ORP5 immunofluorescence, cells were permeabilized with 0.1% saponin/PBS for 30 min, followed by blocking with 3% BSA/0.05% saponin in PBS for 1 h. Cells were incubated with primary antibodies diluted in 1% BSA/PBS for 1 h, followed by incubation with the appropriate conjugated secondary antibodies (Alexa Fluor 488 or Alexa Fluor 568, 1:500) for 1 h. Cells were mounted with ProLong Gold antifade reagent (Invitrogen). Confocal images were acquired at room temperature on a laser-scanning microscope (FV1000; Olympus) and accompanying software (Fluoview version 2.0) using Argon (excitation 488 nm) and HeNe (excitation 543 nm) lasers with 100× 1.4 NA or 63× 1.35 NA oil objective lenses. For filipin-stained cells, the signal was excited with the LD laser 405 nm, and emissions were collected at 450 nm. The manufacturer’s software and Photoshop CS4 (Adobe) were used for data acquisition.
CI-MPR antibody and STxB internalization assays
For CI-MPR internalization assays, monolayers of siRNA-treated HeLa cells grown on coverslips were incubated on ice for 10 min. Cells were chased with mouse monoclonal antibodies to the extracellular domain of CI-MPR in DME (2 µg/ml) for 30 min on ice. After unbound antibodies were removed by PBS washing, antibody-bound CI-MPR was internalized by incubating cells in DME for 60 min at 37°C. Cells were then fixed by 4% paraformaldehyde, and internalized CI-MPR was detected using Alexa Fluor 488–conjugated goat anti–mouse antibodies. The Cy3-conjugated STxB (a gift from P. Gleeson, University of Melbourne, Victoria, Australia) was added to cells grown on coverslips in DME, 1% BSA, and 25 mM Hepes, pH 7.4, for 15 min at 37°C. Cells were washed in PBS and incubated for 1 h at 37°C in complete medium before fixation by 4% paraformaldehyde, followed by processing for immunofluorescence.
Coimmunoprecipitation
Transfected cells grown in 100-mm dishes were harvested, washed with cold PBS, resuspended in 1 ml of cell lysis buffer (50 mM Hepes-KOH, pH 7.4/100 mM NaCl/1.5 mM MgCl2/0.5% [vol/vol] CHAPSO] containing 15 µl protease inhibitor cocktail (Sigma-Aldrich). Cell lysates were passed through a 22-gauge needle 20 times, extracted by rotating for 1 h at 4°C, and clarified by centrifugation at 100,000 g for 15 min at 4°C. Immunoprecipitation of the lysates with a polyclonal antibody against NPC1 or GFP was performed using the Dynabeads Protein G (Invitrogen) according to manufacturer’s instructions. The immunoprecipitated pellets were resuspended in 30 µl of cell lysis buffer. The pellet suspensions and supernatants were mixed with 2× Laemmli buffer (Bio-Rad Laboratories) and then incubated for 15 min at 37°C. The resultant samples were subjected to SDS/PAGE and immunoblotting.
GST pull-down assay
In vitro GST pull-down was performed as described previously (Wang et al., 2005a). In brief, GST-Osh6p fusion protein was expressed in E. coli BL21 and purified by affinity chromatography using glutathione (GSH) agarose beads. Cells expressing GFP-Ncr1p were lysed, and the lysate was incubated on ice for 10 min and then cleared at 100,000 g for 0.5 h. The GST fusion protein-bound GSH beads were mixed with the cleared yeast lysate. After incubation for 2 h at room temperature in the presence of various concentrations of ergosterol, the beads (bound fraction) were spun down and washed three times with ice-cold yeast extraction buffer (0.3 M NaCl and 1 PBS containing 1% Triton X-100). The bound proteins were then eluted from beads with SDS-PAGE sample buffer by heating at 95°C for 3 min, followed by immunoblot analysis.
Immunoblot analysis
Immunoblotting analysis was performed as described previously (Du et al., 2006), with minor modifications. In brief, after electrophoresis, the proteins were transferred to Hybond-C nitrocellulose filters (GE Healthcare). Incubations with primary antibodies were performed at 4°C overnight or RT for 1 h. Secondary antibodies were peroxidase-conjugated AffiniPure donkey anti–rabbit or donkey anti–mouse IgG (H+L; Jackson ImmunoResearch Laboratories, Inc.) used at a 1:5,000 dilution. The bound antibodies were visualized by ECL Western blotting detection reagent (GE Healthcare). The filters were exposed to Hyperfilm ECL (GE Healthcare) for periods of 2 s to 3 min.
Statistical analyses
Statistical analysis between groups was performed using Prism 5 for Windows version 5.02 (GraphPad Software) with a Student’s t test. Data are expressed as mean + SD unless otherwise stated. Significant differences are indicated in the figures.
Online supplemental material
Fig. S1 shows that yeast Ncr1p and Osh6/7p interact in the split-ubiquitin yeast two-hybrid system. Fig. S2 shows that ORP5 knockdown had no effects on lipid droplet formation, the ER structure, and the Golgi complex integrity. Fig. S3 shows that ORP5 overexpression rescues the mislocalization of DsRed-Golgin in ORP5 knockdown cells and that NPC1 is coimmunoprecipitated with ORP5LBD in HEK293 cells. Fig. S4 shows a hypothetical model of how cholesterol can be delivered to the ER from endosomal compartments by ORP5. Fig. S5 displays original images showing the details of filipin staining in NPC1 knockdown and ORP5 knockdown cells, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004142/DC1.
Acknowledgments
We thank Qiong Li and Ika Kristiana for their excellent technical assistance. We also thank Drs. Paul Gleeson, Rohan Teasdale, and Vesa Olkkonen for providing reagents.
This work is jointly supported by research grants from the Ara Parseghian Medical Research Foundation and the National Health and Medical Research Council of Australia (No. 510271). The Institute for Molecular Bioscience is a Special Research Centre of the Australian Research Council. W.A. Prinz and T.A. Schulz were supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Abbreviations used in this paper:
- CI-MPR
- cation-independent mannose-6 receptors
- DHE
- dehydroergosterol
- LDL
- low-density lipoprotein
- LDL-C
- LDL-derived cholesterol
- LE
- late endosome
- LY
- lysosome
- NCS
- newborn calf serum
- NPC
- Niemann Pick type C
- ORD
- OSBP-related domain
- ORP
- OSBP-related protein
- OSBP
- oxysterol-binding protein
- PE
- phosphatidylethanolamine
- PH
- pleckstrin homology
- PIPs
- phosphoinositides
- PM
- plasma membrane
- STxB
- Shiga toxin subunit B
- TMD
- transmembrane domain
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- Beh C.T., Cool L., Phillips J., Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 157:1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N., Woodman P. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Rojas R. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7:568–579 10.1038/nrm1985 [DOI] [PubMed] [Google Scholar]
- Brown A.J., Dean R.T., Jessup W. 1996. Free and esterified oxysterol: formation during copper-oxidation of low density lipoprotein and uptake by macrophages. J. Lipid Res. 37:320–335 [PubMed] [Google Scholar]
- Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B., et al. 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 277:228–231 10.1126/science.277.5323.228 [DOI] [PubMed] [Google Scholar]
- Chang T.Y., Chang C.C., Ohgami N., Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22:129–157 10.1146/annurev.cellbio.22.010305.104656 [DOI] [PubMed] [Google Scholar]
- Davies J.P., Ioannou Y.A. 2000. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 275:24367–24374 10.1074/jbc.M002184200 [DOI] [PubMed] [Google Scholar]
- Du X., Pham Y.H., Brown A.J. 2004. Effects of 25-hydroxycholesterol on cholesterol esterification and sterol regulatory element-binding protein processing are dissociable: implications for cholesterol movement to the regulatory pool in the endoplasmic reticulum. J. Biol. Chem. 279:47010–47016 10.1074/jbc.M408690200 [DOI] [PubMed] [Google Scholar]
- Du X., Kristiana I., Wong J., Brown A.J. 2006. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis. Mol. Biol. Cell. 17:2735–2745 10.1091/mbc.E05-11-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E.R., White I.J., Tsapara A., Futter C.E. 2010. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat. Cell Biol. 12:267–272 [DOI] [PubMed] [Google Scholar]
- Fairn G.D., Curwin A.J., Stefan C.J., McMaster C.R. 2007. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proc. Natl. Acad. Sci. USA. 104:15352–15357 10.1073/pnas.0705571104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., Pfeffer S.R. 2006. Cholesterol accumulation sequesters Rab9 and disrupts late endosome function in NPC1-deficient cells. J. Biol. Chem. 281:17890–17899 10.1074/jbc.M601679200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Wang J., Qi W., Miao H.H., Cao J., Qu Y.X., Li B.L., Song B.L. 2008. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 7:508–519 10.1016/j.cmet.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Goldstein J.L., Basu S.K., Brown M.S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98:241–260 10.1016/0076-6879(83)98152-1 [DOI] [PubMed] [Google Scholar]
- Goldstein J.L., DeBose-Boyd R.A., Brown M.S. 2006. Protein sensors for membrane sterols. Cell. 124:35–46 10.1016/j.cell.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Gruenberg J. 2003. Lipids in endocytic membrane transport and sorting. Curr. Opin. Cell Biol. 15:382–388 10.1016/S0955-0674(03)00078-4 [DOI] [PubMed] [Google Scholar]
- Heese-Peck A., Pichler H., Zanolari B., Watanabe R., Daum G., Riezman H. 2002. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell. 13:2664–2680 10.1091/mbc.E02-04-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., Levine T.P. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220 10.1038/nrm1591 [DOI] [PubMed] [Google Scholar]
- Im Y.J., Raychaudhuri S., Prinz W.A., Hurley J.H. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437:154–158 10.1038/nature03923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante R.E., Wang M.L., Radhakrishnan A., Kwon H.J., Brown M.S., Goldstein J.L. 2008. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 105:15287–15292 10.1073/pnas.0807328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA. 91:10340–10344 10.1073/pnas.91.22.10340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. 2008. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell. 19:2069–2082 10.1091/mbc.E07-08-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D.C., Gordon M.D., Jin J.Y., Scott M.P. 2001. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell. 12:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Beuchat M.H., Lindsay M., Frias S., Palmiter R.D., Sakuraba H., Parton R.G., Gruenberg J. 1999. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1:113–118 10.1038/15666 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiana I., Yang H., Brown A.J. 2008. Different kinetics of cholesterol delivery to components of the cholesterol homeostatic machinery: implications for cholesterol trafficking to the endoplasmic reticulum. Biochim. Biophys. Acta. 1781:724–730 [DOI] [PubMed] [Google Scholar]
- Kwon H.J., Abi-Mosleh L., Wang M.L., Deisenhofer J., Goldstein J.L., Brown M.S., Infante R.E. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137:1213–1224 10.1016/j.cell.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C., Corti M., Goodson H., Cosson P., Cavalli V., Mayran N., Fauré J., Gruenberg J. 2002. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21:1289–1300 10.1093/emboj/21.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M., Laitinen S., Chinetti G., Johansson M., Ehnholm C., Staels B., Ikonen E., Olkkonen V.M. 2001. The OSBP-related protein family in humans. J. Lipid Res. 42:1203–1213 [PubMed] [Google Scholar]
- Lieu Z.Z., Gleeson P.A. 2010. Identification of different itineraries and retromer components for endosome-to-Golgi transport of TGN38 and Shiga toxin. Eur. J. Cell Biol. 89:379–393 10.1016/j.ejcb.2009.10.021 [DOI] [PubMed] [Google Scholar]
- Liscum L., Ruggiero R.M., Faust J.R. 1989. The intracellular transport of low density lipoprotein-derived cholesterol is defective in Niemann-Pick type C fibroblasts. J. Cell Biol. 108:1625–1636 10.1083/jcb.108.5.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., Levine T.P. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K., Higaki K., Tinkelenberg A.H., Balderes D.A., Almanzar-Paramio D., Wilcox L.J., Erdeniz N., Redican F., Padamsee M., Liu Y., et al. 2004. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C–related protein reveals a primordial role in subcellular sphingolipid distribution. J. Cell Biol. 164:547–556 10.1083/jcb.200310046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438:612–621 10.1038/nature04399 [DOI] [PubMed] [Google Scholar]
- Mayor S., Sabharanjak S., Maxfield F.R. 1998. Cholesterol-dependent retention of GPI-anchored proteins in endosomes. EMBO J. 17:4626–4638 10.1093/emboj/17.16.4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran N., Parton R.G., Gruenberg J. 2003. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 22:3242–3253 10.1093/emboj/cdg321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B., Maxfield F.R. 2009. Intracellular sterol dynamics. Biochim. Biophys. Acta. 1791:636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard E.E., Gale S.E., Dudley N., Zhang J., Schaffer J.E., Ory D.S. 2005. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 280:28581–28590 10.1074/jbc.M414024200 [DOI] [PubMed] [Google Scholar]
- Ngo M., Ridgway N.D. 2009. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell. 20:1388–1399 10.1091/mbc.E08-09-0905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami N., Ko D.C., Thomas M., Scott M.P., Chang C.C., Chang T.Y. 2004. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 101:12473–12478 10.1073/pnas.0405255101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Sugimoto Y., Suzuki M., Hosokawa H., Yoshimori T., Davies J.P., Ioannou Y.A., Vanier M.T., Ohno K., Ninomiya H. 2006. Cholesterol depletion facilitates ubiquitylation of NPC1 and its association with SKD1/Vps4. J. Cell Sci. 119:2643–2653 10.1242/jcs.02993 [DOI] [PubMed] [Google Scholar]
- Prinz W.A. 2007. Non-vesicular sterol transport in cells. Prog. Lipid Res. 46:297–314 10.1016/j.plipres.2007.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y.J., Hurley J.H., Prinz W.A. 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173:107–119 10.1083/jcb.200510084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway N.D., Dawson P.A., Ho Y.K., Brown M.S., Goldstein J.L. 1992. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116:307–319 10.1083/jcb.116.2.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G., Ortlund E.A., Tyeryar K.R., Mousley C.J., Ile K.E., Garrett T.A., Ren J., Woolls M.J., Raetz C.R., Redinbo M.R., Bankaitis V.A. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29:191–206 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.A., Choi M.G., Raychaudhuri S., Mears J.A., Ghirlando R., Hinshaw J.E., Prinz W.A. 2009. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 187:889–903 10.1083/jcb.200905007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J., Xu Z. 2009. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim. Biophys. Acta. 1791:671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam V.N., Loh E., Hong W. 1997. N-Ethylmaleimide-sensitive factor (NSF) and alpha-soluble NSF attachment proteins (SNAP) mediate dissociation of GS28-syntaxin 5 Golgi SNAP receptors (SNARE) complex. J. Biol. Chem. 272:25441–25444 10.1074/jbc.272.41.25441 [DOI] [PubMed] [Google Scholar]
- Suchanek M., Hynynen R., Wohlfahrt G., Lehto M., Johansson M., Saarinen H., Radzikowska A., Thiele C., Olkkonen V.M. 2007. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 405:473–480 10.1042/BJ20070176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Watanabe H., Murphy S.R., Shibuya Y., Geng Y., Peden A.A., Chang C.C., Chang T.Y. 2008. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc. Natl. Acad. Sci. USA. 105:16513–16518 10.1073/pnas.0807450105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Malcolm B.A. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 26:680–682 [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang Y., Li H., Chieu H.K., Munn A.L., Yang H. 2005a. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO J. 24:2989–2999 10.1038/sj.emboj.7600764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.Y., Weng J., Anderson R.G. 2005b. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 307:1472–1476 10.1126/science.1107710 [DOI] [PubMed] [Google Scholar]
- Wojtanik K.M., Liscum L. 2003. The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J. Biol. Chem. 278:14850–14856 10.1074/jbc.M300488200 [DOI] [PubMed] [Google Scholar]
- Wyles J.P., McMaster C.R., Ridgway N.D. 2002. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 277:29908–29918 10.1074/jbc.M201191200 [DOI] [PubMed] [Google Scholar]
- Yan D., Olkkonen V.M. 2008. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265:253–285 10.1016/S0074-7696(07)65007-4 [DOI] [PubMed] [Google Scholar]
- Yan D., Mäyränpää M.I., Wong J., Perttilä J., Lehto M., Jauhiainen M., Kovanen P.T., Ehnholm C., Brown A.J., Olkkonen V.M. 2008. OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J. Biol. Chem. 283:332–340 10.1074/jbc.M705313200 [DOI] [PubMed] [Google Scholar]
- Yang H. 2006. Nonvesicular sterol transport: two protein families and a sterol sensor? Trends Cell Biol. 16:427–432 10.1016/j.tcb.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Zhang S., Ren J., Li H., Zhang Q., Armstrong J.S., Munn A.L., Yang H. 2004. Ncr1p, the yeast ortholog of mammalian Niemann Pick C1 protein, is dispensable for endocytic transport. Traffic. 5:1017–1030 10.1111/j.1600-0854.2004.00241.x [DOI] [PubMed] [Google Scholar]