Abstract
Mitochondria are dynamic organelles whose functional integrity requires a coordinated supply of proteins and phospholipids. Defined functions of specific phospholipids, like the mitochondrial signature lipid cardiolipin, are emerging in diverse processes, ranging from protein biogenesis and energy production to membrane fusion and apoptosis. The accumulation of phospholipids within mitochondria depends on interorganellar lipid transport between the endoplasmic reticulum (ER) and mitochondria as well as intramitochondrial lipid trafficking. The discovery of proteins that regulate mitochondrial membrane lipid composition and of a multiprotein complex tethering ER to mitochondrial membranes has unveiled novel mechanisms of mitochondrial membrane biogenesis.
Introduction
Mitochondria are engaged in a plethora of cellular processes and are therefore of utmost importance for cell viability. Mitochondria are not static entities but are highly dynamic and require that supplies of proteins and membrane lipids be coordinated and adjusted to meet physiological and functional demands. Although an increasingly detailed structural and mechanistic picture is emerging for the biogenesis, sorting, and compartmentation of mitochondrial proteins (Schmidt et al., 2010), much less is known about mechanisms regulating the supply of phospholipids and the maintenance of mitochondrial membrane integrity. The mitochondrial phospholipid composition varies little among different cells, suggesting that major changes cannot be tolerated. Indeed, both altered phospholipid levels and phospholipid damage have been linked to a variety of human disease states (Chicco and Sparagna, 2007; Joshi et al., 2009). Phospholipids like cardiolipin (CL) have long been known to affect the stability and catalytic activity of mitochondrial membrane proteins (Schlame and Ren, 2009). However, considering phospholipids merely as the fabric that keeps mitochondria together vastly underestimates their contribution toward the functional integrity of these organelles.
In this article, we summarize recent findings that highlight distinct functions of mitochondrial phospholipids in diverse mitochondria-associated processes such as mitochondrial fusion, protein import into mitochondria, and apoptosis. We will focus on phosphatidylethanolamine (PE) and the mitochondria-specific dimeric glycerophospholipid CL. Both PE and CL are non–bilayer-forming phospholipids, a feature best explained by their shape (Fig. 1; van den Brink-van der Laan et al., 2004). Bilayer-forming phospholipids like phosphatidylcholine (PC) are cylindrically shaped with the fatty acid portions defining extended hydrophobic domains and the polar head groups defining the short hydrophilic domains along the length of the cylinder. The nearly equivalent diameters of the cylinder in both domains allow molecular packing that favors bilayers. The non–bilayer-forming lipids PE and CL are more conical shaped with a smaller hydrophilic head group diameter and a relatively larger hydrophobic domain diameter. This shape allows the formation of hexagonal phases that can be observed for isolated lipids depending on the pH and ionic strength (Ortiz et al., 1999). PE and CL are thought to be present mainly in bilayer structures in vivo, but their tendency to form hexagonal phases can create tension in membranes that is likely of functional importance to various mitochondrial processes like membrane fusion or the movement of proteins or solutes across membranes (van den Brink-van der Laan et al., 2004). The functional importance of non–bilayer-forming lipids is highlighted by the fact that yeast and bacteria cannot tolerate simultaneous reduction of PE and CL (Rietveld et al., 1993; Gohil et al., 2005). The biosynthesis of PE and CL occurs, at least in part, within mitochondria and relies on an intricate exchange of precursor forms between the membrane of the ER and the mitochondrial outer membrane at distinct contact sites, whose structural basis we are just beginning to understand. We will highlight recent advances and unresolved questions regarding this interorganellar communication and the intramitochondrial trafficking of phospholipids.
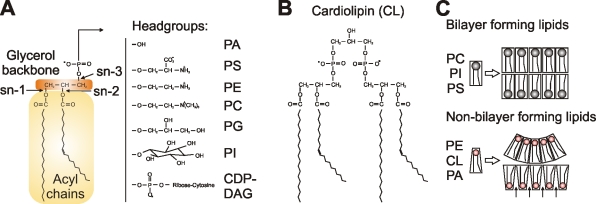
Figure 1.
Phospholipids in mitochondrial membranes. (A) The central structural element of phospholipids is a glycerol backbone. Acyl chains that can vary in length and saturation are attached to the sn-1 and sn-2 hydroxyl groups. Distinct hydrophilic head groups can be attached to the sn-3 position of the glycerol backbone via a phosphodiester bond and confer unique biophysical properties that distinguish the different phospholipid classes: PA, PS, PE, PC, PG, PI, and CDP-DAG. CDP-DAG is an intermediate that does not accumulate in significant amounts in mitochondrial membranes under normal conditions. (B) CL is a lipid unique to mitochondria, which consists of two PA moieties covalently linked to each other by a glycerol bridge, with the phosphodiester bonds at the sn-1 and sn-3 positions of the bridging glycerol. (C) Bilayer and non-bilayer phospholipids have different shapes. The conical shape of non-bilayer lipids induces membrane curvature or creates a unique biochemical microenvironment in a planar bilayer, where the hydrophobic parts are exposed between neighboring phospholipids (marked with arrows).
Mitochondrial phospholipids and membrane domains
The phospholipid composition of mitochondrial membranes has been determined in yeast and mammalian cells. Although the exact composition determined in different studies varies, most likely because of differences in the growth conditions or the purity of the analyzed fraction, the relative abundance of different phospholipids remains within a relatively narrow range. PC and PE are the most abundant phospholipids and comprise ∼40% and ∼30% of total mitochondrial phospholipids, respectively. CL and phosphatidylinositol (PI) account for ∼10–15% of phospholipids, whereas phosphatidic acid (PA) and phosphatidylserine (PS) comprise ∼5% of the total mitochondrial phospholipids (Colbeau et al., 1971; Zinser and Daum, 1995). The lipids CDP-DAG, phosphatidylglycerol (PG) phosphate (PGP), and PG are important intermediates for the synthesis of the abundant phospholipid species but do not accumulate in mitochondria under normal conditions. However, it has to be noted that PG, which accumulates in mitochondria in the absence of the CL synthase, can partially compensate for several cellular functions of CL (Jiang et al., 2000). In mammalian cells, mutations in PGP synthase eliminate PG and CL pools, resulting in altered mitochondrial structure and function (Ohtsuka et al., 1993a,b). Other membrane lipids, like sphingolipids and sterols, which are important structural lipids that significantly contribute to the composition of the plasma membrane, the membrane of the Golgi apparatus, and the lysosomal compartments, are only found in trace amounts in mitochondrial membranes (van Meer et al., 2008). Notable exceptions are mitochondria of steroidogenic cells that are involved in the biosynthesis of hormones and consequently have a higher content of sterols (Strauss et al., 2003).
The diversity of mitochondrial membrane lipids is also a consequence of the variation in chain length and degree of unsaturation of fatty acids present within each class of phospholipid. Acyl chains are important determinants for the biophysical properties of cellular membranes. With the exception of the acyl chain remodeling of CL, which has been studied in some detail (Houtkooper et al., 2009; see Mitochondrial synthesis of CL), the regulation of the acyl chain composition of mitochondrial lipids and their functional importance for mitochondrial processes are poorly understood.
Perhaps the most significant difference in the relative abundance of phospholipids between the outer and the inner mitochondrial membrane is observed for CL. It has long remained controversial whether CL is even present at all in the outer mitochondrial membrane. However, a recent study in yeast has now convincingly demonstrated that a purified mitochondrial outer membrane fraction from yeast indeed contains ∼25% of the CL of total mitochondrial membranes (Gebert et al., 2009).
Little is currently known about the lateral distribution of phospholipids in mitochondrial membranes. The non–bilayer-forming lipids PE and CL laterally segregate into distinct domains in bacterial membranes, which, similar to mitochondria, contain CL but lack sterols and sphingolipids (Mileykovskaya and Dowhan, 2000; Kawai et al., 2004; Nishibori et al., 2005). A spatially defined lipid distribution may also affect mitochondrial processes, such as fusion or fission, as well as the insertion or extraction of membrane proteins. The high membrane curvature at cristae tips may impose geometric constraints that could lead to an enrichment of non–bilayer-forming lipids. Membrane domains may self-assemble to some extent, but it is conceivable that scaffolding proteins assist in their formation and maintenance. This might be of particular relevance in the mitochondrial inner membrane, which is considered to be the most protein-rich cellular membrane. Prohibitins, which are evolutionarily conserved proteins forming ring complexes in the mitochondrial inner membrane (Tatsuta et al., 2005), were proposed to act as membrane scaffolds that recruit proteins to lipid domains enriched in PE and CL in the mitochondrial inner membrane (Fig. 2 A; Osman et al., 2009a,b). Bacterial flotillins, scaffolding proteins of the SPFH family distantly related to prohibitins, have recently been found to associate with negatively charged phospholipids (Donovan and Bramkamp, 2009). Similarly, prohibitins may enrich PE and CL within the ring complexes. This could explain genetic evidence in yeast demonstrating that prohibitins are essential for the survival of yeast cells containing reduced levels of mitochondrial PE or CL (Osman et al., 2009a). Accordingly, a perturbed membrane organization could cause the pleiotropic mitochondrial deficiencies observed in prohibitin-deficient cells, but direct experimental evidence in support of a lipid scaffolding function of prohibitin complexes remains elusive.
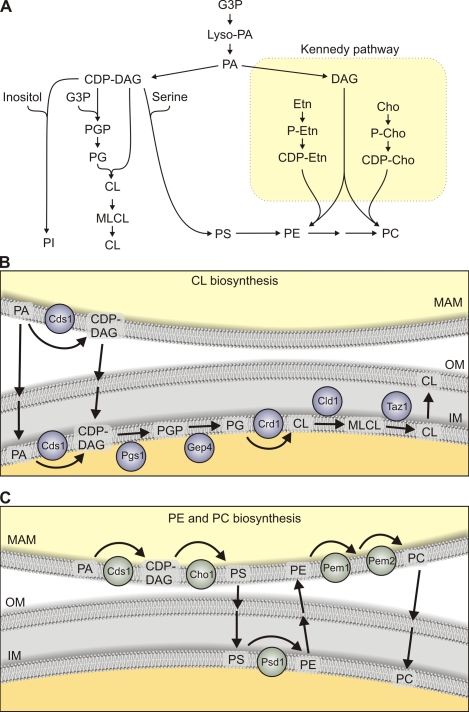
Figure 2.
Mitochondria and the synthesis of phospholipids. (A) Schematic summary of phospholipid biosynthesis. Cleavage of the pyrophosphate bond in CDP-DAG provides the energetic driving force to catalytically replace CMP with inositol, G3P, or serine to form PI, PGP, or PS, respectively, using specific synthetic enzymes. PGP is dephosphorylated to produce PG. CL is synthesized from PG and CDP-DAG substrates with the catalytic cleavage of the pyrophosphate bond in the latter substrate providing the chemical energy to transfer the PA moiety to the vacant primary hydroxyl of PG. PS can be decarboxylated to PE, which in turn can be methylated to yield PC. Alternatively, PE and PC can be synthesized via an enzymatic cascade known as the Kennedy pathway. See Mitochondrial phospholipid biosynthesis for further details. Cho, choline; Etn, ethanolamine; MLCL, monolyso-CL; P-Cho, phosphocholine; P-Etn, phosphoethanolamine. (B and C) Membrane topology and lipid transport events in the synthesis of CL (B) and aminoglycerophospholipids (PE and PC; C). Yeast biosynthetic enzymes are indicated. PA synthesized in the ER or mitochondria drive biosynthetic reactions. CDP-DAG may derive from the ER/MAM or be generated at the mitochondrial inner membrane by the action of CDP-DAG synthase (Cds1; Kuchler et al., 1986). IM, mitochondrial inner membrane; OM, mitochondrial outer membrane.
Mitochondrial phospholipid biosynthesis
The maintenance of a defined lipid composition within mitochondria depends on their capacity to synthesize phospholipids such as CL, PE, PG, and PA, whereas PI, PC, and PS are primarily synthesized in the ER and must be imported into the organelle for use as a finished end product or precursors for other lipids (Fig. 2). The biochemical steps in the synthesis of all phospholipids commence with the acylation of the sn-1 position of glycerol-3-phosphate (G3P) or dihydroxyacetone phosphate by acyltransferases (G3P acyltransferases [GPATs]) producing lyso-PA (Fig. 2 A). The yeast GPATs are associated with the ER and lipid particles, whereas the mammalian GPATs are localized to multiple organelles, including mitochondria (Wendel et al., 2009). Several lyso-PA acyltransferases (LPAATs) then convert lyso-PA to PA, which serves as a crucial intermediate supplying two independent cellular pathways for the synthesis of phospholipids (Fig. 2 A). One branch of the pathway converts PA to DAG catalyzed by the phosphatase Pah1 (Han et al., 2006) and eventually produces the zwitterionic lipids PE and PC in an enzymatic cascade known as the Kennedy pathway (Daum et al., 1998). The other branch of the pathway leads to the synthesis of CDP-DAG catalyzed by Cds1 (Shen et al., 1996) and produces the acidic phospholipids PS, PI, PG, and CL as its principal products (Fig. 2 A).
Mitochondrial synthesis of CL.
A multienzyme cascade in the mitochondrial inner membrane synthesizes CL from CDP-DAG (Fig. 2 B; Joshi et al., 2009; Schlame and Ren, 2009) by the stepwise formation of PGP catalyzed by Pgs1 (Chang et al., 1998; Dzugasová et al., 1998) and its subsequent dephosphorylation catalyzed by the recently identified yeast PGP phosphatase Gep4 (Osman et al., 2010). Gep4 localizes to the matrix side of the inner membrane (Osman et al., 2010), which is also the predicted location for Pgs1. The localization of both enzymes in yeast mitochondria is in agreement with the proposed initiation of CL synthesis on the matrix-exposed leaflet of the inner membrane (Joshi et al., 2009; Schlame and Ren, 2009). How newly synthesized CL molecules are then redistributed within mitochondria remains to be examined. Although CL synthase generates CL from PG and CDP-DAG on the matrix side of the membrane (Schlame and Haldar, 1993), later acyl chain remodeling steps appear to occur on the outer leaflet of the inner membrane (Claypool et al., 2006). The acyl chain composition of nascent CL species is remodeled by the sequential action of a phospholipase A (Cld1 in yeast) and a transacylation reaction catalyzed by Taz1 (Xu et al., 2006; Beranek et al., 2009). In humans, mutations in Taz1 cause cardiomyopathy and Barth syndrome, underscoring the physiological importance of CL and its remodeling for mitochondrial homeostasis and function (Bione et al., 1996; Houtkooper et al., 2009).
Although enzymes involved in CL biosynthesis from CDP-DAG are localized at the mitochondrial inner membrane, it is currently not clear how much CL synthesis depends on the transport of precursor lipids from extramitochondrial sources. The de novo synthesis of PA occurs in the ER, but PA may also be generated within mitochondria by phospholipases like MitoPLD (Choi et al., 2006). Thus, mitochondria may use both extrinsic and intrinsic sources of phospholipid precursors for CL formation.
Mitochondrial synthesis of PE.
Extramitochondrial PS formed in the ER or specialized domains of the ER that are tightly associated with mitochondria serve as a precursor for mitochondrial PE in both yeast and mammalian cells (Fig. 2 C). This PS is synthesized from a CDP-DAG substrate in yeast (Letts et al., 1983; Nikawa and Yamashita, 1984; Kuchler et al., 1986) or by base exchange enzymes in mammalian cells (Kuge and Nishijima, 1997; Vance, 2008). The imported PS is a substrate for Psd1 (PS decarboxylase 1) located in the mitochondrial inner membrane (Clancey et al., 1993; Trotter et al., 1993). Although a second decarboxylase (Psd2) is present outside of mitochondria in yeast (but not in mammals; Trotter and Voelker, 1995), the majority of the catalytic activity occurs within mitochondria. PE produced via the Kennedy pathway or by the action of Psd2 is poorly assimilated into mitochondria and insufficient to meet the requirements for respiration. The PE produced in mitochondria is actively exported to other organelles (Voelker, 1984).
One major consequence of this PE export is the synthesis of PC in the ER by the sequential methylation of the primary amine of PE, catalyzed by the yeast methyltransferases Pem1 and Pem2 (originally named Cho2 and Opi3; Kuchler et al., 1986; Kodaki and Yamashita, 1987, 1989). In the majority of mammalian tissues, PC is produced via the Kennedy pathway (Fig. 2), but in the liver, PE methyltransferase activity is significant and can provide adequate levels of PC during periods of choline deficiency (Li and Vance, 2008).
In many eukaryotes, the aminoglycerophospholipids PS, PE, and PC comprise 75–80% of the total glycerophospholipids found within the cell (van Meer et al., 2008). As mitochondria have the synthetic capacity to synthesize the entire PE pool required for cell growth (Birner et al., 2001), the flux of PS into the mitochondria, and its subsequent decarboxylation and export as PE, can account for the biosynthesis of the majority of the glycerophospholipids present in all cellular membranes. This dynamic role of mitochondria as a major source of phospholipids is widely underappreciated. The role of mitochondria in exporting phospholipids is true for eukaryotes other than yeast. Mammalian cells can also produce the majority of all PE via the mitochondrial pathway (Voelker, 1984).
Mitochondrial phospholipid trafficking
The differential localization of enzymes of phospholipid biosynthetic pathways among different organelles and different membrane compartments within one organelle implicitly defines a requirement for extensive intracellular lipid trafficking (Fig. 2, B and C). Specific mechanisms must exist to ensure the transport of phospholipids from the ER to mitochondria and between outer and inner mitochondrial membranes. However, we are only beginning to understand how these transport processes occur and how they are regulated.
Phospholipid transport to and within mitochondria appears to proceed via close membrane contacts rather than vesicular pathways. A close apposition of two membranes may facilitate direct lipid flipping between bilayers at regions of positive membrane curvature or may allow lipid trafficking by yet to be identified soluble lipid carriers or by protein complexes that bridge both membranes (Voelker, 2009). Intermembrane lipid exchange might also be mediated via a stabilized hemifusion state, which would result in continuity between leaflets of both membranes, but evidence for such a mechanism is still lacking.
Tethering of ER and mitochondrial membranes.
Transport of phospholipids between membranes of the ER and mitochondria occurs at specialized fractions of the ER that are tightly associated with mitochondria (Voelker, 1990) and were therefore termed mitochondria-associated membranes (MAMs; Vance, 1990; Ardail et al., 1993; Gaigg et al., 1995; Shiao et al., 1995). MAMs are enriched in certain lipids and various phospholipid biosynthetic enzymes, including PSS-1 (PS synthase-1), FACL4 (long-chain fatty acid-CoA ligase type 4; Vance, 1990; Rusiñol et al., 1994; Gaigg et al., 1995), and Ale1 acyltransferase (Riekhof et al., 2007). Direct evidence that phospholipid transport involves MAMs came from in vitro assays that showed that transport of PS from MAMs to mitochondria occurs more efficiently when MAMs, rather than bulk ER membranes, are mixed with mitochondria (Gaigg et al., 1995). Although independent of ATP, transport appears to be regulated by ubiquitination. A genetic screen in yeast for mutants affecting PS transport into mitochondria led to the identification of the F-box protein Met30, an E3 ubiquitin ligase (Schumacher et al., 2002). Met30 ubiquitinates and thereby inactivates the transcription factor Met4, leading to an increased transport of PS from MAMs to mitochondria (Schumacher et al., 2002; Voelker, 2009). However, the downstream targets of Met4 remain elusive.
Phospholipid transport from MAM-derived vesicles to mitochondria proved to be partially protease sensitive, indicating that membrane proteins of the ER or mitochondria exposed to the cytosol mediate the interaction between both organelles (Vance, 1991; Achleitner et al., 1999). Electron tomography of intact cells revealed close appositions of ER membranes and mitochondria with a relatively defined, separating distance of ∼10–25 nm (Csordás et al., 2006). Several proteins were proposed to be involved in ER–mitochondria membrane tethering in mammalian cells (Szabadkai et al., 2006; de Brito and Scorrano, 2008), but evidence for a direct role in phospholipid trafficking has not yet been reported for any of these proteins. In contrast, direct evidence supporting the role of a macromolecular protein bridge for interorganellar phospholipid transport was recently obtained in yeast (Fig. 3). A synthetic biology approach using an artificial membrane tethering protein led to the identification of Mdm12 as an essential component for the interaction of ER and mitochondria (Kornmann et al., 2009). Mdm12 is associated with the outer membrane of mitochondria (Berger et al., 1997; Kornmann et al., 2009) and assembles with Mmm1, a glycosylated ER membrane protein, and the mitochondrial outer membrane proteins Mdm10 and Mdm34 into a complex (Boldogh et al., 2003; Youngman et al., 2004; Kornmann et al., 2009). Strikingly, cells lacking individual subunits of this complex, which was termed ER–mitochondria encounter structure (ERMES) complex (Kornmann et al., 2009), show reduced levels of mitochondrial PE and CL, suggesting that the ERMES structure is required for the exchange of phospholipids at ER–mitochondria contact sites. Consistently, the conversion of PS to PE and PC was slowed down in cells lacking ERMES components (Kornmann et al., 2009). It will be of interest to determine whether the ERMES complex only functions exclusively as a membrane tether ensuring the close apposition of ER and mitochondrial membranes or whether components of this complex actively contribute to the transport of phospholipids.
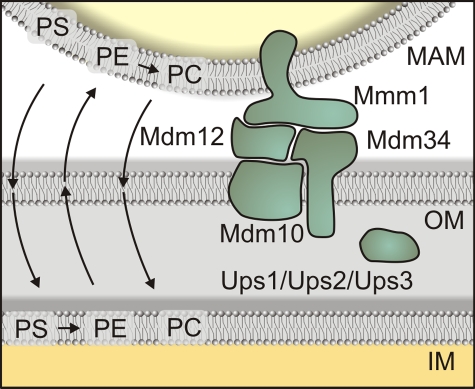
Figure 3.
A multiprotein complex involved in lipid movement and metabolism between the ER and mitochondria in yeast. A tethering complex composed of an integral ER glycoprotein (Mmm1) and three mitochondria-associated proteins (Mdm34, Mdm10, and Mdm12) promotes and stabilizes interactions between the two membranes affecting import of PS into mitochondria and the export of PE from the mitochondria. Ups1/PRELI-like proteins (Ups1, Ups2, and Ups3) regulate the accumulation of CL and PE within mitochondria and might be involved in intramitochondrial lipid movements. IM, mitochondrial inner membrane; OM, mitochondrial outer membrane.
Notably, the role of the ERMES complex for ER–mitochondrial juxtaposition raises questions about functions previously associated with subunits of this complex (Boldogh et al., 2003; Meeusen and Nunnari, 2003; Meisinger et al., 2004). All components were originally reported to be required for mitochondrial inheritance and the maintenance of mitochondrial morphology. It is conceivable that these phenotypes are caused by disturbances in the levels of mitochondrial phospholipids, which affect mitochondrial structure and transport. Similarly, ER–mitochondria contact sites appear to control other mitochondrial functions such as mitochondrial DNA (mtDNA) stability. The localization of the ER-localized ERMES subunit Mmm1 overlaps with that of mtDNA nucleoids (Hobbs et al., 2001), and cells lacking the ERMES complex lose mtDNA (Meeusen and Nunnari, 2003). However, it should be noted that subunits of the ERMES complex can be part of other protein complexes and exert independent functions. Indeed, Mdm10 has also been found as a constituent of the sorting and assembly machinery (SAM) complex that mediates the insertion of β-barrel proteins in the mitochondrial outer membrane (Meisinger et al., 2004). The presence of Mdm10 in both ERMES and SAM complexes may provide the means to balance the accumulation of phospholipids and protein biogenesis in mitochondria.
Intramitochondrial lipid trafficking.
Relatively little is known about how newly imported phospholipids or lipid precursors are transported within mitochondria. As phospholipids are either imported from the ER or synthesized at the outer or inner surface of the inner membrane, mechanisms must exist allowing trans-bilayer movements from one leaflet to the other. These movements, which are energetically disfavored because of the presence of polar head groups, are generally facilitated by dedicated enzymes commonly referred to as flippases. However, the only known mitochondrial flippase is PLS3 (phospholipid scramblase 3; Liu et al., 2003), which catalyzes trans-bilayer flipping of CL in vitro (Liu et al., 2008). PLS3 modulates CL levels exposed at the mitochondrial surface and may play an important role during the apoptotic response (Fig. 4 C; Liu et al., 2008; Ndebele et al., 2008; see CL and apoptosis).
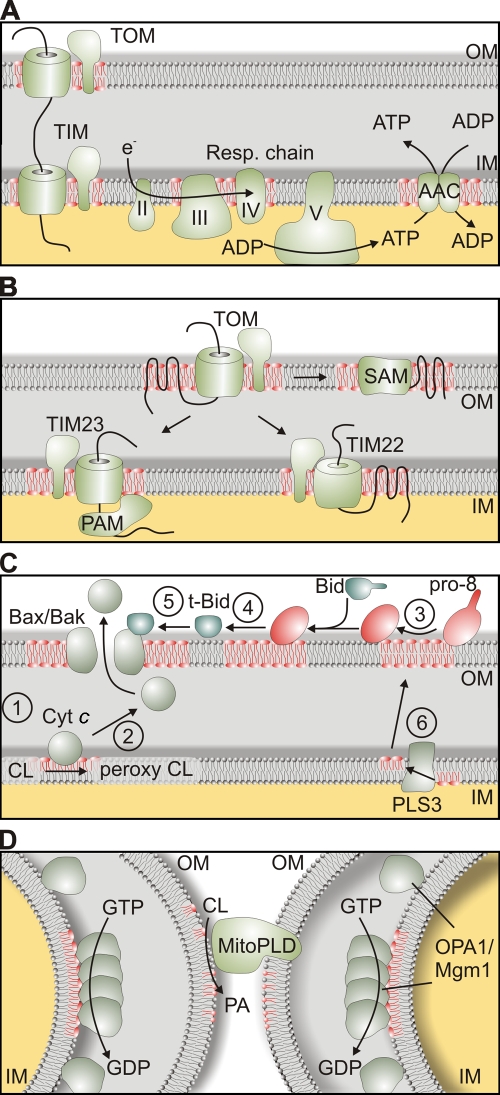
Figure 4.
The role of CL in mitochondrial processes. (A) CL (depicted in red) affects mitochondrial energy production and is required for dimerization and optimal activity of the AAC and the formation of respiratory chain supercomplexes. (B) Assembly and activity of protein translocases in the outer (TOM) and inner membrane (TIM22 and TIM23 complexes), the SAM complex in the outer membrane, and the assembly of TIM23 complex with the mitochondrial import motor (PAM complex) is supported by CL. (C) Various roles of CL during apoptosis. (1) Cytochrome c (Cyt c) binds to CL in the inner membrane. (2) Release of cytochrome c upon oxidation of CL. (3) Pro–caspase-8 (pro-8) binds to the surface of mitochondria, oligomerizes, and undergoes autocatalytic processing in a CL-dependent manner. (4 and 5) Bid cleavage to truncated Bid (t-Bid) by pro–caspase-8 (4) and activation and oligomerization of Bax/Bak is stimulated by CL (5). (6) PLS3 allows export of CL from the inner to the outer mitochondrial membrane. (D) CL affects fusion of mitochondrial outer and inner membranes. The phospholipase MitoPLD converts in trans CL into PA (depicted in red), triggering the fusion of outer membranes. CL in the inner membrane stimulates oligomerization and GTP hydrolysis of short Mgm1/OPA1 isoforms. IM, mitochondrial inner membrane; OM, mitochondrial outer membrane.
Phospholipid transport between the outer and inner mitochondrial membranes has been proposed to occur, similar to protein transport, at contact sites between mitochondrial inner and outer membranes (Ardail et al., 1991; Simbeni et al., 1991). Experiments with CHO cell mutants have identified a variant with a lesion in PS transport between the outer and inner mitochondrial membranes, but the gene responsible for this defect has yet to be identified (Emoto et al., 1999). Two proteins, mitochondrial creatine kinase (MtCK) and nucleotide diphosphate kinase (NDPK-D) facilitate CL transport between liposomes with a lipid composition resembling those of contact sites (Epand et al., 2007). However, the in vivo relevance of this pathway remains to be established.
The recent identification of conserved proteins in the intermembrane space, which regulate the accumulation of CL and PE in mitochondria, may provide new clues about the mechanism of phospholipid transport across this compartment. Ups1 was originally identified to affect the processing of the dynamin-like GTPase Mgm1 in yeast (Sesaki et al., 2006) and later shown to regulate CL level in mitochondria (Osman et al., 2009a; Tamura et al., 2009). Ups1 belongs to the conserved Ups1/PRELI protein family, which is characterized by the presence of a conserved MSF´ domain (originally identified in yeast Msf´) of unknown function (Dee and Moffat, 2005). A homologue of Ups1, termed Ups2 or Gep1, regulates the accumulation of PE within mitochondria (Osman et al., 2009a; Tamura et al., 2009). Although PE levels are decreased in the absence of Ups2, overexpression of Ups2 reduces CL, pointing to a coordinated regulation of PE and CL by these conserved regulatory proteins. Consistently, deletion of UPS2 restores normal CL levels in Ups1-deficient yeast cells. Two recent studies in yeast identified Mdm35 as a common binding partner of both Ups1 and Ups2 in the intermembrane space, providing a molecular explanation for the coordinated regulation of CL and PE within mitochondria (Potting et al., 2010; Tamura et al., 2010). Mdm35 binding ensures mitochondrial import of Ups1 and Ups2 and protects both proteins against proteolysis. Notably, both Ups1 and Ups2 are intrinsically unstable proteins and are degraded by the i-AAA protease Yme1 and Atp23 in wild-type cells even under normal growth conditions (Potting et al., 2010). It is therefore conceivable that the mitochondrial quality control system affects the accumulation of CL and PE within mitochondria by regulating the stability of Ups1-like proteins. The strong conservation of all components of the regulatory circuit and the altered PE levels in i-AAA protease-deficient mitochondria (Nebauer et al., 2007) point in this direction.
However, the molecular function of Ups1 and Ups2 remains speculative. Because reduced mitochondrial PE levels in the absence of Ups2 were caused by decreased stability rather than altered synthesis of PE (Osman et al., 2009a), Ups2 might regulate the export of PE from mitochondria. It is therefore an intriguing possibility that lipid trafficking between inner and outer mitochondrial membrane controlled by Ups1/PRELI-like proteins determines the phospholipid composition of mitochondrial membranes.
The role of CL in mitochondria
Although studies examining functional roles of phospholipids within mitochondria are generally hampered by their broad distribution among different cellular membranes, the predominant localization of CL in mitochondria has enabled the identification of an increasing number of mitochondrial processes dependent on this lipid, and the assignment of pathologies associated with alterations in the CL metabolism to mitochondrial dysfunction (Chicco and Sparagna, 2007; Joshi et al., 2009). The unique, dimerically cross-linked phospholipid structure of CL affects the stability and activity of various membrane protein complexes and metabolite carriers (Fig. 4; Houtkooper and Vaz, 2008). CL molecules are present in crystal structures of the ATP/ADP carrier (AAC) and the respiratory complexes III and IV and have been proposed to fulfill important structural roles (Lange et al., 2001; Pebay-Peyroula et al., 2003; Shinzawa-Itoh et al., 2007). Indeed, respiratory supercomplexes consisting of complexes III and IV are destabilized in mitochondria lacking CL (Pfeiffer et al., 2003; Claypool et al., 2008b). Similarly, dimers of AAC and other AAC-containing complexes dissociate in CL-deficient mitochondria (Claypool et al., 2008b). These examples illustrate the importance of CL for bioenergetic functions; but in addition, recent studies are now revealing that CL has a much broader impact on mitochondrial physiology.
CL and protein import into mitochondria.
The vast majority of mitochondrial proteins are nuclear encoded and imported into the organelle via heterooligomeric protein translocases residing in the mitochondrial inner and outer membranes (Schmidt et al., 2010). Several independent studies revealed that the assembly and function of these TIM (translocase of the inner mitochondrial membrane) and TOM (translocase of the outer mitochondrial membrane) complexes depend on CL (Fig. 4 B).
Tam41 (translocator assembly and maintenance protein 41) was identified as a novel mitochondrial matrix protein, which is required for the integrity of the TIM23 complex in the inner membrane and its functional interaction with the mitochondrial import motor PAM (presequence translocase-associated motor; Gallas et al., 2006; Tamura et al., 2006). A later study attributed these deficiencies to the loss of CL in the absence of Tam41 (Kutik et al., 2008). Similarly, the interaction of TIM and PAM complexes is affected in mitochondria that lack the CL synthase Crd1 or Ups1 (Kutik et al., 2008; Tamura et al., 2009). Interestingly, an altered electrophoretic mobility of another protein translocase of the inner membrane, the TIM22 complex mediating the membrane insertion of metabolite carrier proteins, was observed when Δcrd1 and Δtam41 mitochondria were analyzed, which may point to an altered assembly of the translocase or to a specific association of CL molecules with this complex (Kutik et al., 2008). Regardless, it appears from these studies that the reduced protein import into CL-deficient mitochondria is not simply the consequence of altered bioenergetics and a reduced membrane potential across the inner membrane, but rather reflects the specific requirement of CL for the functional integrity of the mitochondrial import machinery.
This view is supported by the recent observation that CL levels also regulate protein translocases in the outer membrane (Gebert et al., 2009), reconciling earlier observations that protein import into mitochondria can be inhibited by drugs binding to acidic phospholipids (Eilers et al., 1989) and is impaired in CL-deficient yeast cells (Jiang et al., 2000). The assembly of the import receptor Tom20 with the TOM complex as well as the organization of the SAM complex that mediates the assembly of β-barrel proteins in the outer membrane are altered in CL-deficient mitochondria (Fig. 4 B; Gebert et al., 2009). As a consequence, the biogenesis of β-barrel proteins in the outer membrane as well as that of proteins located in other mitochondrial subcompartments is impaired.
CL and apoptosis.
Further support for a functional role of CL in the mitochondrial outer membrane came from studies on the role of mitochondria during apoptosis, which revealed that CL regulates multiple steps of the apoptotic program (Fig. 4 C). Apoptosis can be induced by activation of the death receptor (Fas receptor) in the plasma membrane. Ligand-bound Fas receptor oligomerizes and recruits pro–caspase-8, which in response undergoes an autocatalytic processing step resulting in its activation. However, activation of caspase-8 at the plasma membrane was found to be insufficient for triggering apoptosis in some cells, and thus completion of the apoptotic program required a mitochondria-dependent feedback loop (Scaffidi et al., 1998). A recent study revealed that CL in the mitochondrial outer membrane provides an anchor and activating platform for caspase-8, which is processed and translocates to mitochondria upon Fas receptor activation (Gonzalvez et al., 2008).
Caspase-8–mediated cleavage of the BH3-only BID protein leads to its translocation to mitochondria. Truncated BID triggers activation of Bax and Bak, members of the Bcl2-family which induce outer membrane permeabilization and release of cytochrome c (Lovell et al., 2008). CL together with the major facilitator protein MTCH2/MIMP in the outer membrane regulates truncated BID recruitment to contact sites between the inner and outer membranes (Lutter et al., 2000; Lucken-Ardjomande et al., 2008; Sani et al., 2009; Zaltsman et al., 2010). Similarly, membrane insertion of Bax and its oligomerization were found to proceed more efficiently in the presence of CL (Lutter et al., 2000; Lucken-Ardjomande et al., 2008; Sani et al., 2009).
Finally, CL affects the release of cytochrome c from mitochondria during apoptosis. It binds directly to cytochrome c, retaining it within the cristae (Choi et al., 2007; Sinibaldi et al., 2008). The interaction between cytochrome c and CL is weakened upon peroxidation of the unsaturated acyl chains of CL (Nomura et al., 2000). The release of cytochrome c during apoptosis has been proposed to be further facilitated by remodeling of the mitochondrial cristae that facilitates the redistribution of cytochrome c molecules from the cristae lumen (Scorrano et al., 2002). Cristae morphology is controlled by the dynamin-like GTPase OPA1, a central component of the mitochondrial fusion machinery, whose activity is affected by CL (see next section). Thus, CL plays multiple roles during apoptosis in both mitochondrial membranes and may serve as a factor that coordinates the sequence of apoptotic events in mitochondria.
CL and mitochondrial dynamics.
Early studies with model membranes demonstrated that the formation of hexagonal structures induce membrane fusion and suggested a crucial role of non-bilayer lipids such as CL or PE for membrane fusion in vivo (Cullis and de Kruijff, 1979). Indeed, reductions of mitochondrial PE and CL levels were reported to result in abnormal mitochondrial morphology (Kawasaki et al., 1999; Steenbergen et al., 2005; Choi et al., 2006; Claypool et al., 2008a) and high frequency generation of respiratory deficient mitochondria (Birner et al., 2003; Zhong et al., 2004). Membrane fusion is mediated by evolutionarily conserved dynamin-like GTPases present in both mitochondrial membranes (Hoppins et al., 2007). In the inner membrane, OPA1 (or Mgm1 in yeast) is proteolytically processed, resulting in the balanced accumulation of long and short protein isoforms within mitochondria, both of which are required for mitochondrial fusion and cristae morphogenesis (Herlan et al., 2003; Ishihara et al., 2003; Song et al., 2007). Processing of yeast Mgm1 was affected in the absence of Ups1 or Ups2, which regulate the accumulation of CL and PE within mitochondria, respectively (Sesaki et al., 2006; Osman et al., 2009a). Impaired processing of Mgm1 could explain the aberrant morphology of mitochondria with an altered membrane lipid composition, but Mgm1 cleavage has so far not been analyzed in other CL-deficient cells, and other scenarios are conceivable. The short forms of both Mgm1 and OPA1 bind to negatively charged phospholipids, in particular CL, that stimulate its oligomerization and its GTPase activity (Fig. 4 D; DeVay et al., 2009; Meglei and McQuibban, 2009; Rujiviphat et al., 2009; Ban et al., 2010). It is possible that interaction with CL restricts the function of Mgm1/OPA1 to specific membrane domains, like contact sites between both mitochondrial membranes, which are known to be enriched in CL (Ardail et al., 1990).
These contact sites have been proposed to be the site of action of a phospholipase D, termed MitoPLD, which converts CL in the outer membrane to PA (Fig. 4 D; Choi et al., 2006). MitoPLD is required for mitochondrial fusion in vitro, and modulation of its expression in vivo causes morphological abnormalities (Choi et al., 2006). The formation of PA may allow the recruitment of additional fusion components or render membranes fusogenic. Such a role of PA would be reminiscent of SNARE-mediated fusion (Huang et al., 2005) and could point to a crucial role of local membrane lipid alterations in seemingly unrelated membrane fusion processes.
Perspectives
Recent discoveries have brought about significant progress in our understanding of the metabolism of mitochondrial phospholipids. This development was accompanied by a drastically altered view of the role that specific phospholipids play in various mitochondrial processes and the role of mitochondria in broader aspects of the biogenesis of nonmitochondrial membranes. Defined molecular functions of specific phospholipids, like CL, have been recognized, and the accumulation of these lipids in specific membrane domains is emerging as an important property of mitochondrial membranes. The recent identification of novel genes in yeast affecting the phospholipid composition of mitochondria, many of them conserved in mammals, now promises to provide insight into some of the mysteries of mitochondrial phospholipid metabolism and trafficking. Mitochondria may prove once again to be an excellent model to unravel basic cell biological processes that will be relevant to other membrane systems. Undoubtedly, exciting discoveries are just around the corner.
Acknowledgments
We thank members of our laboratories for helpful discussions and Dr. T. Wai for critically reading the manuscript.
This work was supported by a National Institutes of Health grant (5R37-GM32453) to D.R. Voelker and grants from the Deutsche Forschungsgemeinschaft (SFB635) and the European Research Council (ERC-AdG-233078) to T. Langer.
Footnotes
Abbreviations used in this paper:
- AAC
- ATP/ADP carrier
- CL
- cardiolipin
- ERMES
- ER–mitochondria encounter structure
- G3P
- glycerol-3-phosphate
- GPAT
- G3P acyltransferase
- MAM
- mitochondria-associated membrane
- mtDNA
- mitochondrial DNA
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PGP
- PG phosphate
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- SAM
- sorting and assembly machinery
References
- Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S.D., Perktold A., Zellnig G., Daum G. 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264:545–553 10.1046/j.1432-1327.1999.00658.x [DOI] [PubMed] [Google Scholar]
- Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265:18797–18802 [PubMed] [Google Scholar]
- Ardail D., Lerme F., Louisot P. 1991. Involvement of contact sites in phosphatidylserine import into liver mitochondria. J. Biol. Chem. 266:7978–7981 [PubMed] [Google Scholar]
- Ardail D., Gasnier F., Lermé F., Simonot C., Louisot P., Gateau-Roesch O. 1993. Involvement of mitochondrial contact sites in the subcellular compartmentalization of phospholipid biosynthetic enzymes. J. Biol. Chem. 268:25985–25992 [PubMed] [Google Scholar]
- Ban T., Heymann J.A., Song Z., Hinshaw J.E., Chan D.C. 2010. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19:2113–2122 10.1093/hmg/ddq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A., Rechberger G., Knauer H., Wolinski H., Kohlwein S.D., Leber R. 2009. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J. Biol. Chem. 284:11572–11578 10.1074/jbc.M805511200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.H., Sogo L.F., Yaffe M.P. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136:545–553 10.1083/jcb.136.3.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S., D’Adamo P., Maestrini E., Gedeon A.K., Bolhuis P.A., Toniolo D. 1996. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12:385–389 10.1038/ng0496-385 [DOI] [PubMed] [Google Scholar]
- Birner R., Bürgermeister M., Schneiter R., Daum G. 2001. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell. 12:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Nebauer R., Schneiter R., Daum G. 2003. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell. 14:370–383 10.1091/mbc.E02-05-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I.R., Nowakowski D.W., Yang H.C., Chung H., Karmon S., Royes P., Pon L.A. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 14:4618–4627 10.1091/mbc.E03-04-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.C., Heacock P.N., Clancey C.J., Dowhan W. 1998. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273:9829–9836 10.1074/jbc.273.16.9829 [DOI] [PubMed] [Google Scholar]
- Chicco A.J., Sparagna G.C. 2007. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292:C33–C44 10.1152/ajpcell.00243.2006 [DOI] [PubMed] [Google Scholar]
- Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8:1255–1262 10.1038/ncb1487 [DOI] [PubMed] [Google Scholar]
- Choi S.Y., Gonzalvez F., Jenkins G.M., Slomianny C., Chretien D., Arnoult D., Petit P.X., Frohman M.A. 2007. Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell Death Differ. 14:597–606 10.1038/sj.cdd.4402020 [DOI] [PubMed] [Google Scholar]
- Clancey C.J., Chang S.C., Dowhan W. 1993. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268:24580–24590 [PubMed] [Google Scholar]
- Claypool S.M., McCaffery J.M., Koehler C.M. 2006. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174:379–390 10.1083/jcb.200605043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., Boontheung P., McCaffery J.M., Loo J.A., Koehler C.M. 2008a. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell. 19:5143–5155 10.1091/mbc.E08-09-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., Oktay Y., Boontheung P., Loo J.A., Koehler C.M. 2008b. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182:937–950 10.1083/jcb.200801152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeau A., Nachbaur J., Vignais P.M. 1971. Enzymic characterization and lipid composition of rat liver subcellular membranes. Biochim. Biophys. Acta. 249:462–492 10.1016/0005-2736(71)90123-4 [DOI] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., Hajnóczky G. 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174:915–921 10.1083/jcb.200604016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis P.R., de Kruijff B. 1979. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta. 559:399–420 [DOI] [PubMed] [Google Scholar]
- Daum G., Lees N.D., Bard M., Dickson R. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 14:1471–1510 [DOI] [PubMed] [Google Scholar]
- de Brito O.M., Scorrano L. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- Dee C.T., Moffat K.G. 2005. A novel family of mitochondrial proteins is represented by the Drosophila genes slmo, preli-like and real-time. Dev. Genes Evol. 215:248–254 10.1007/s00427-005-0470-4 [DOI] [PubMed] [Google Scholar]
- DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H., Nunnari J. 2009. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186:793–803 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C., Bramkamp M. 2009. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology. 155:1786–1799 10.1099/mic.0.025312-0 [DOI] [PubMed] [Google Scholar]
- Dzugasová V., Obernauerová M., Horváthová K., Vachová M., Záková M., Subík J. 1998. Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Curr. Genet. 34:297–302 10.1007/s002940050399 [DOI] [PubMed] [Google Scholar]
- Eilers M., Endo T., Schatz G. 1989. Adriamycin, a drug interacting with acidic phospholipids, blocks import of precursor proteins by isolated yeast mitochondria. J. Biol. Chem. 264:2945–2950 [PubMed] [Google Scholar]
- Emoto K., Kuge O., Nishijima M., Umeda M. 1999. Isolation of a Chinese hamster ovary cell mutant defective in intramitochondrial transport of phosphatidylserine. Proc. Natl. Acad. Sci. USA. 96:12400–12405 10.1073/pnas.96.22.12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R.F., Schlattner U., Wallimann T., Lacombe M.L., Epand R.M. 2007. Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys. J. 92:126–137 10.1529/biophysj.106.092353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaigg B., Simbeni R., Hrastnik C., Paltauf F., Daum G. 1995. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta. 1234:214–220 10.1016/0005-2736(94)00287-Y [DOI] [PubMed] [Google Scholar]
- Gallas M.R., Dienhart M.K., Stuart R.A., Long R.M. 2006. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell. 17:4051–4062 10.1091/mbc.E06-04-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N., Joshi A.S., Kutik S., Becker T., McKenzie M., Guan X.L., Mooga V.P., Stroud D.A., Kulkarni G., Wenk M.R., et al. 2009. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr. Biol. 19:2133–2139 10.1016/j.cub.2009.10.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil V.M., Thompson M.N., Greenberg M.L. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280:35410–35416 10.1074/jbc.M505478200 [DOI] [PubMed] [Google Scholar]
- Gonzalvez F., Schug Z.T., Houtkooper R.H., MacKenzie E.D., Brooks D.G., Wanders R.J., Petit P.X., Vaz F.M., Gottlieb E. 2008. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 183:681–696 10.1083/jcb.200803129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., Wu W.I., Carman G.M. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281:9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhövd C., Neupert W., Reichert A.S. 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278:27781–27788 10.1074/jbc.M211311200 [DOI] [PubMed] [Google Scholar]
- Hobbs A.E., Srinivasan M., McCaffery J.M., Jensen R.E. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410 10.1083/jcb.152.2.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76:751–780 10.1146/annurev.biochem.76.071905.090048 [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Vaz F.M. 2008. Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 65:2493–2506 10.1007/s00018-008-8030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Turkenburg M., Poll-The B.T., Karall D., Pérez-Cerdá C., Morrone A., Malvagia S., Wanders R.J., Kulik W., Vaz F.M. 2009. The enigmatic role of tafazzin in cardiolipin metabolism. Biochim. Biophys. Acta. 1788:2003–2014 10.1016/j.bbamem.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Huang P., Altshuller Y.M., Hou J.C., Pessin J.E., Frohman M.A. 2005. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol. Biol. Cell. 16:2614–2623 10.1091/mbc.E04-12-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Jofuku A., Eura Y., Mihara K. 2003. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 301:891–898 10.1016/S0006-291X(03)00050-0 [DOI] [PubMed] [Google Scholar]
- Jiang F., Ryan M.T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M.L. 2000. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275:22387–22394 10.1074/jbc.M909868199 [DOI] [PubMed] [Google Scholar]
- Joshi A.S., Zhou J., Gohil V.M., Chen S., Greenberg M.L. 2009. Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta. 1793:212–218 10.1016/j.bbamcr.2008.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F., Shoda M., Harashima R., Sadaie Y., Hara H., Matsumoto K. 2004. Cardiolipin domains in Bacillus subtilis marburg membranes. J. Bacteriol. 186:1475–1483 10.1128/JB.186.5.1475-1483.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., Kuge O., Chang S.C., Heacock P.N., Rho M., Suzuki K., Nishijima M., Dowhan W. 1999. Isolation of a chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J. Biol. Chem. 274:1828–1834 10.1074/jbc.274.3.1828 [DOI] [PubMed] [Google Scholar]
- Kodaki T., Yamashita S. 1987. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262:15428–15435 [PubMed] [Google Scholar]
- Kodaki T., Yamashita S. 1989. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur. J. Biochem. 185:243–251 10.1111/j.1432-1033.1989.tb15109.x [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Daum G., Paltauf F. 1986. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165:901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O., Nishijima M. 1997. Phosphatidylserine synthase I and II of mammalian cells. Biochim. Biophys. Acta. 1348:151–156 [DOI] [PubMed] [Google Scholar]
- Kutik S., Rissler M., Guan X.L., Guiard B., Shui G., Gebert N., Heacock P.N., Rehling P., Dowhan W., Wenk M.R., et al. 2008. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J. Cell Biol. 183:1213–1221 10.1083/jcb.200806048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Nett J.H., Trumpower B.L., Hunte C. 2001. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20:6591–6600 10.1093/emboj/20.23.6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts V.A., Klig L.S., Bae-Lee M., Carman G.M., Henry S.A. 1983. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc. Natl. Acad. Sci. USA. 80:7279–7283 10.1073/pnas.80.23.7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vance D.E. 2008. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 49:1187–1194 10.1194/jlr.R700019-JLR200 [DOI] [PubMed] [Google Scholar]
- Liu J., Dai Q., Chen J., Durrant D., Freeman A., Liu T., Grossman D., Lee R.M. 2003. Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol. Cancer Res. 1:892–902 [PubMed] [Google Scholar]
- Liu J., Epand R.F., Durrant D., Grossman D., Chi N.W., Epand R.M., Lee R.M. 2008. Role of phospholipid scramblase 3 in the regulation of tumor necrosis factor-alpha-induced apoptosis. Biochemistry. 47:4518–4529 10.1021/bi701962c [DOI] [PubMed] [Google Scholar]
- Lovell J.F., Billen L.P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D.W. 2008. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 135:1074–1084 10.1016/j.cell.2008.11.010 [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S., Montessuit S., Martinou J.C. 2008. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 15:929–937 10.1038/cdd.2008.9 [DOI] [PubMed] [Google Scholar]
- Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. 2000. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2:754–761 10.1038/35036395 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Nunnari J. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglei G., McQuibban G.A. 2009. The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion. Biochemistry. 48:1774–1784 10.1021/bi801723d [DOI] [PubMed] [Google Scholar]
- Meisinger C., Rissler M., Chacinska A., Szklarz L.K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H.E., Yaffe M.P., et al. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 7:61–71 10.1016/j.devcel.2004.06.003 [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E., Dowhan W. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182:1172–1175 10.1128/JB.182.4.1172-1175.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndebele K., Gona P., Jin T.G., Benhaga N., Chalah A., Degli-Esposti M., Khosravi-Far R. 2008. Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) induced mitochondrial pathway to apoptosis and caspase activation is potentiated by phospholipid scramblase-3. Apoptosis. 13:845–856 10.1007/s10495-008-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebauer R., Schuiki I., Kulterer B., Trajanoski Z., Daum G. 2007. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J. 274:6180–6190 10.1111/j.1742-4658.2007.06138.x [DOI] [PubMed] [Google Scholar]
- Nikawa J., Yamashita S. 1984. Molecular cloning of the gene encoding CDPdiacylglycerol-inositol 3-phosphatidyl transferase in Saccharomyces cerevisiae. Eur. J. Biochem. 143:251–256 10.1111/j.1432-1033.1984.tb08366.x [DOI] [PubMed] [Google Scholar]
- Nishibori A., Kusaka J., Hara H., Umeda M., Matsumoto K. 2005. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187:2163–2174 10.1128/JB.187.6.2163-2174.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Imai H., Koumura T., Kobayashi T., Nakagawa Y. 2000. Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem. J. 351:183–193 10.1042/0264-6021:3510183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Nishijima M., Akamatsu Y. 1993a. A somatic cell mutant defective in phosphatidylglycerophosphate synthase, with impaired phosphatidylglycerol and cardiolipin biosynthesis. J. Biol. Chem. 268:22908–22913 [PubMed] [Google Scholar]
- Ohtsuka T., Nishijima M., Suzuki K., Akamatsu Y. 1993b. Mitochondrial dysfunction of a cultured Chinese hamster ovary cell mutant deficient in cardiolipin. J. Biol. Chem. 268:22914–22919 [PubMed] [Google Scholar]
- Ortiz A., Killian J.A., Verkleij A.J., Wilschut J. 1999. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys. J. 77:2003–2014 10.1016/S0006-3495(99)77041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Haag M., Potting C., Rodenfels J., Dip P.V., Wieland F.T., Brügger B., Westermann B., Langer T. 2009a. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184:583–596 10.1083/jcb.200810189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Merkwirth C., Langer T. 2009b. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122:3823–3830 10.1242/jcs.037655 [DOI] [PubMed] [Google Scholar]
- Osman C., Haag M., Wieland F.T., Brügger B., Langer T. 2010. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29:1976–1987 10.1038/emboj.2010.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G.J., Brandolin G. 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 426:39–44 10.1038/nature02056 [DOI] [PubMed] [Google Scholar]
- Pfeiffer K., Gohil V., Stuart R.A., Hunte C., Brandt U., Greenberg M.L., Schägger H. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278:52873–52880 10.1074/jbc.M308366200 [DOI] [PubMed] [Google Scholar]
- Potting C., Wilmes C., Engmann T., Osman C., Langer T. 2010. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 29:2888–2898 10.1038/emboj.2010.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof W.R., Wu J., Gijón M.A., Zarini S., Murphy R.C., Voelker D.R. 2007. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 282:36853–36861 10.1074/jbc.M706718200 [DOI] [PubMed] [Google Scholar]
- Rietveld A.G., Killian J.A., Dowhan W., de Kruijff B. 1993. Polymorphic regulation of membrane phospholipid composition in Escherichia coli. J. Biol. Chem. 268:12427–12433 [PubMed] [Google Scholar]
- Rujiviphat J., Meglei G., Rubinstein J.L., McQuibban G.A. 2009. Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284:28682–28686 10.1074/jbc.M109.044933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol A.E., Cui Z., Chen M.H., Vance J.E. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269:27494–27502 [PubMed] [Google Scholar]
- Sani M.A., Dufourc E.J., Gröbner G. 2009. How does the Bax-alpha1 targeting sequence interact with mitochondrial membranes? The role of cardiolipin. Biochim. Biophys. Acta. 1788:623–631 10.1016/j.bbamem.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K.J., Debatin K.M., Krammer P.H., Peter M.E. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675–1687 10.1093/emboj/17.6.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M., Haldar D. 1993. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 268:74–79 [PubMed] [Google Scholar]
- Schlame M., Ren M. 2009. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta. 1788:2080–2083 10.1016/j.bbamem.2009.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O., Pfanner N., Meisinger C. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11:655–667 10.1038/nrm2959 [DOI] [PubMed] [Google Scholar]
- Schumacher M.M., Choi J.Y., Voelker D.R. 2002. Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J. Biol. Chem. 277:51033–51042 10.1074/jbc.M205301200 [DOI] [PubMed] [Google Scholar]
- Scorrano L., Ashiya M., Buttle K., Weiler S., Oakes S.A., Mannella C.A., Korsmeyer S.J. 2002. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2:55–67 10.1016/S1534-5807(01)00116-2 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Dunn C.D., Iijima M., Shepard K.A., Yaffe M.P., Machamer C.E., Jensen R.E. 2006. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J. Cell Biol. 173:651–658 10.1083/jcb.200603092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Heacock P.N., Clancey C.J., Dowhan W. 1996. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271:789–795 10.1074/jbc.271.2.789 [DOI] [PubMed] [Google Scholar]
- Shiao Y.J., Lupo G., Vance J.E. 1995. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270:11190–11198 10.1074/jbc.270.19.11190 [DOI] [PubMed] [Google Scholar]
- Shinzawa-Itoh K., Aoyama H., Muramoto K., Terada H., Kurauchi T., Tadehara Y., Yamasaki A., Sugimura T., Kurono S., Tsujimoto K., et al. 2007. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26:1713–1725 10.1038/sj.emboj.7601618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbeni R., Pon L., Zinser E., Paltauf F., Daum G. 1991. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 266:10047–10049 [PubMed] [Google Scholar]
- Sinibaldi F., Fiorucci L., Patriarca A., Lauceri R., Ferri T., Coletta M., Santucci R. 2008. Insights into cytochrome c-cardiolipin interaction. Role played by ionic strength. Biochemistry. 47:6928–6935 10.1021/bi800048v [DOI] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178:749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen R., Nanowski T.S., Beigneux A., Kulinski A., Young S.G., Vance J.E. 2005. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280:40032–40040 10.1074/jbc.M506510200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.F., III, Kishida T., Christenson L.K., Fujimoto T., Hiroi H. 2003. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol. Cell. Endocrinol. 202:59–65 [DOI] [PubMed] [Google Scholar]
- Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M.R., Cavagna D., Nagy A.I., Balla T., Rizzuto R. 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175:901–911 10.1083/jcb.200608073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Harada Y., Yamano K., Watanabe K., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. 2006. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174:631–637 10.1083/jcb.200603087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Endo T., Iijima M., Sesaki H. 2009. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185:1029–1045 10.1083/jcb.200812018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Iijima M., Sesaki H. 2010. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 29:2875–2887 10.1038/emboj.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Model K., Langer T. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell. 16:248–259 10.1091/mbc.E04-09-0807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter P.J., Voelker D.R. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062–6070 10.1074/jbc.270.11.6062 [DOI] [PubMed] [Google Scholar]
- Trotter P.J., Pedretti J., Voelker D.R. 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268:21416–21424 [PubMed] [Google Scholar]
- van den Brink-van der Laan E., Killian J.A., de Kruijff B. 2004. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 1666:275–288 10.1016/j.bbamem.2004.06.010 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248–7256 [PubMed] [Google Scholar]
- Vance J.E. 1991. Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J. Biol. Chem. 266:89–97 [PubMed] [Google Scholar]
- Vance J.E. 2008. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J. Lipid Res. 49:1377–1387 10.1194/jlr.R700020-JLR200 [DOI] [PubMed] [Google Scholar]
- Voelker D.R. 1984. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. USA. 81:2669–2673 10.1073/pnas.81.9.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker D.R. 1990. Characterization of phosphatidylserine synthesis and translocation in permeabilized animal cells. J. Biol. Chem. 265:14340–14346 [PubMed] [Google Scholar]
- Voelker D.R. 2009. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu. Rev. Biochem. 78:827–856 10.1146/annurev.biochem.78.081307.112144 [DOI] [PubMed] [Google Scholar]
- Wendel A.A., Lewin T.M., Coleman R.A. 2009. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim. Biophys. Acta. 1791:501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Malhotra A., Ren M., Schlame M. 2006. The enzymatic function of tafazzin. J. Biol. Chem. 281:39217–39224 10.1074/jbc.M606100200 [DOI] [PubMed] [Google Scholar]
- Youngman M.J., Hobbs A.E., Burgess S.M., Srinivasan M., Jensen R.E. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164:677–688 10.1083/jcb.200308012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman Y., Shachnai L., Yivgi-Ohana N., Schwarz M., Maryanovich M., Houtkooper R.H., Vaz F.M., De Leonardis F., Fiermonte G., Palmieri F., et al. 2010. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat. Cell Biol. 12:553–562 10.1038/ncb2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Gohil V.M., Ma L., Greenberg M.L. 2004. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279:32294–32300 10.1074/jbc.M403275200 [DOI] [PubMed] [Google Scholar]
- Zinser E., Daum G. 1995. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 11:493–536 10.1002/yea.320110602 [DOI] [PubMed] [Google Scholar]