Polychaetoid coordinates receptor trafficking and signaling with adherens junction organization.
Abstract
Zona occludens (ZO) proteins are molecular scaffolds localized to cell junctions, which regulate epithelial integrity in mammals. Using newly generated null alleles, we demonstrate that polychaetoid (pyd), the unique Drosophila melanogaster ZO homologue, regulates accumulation of adherens junction–localized receptors, such as Notch, although it is dispensable for epithelial polarization. Pyd positively regulates Notch signaling during sensory organ development but acts negatively on Notch to restrict the ovary germline stem cell niche. In both contexts, we identify a core antagonistic interaction between Pyd and the WW domain E3 ubiquitin ligase Su(dx). Pyd binds Su(dx) directly, in part through a noncanonical WW-binding motif. Pyd also restricts epithelial wing cell numbers to control adult wing shape, a function associated with the FERM protein Expanded and independent of Su(dx). As both Su(dx) and Expanded regulate trafficking, we propose that a conserved role of ZO proteins is to coordinate receptor trafficking and signaling with junctional organization.
Introduction
Epithelial cells are polarized along their apicobasal axis and contact their neighbors through adherens junctions (AJs), which are mediated by homophylic interactions of E-cadherin (E-Cad) molecules on the adjacent cells. Other cell junctions, such as the tight junctions (TJs) in vertebrates and septate junctions (SJs) in invertebrates, are important in ensuring the impermeability of the epithelial sheet (Shin et al., 2006).
Multiple signaling pathway receptors and scaffold proteins are associated with these junctions, linking the adhesion complexes to different types of signaling components and to the cytoskeleton. The resulting protein network is important in the control of junctional dynamics, proliferation, and polarity. Zona occludens (ZO) proteins (ZO-1, -2, and -3) are scaffold proteins localized predominantly at TJs in mammalian epithelial cells (Wittchen et al., 1999; Fanning et al., 2007; Ikenouchi et al., 2007; Hartsock and Nelson, 2008). As well as having a well-described structural role in stabilizing Claudin clustering at the TJ (Shin et al., 2006), ZO proteins have been implicated in the regulation of cell cycle (Balda and Matter, 2000) and in the control of exocytosis (Köhler and Zahraoui, 2005), which remain poorly understood.
Drosophila melanogaster has a single ZO-1 homologue, Polychaetoid (Pyd), which has roles in junction remodeling during tracheal morphogenesis and pupal eye development (Jung et al., 2006; Seppa et al., 2008). The main isoform of Pyd appears to localize to AJs (Wei and Ellis, 2001), but it remains unclear whether Pyd contributes to epithelial polarity maintenance (Chen et al., 1996; Takahisa et al., 1996; Wei and Ellis, 2001). In addition, pyd alleles result in extra sensory bristles (macrochaetae) on the body of the adult fly, which is indicative of defects in the selection of sensory organ precursors (SOPs; Chen et al., 1996; Takahisa et al., 1996).
To gain insight into the different roles of ZO proteins, we generated null pyd alleles and used these to investigate developmental functions. Our results show that pyd is not essential for cell viability or for the maintenance of epithelial polarity but is important in regulating the apical domain. We also reveal new roles for Pyd in regulating niche and stem cell numbers in the ovaries and the wing shape. We find that the acidic and the C-terminal Pro-Rich domains of Pyd directly interact with the E3 ubiquitin ligase Su(dx), which regulates Notch trafficking (Cornell et al., 1999; Wilkin et al., 2004; Chastagner et al., 2008). Through these direct interactions, Pyd recruits Su(dx), and genetic assays indicate that the two proteins act antagonistically and that the Pyd–Su(dx) interaction impinges on Notch activity in SOP selection and niche regulation. In contrast, Pyd effects on wing shape are independent of Su(dx) and involve Expanded, a regulator of the Salvador–Warts–Hippo growth pathway.
Results
Null alleles of Pyd are viable but affect the levels of Notch and other AJ proteins
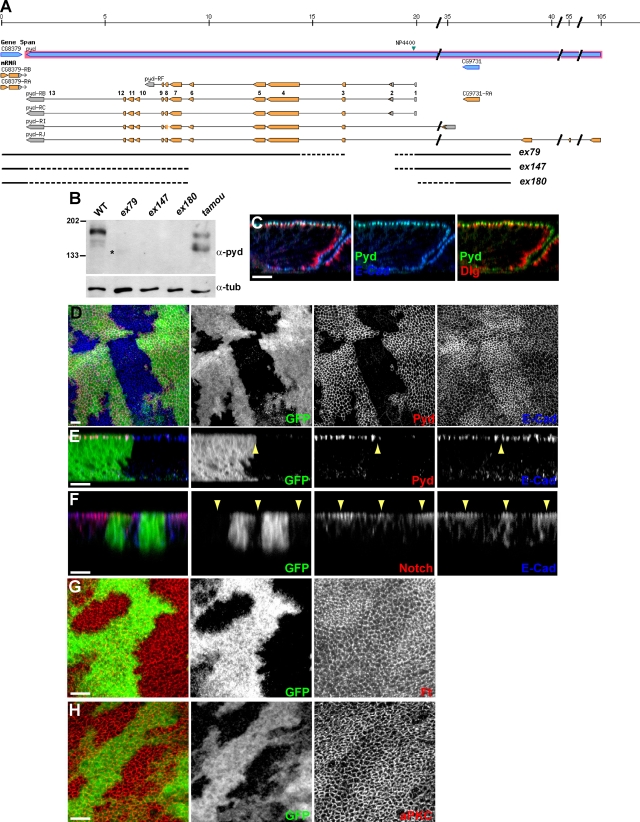
Many studies of pyd function have used the hypomorphic allele pydtamou. Of the other reported pyd alleles, several appear to have confounding second mutations on the chromosome and/or have little or no phenotype in combination with deficiencies removing the locus (Chen et al., 1996; Wei and Ellis, 2001; Jung et al., 2006). We therefore first generated new alleles of pyd by mobilizing the viable P element NP4400 located in the 5′ region of the gene. Two of the resulting three alleles (pydex147 and pydex180) have deletions that encompass exon 3, a coding exon shared by all the pyd isoforms (Fig. 1 A). In neither case is residual Pyd detectable in fly extracts (Fig. 1 B), suggesting these are null mutations. All three mutants recovered are viable as homozygotes and in trans to deficiencies uncovering the pyd locus. The viability and fertility of pydex147 and pydex180 imply that the Drosophila ZO-1 homologue is not essential for cell survival.
Figure 1.
AJ proteins accumulate in pyd mutant cells. (A) pyd locus (adapted from Flybase GBrowse) showing novel pyd alleles. Three (ex79, ex147, and ex180) deletions generated by mobilizing the viable NP4400 P element (green arrowhead) were mapped by PCR and are represented by interrupted lines (DNA absent; dotted regions indicate regions of uncertainty). pyd transcripts are represented by shaded boxes (salmon, coding; and gray, noncoding), and exons are numbered with respect to pyd-RB. (B) Western blot analysis of total protein extracts from larval brains and discs. (top) Western blot with polyclonal anti-Pyd showing that some residual Pyd polypeptide can be detected in pydex79 (*) and pydtamou, whereas no Pyd protein can be detected in pydex147 and pydex180. pydex79 is therefore likely to be a hypomorphic pyd allele, like pydtamou, whereas pydex147 and pydex180 are protein null alleles. (bottom) Western blot using a tubulin antibody as a loading control. All alleles were in trans with Df(3R)p-XT103 deleting the entire pyd locus. Molecular masses are given in kilodaltons. WT, wild type. (C) In imaginal disc epithelial cells, Pyd colocalizes with E-Cad at the AJ level, which is apical to an SJ marker, Dlg. Confocal z section. (D–H) In pyd mutant clones (arrowheads) marked by the absence of GFP, Pyd protein is absent (D and E), and the levels of E-Cad (D–F), Notch (F), Fat (Ft, G), and aPKC (H) are elevated. xy apical (D, G, and H) and z (E and F) confocal sections. Bars, 10 µm.
In the wing disc epithelium (and also in follicular epithelia; not depicted), Pyd is found at the AJ colocalizing with E-Cad and apical to the SJ marker Dlg (Fig. 1 C). The overall epithelial organization was largely unaffected in pyd mutant clones, as distinct AJs and SJs were still detectable in pyd mutant clones, and the levels and localization of the SJ proteins Scrib and Dlg were as in wild type (Fig. S1). However, the levels of several AJ-localized proteins, including E-Cad, Arm, Notch, and Fat, were elevated (Fig. 1, D–G; similar effects have been reported for E-Cad in the pupal retina; Seppa et al., 2008). Elevated levels of E-Cad and Notch were detected even with nonpermeabilized tissues, showing that the proteins accumulate at the membrane rather than inside the cell (Fig. S1). As Notch accumulation is unchanged under conditions in which E-Cad is expressed ectopically to cause expansion of the AJ (Fig. S1), it is unlikely that the accumulation of Notch and of the other AJ proteins is an indirect effect mediated via E-Cad. Furthermore, pyd mutant epithelial cells showed elevated accumulation of atypical PKC (aPKC), a marker of the subapical domain (Fig. 1 F), and a subtle expansion of the apical domain evident as an increase in apical membrane processes in transmission electron microscopy analysis (Fig. S1). Thus, pyd mutations affect the membrane accumulation of apical markers and AJ proteins, including aPKC, Notch, and Fat, although the overall structure and polarization of the epithelium appear unaltered. These phenotypes are reminiscent of, although weaker than, those observed for mutations affecting the Hippo pathway, which has been shown to regulate the apical domains and growth of epithelia (Genevet et al., 2009; Hamaratoglu et al., 2009). They are also in agreement with recent results suggesting that overall polarity is only marginally affected by ablation of all three ZO proteins in mammalian epithelial cells (Umeda et al., 2006).
Pyd is required for wing shape and ovarian niche size as well as SOP selection
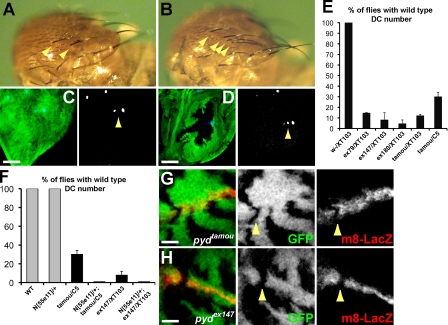
Although viable, the pyd mutant flies exhibit several phenotypes. First, as documented previously, pyd alleles display supernumerary chordotonal organs and extra macrochaetae (Chen et al., 1996; Takahisa et al., 1996). For example, 70% of pydtamou/pydC5 flies and >90% of pydex147 flies have extra (more than four) dorsocentral (DC) macrochaetae (Fig. 2, A, B, and E). This correlates with the presence of ectopic SOPs in imaginal discs containing clones of pyd mutant cells (Fig. 2, C and D). In addition, the extra macrochaetae phenotype of pydtamou/pydC5 and of the null pydex147/Df(3R)p-XT103 were enhanced by a reduction in Notch (N/+) so that 100% of flies emerged with extra DC macrochaetae (Fig. 2 F). Furthermore, expression of the E(spl)m8–lacZ Notch pathway reporter was decreased both in pydtamou and in pydex147 mutant cells, although the effects were not fully penetrant (seen in 2/4 pydex147 clones and 1/4 pydtamou clones that crossed the wing dorsoventral boundary; Fig. 2 G). Nevertheless, this effect and the genetic interactions suggest that Pyd has a positive influence on Notch signaling in this context, acting either upstream or in parallel to Notch.
Figure 2.
Notch pathway activity is impaired in pyd mutant proneural clusters. (A–D) Extra sensory bristles in pyd mutants. Wild-type (WT) flies have four dorsocentral (DC) macrochaetae (A, arrowheads indicate two left DCs), and pyd mutants have greater than four DCs (B, pydex147/Df(3R)p-XT103, arrowheads). DC sensory bristle precursors are detected by anti-Senseless (C and D, blue and arrowheads). Two are detected in a wild-type wing imaginal disc (C); extra Senseless-positive cells are detected in pyd mutant clones (pydex147 marked by the absence of GFP; D, green). Bars, 25 µm. (E) Percentage of wild-type (w-) and pyd mutant flies with the normal complement of four DC macrochaetae (indicated pyd alleles were in trans to Df(3R)p-XT103). Subsequent experiments used the hypomorphic combination pydtamou/pydC5 in which ∼30% of pydtamou/pydC5 flies have four DC macrochaetae. n ≥ 4. (F) Percentage of flies with four DC macrochaetae in the indicated genotypes. Gray bars, control genotypes; 100% have four DC macrochaetae. Black bars, pyd mutant flies ± reduction in Notch. A pyd DC phenotype is manifested by fewer flies having four macrochaetae and is strongly enhanced by removing one copy of Notch (P < 0.0001). n = 4. (E and F) Error bars represent SD. (G and H) Expression of the Notch reporter E(spl)m8–lacZ (red) is lost in several hypomorphic pydtamou (G) or null pydex147 (H) pyd mutant clones that cross the dorsoventral boundary (e.g., arrowheads). Confocal xy section, pyd clones marked by the absence of GFP. Bars, 10 µm.
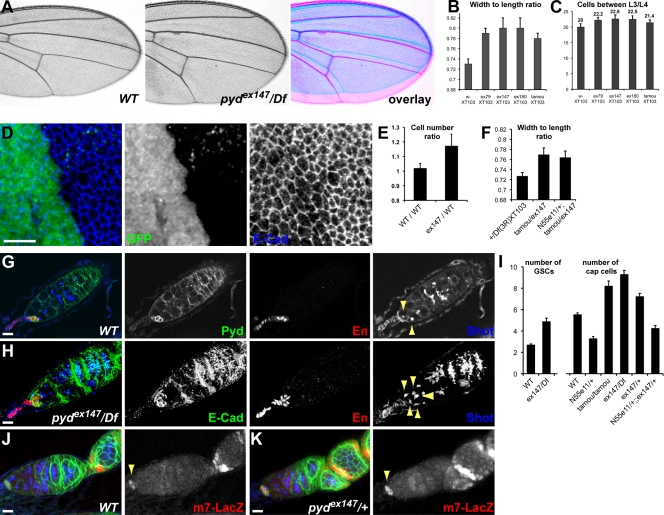
Second, we noted that pyd-null mutants have broader wings than wild type (Fig. 3 A). To quantify the phenotype, we measured the width and the length of the wings and assessed whether there was a difference in this ratio (allowing us to control for overall body size). All pyd alleles, including the hypomorphic pydtamou, had an increased ratio indicative of broader wings (Fig. 3 B). This correlated with a 10% increase in the number of cells found between the L3 and L4 veins (Fig. 3 C). In the imaginal disc, counting cell apices in equivalent areas of adjacent heterozygous and homozygous cells, the homozygous mutant pyd cells appeared smaller, and there were ∼15% more cells per surface unit (Fig. 3, D and E). In contrast to the sensory organ context, the effect of pyd on wing shape was not modified by changes in the dosage of Notch (Fig. 3 F).
Figure 3.
pyd mutants have broader wings and enlarged niche of the female germline stem cells (GSCs). (A) pyd mutant flies have broader wings than wild type. An overlay is presented on the right. (B and C) Quantification of a pyd broad-wing phenotype in females bearing the indicated pyd alleles in trans with Df(3R)p-XT103. (B) Wing width/length ratio. This ratio normalizes for differences in whole body size; the bigger the ratio, the broader the wings (P < 0.0001). n ≥ 40. (C) Number of cells found between L3 and L4 veins. As each cell forms one trichome, the number of trichomes was counted along a defined segment between the L3 and L4 veins extending from the intersection of the posterior cross vein (P < 0.0001). n = 20. (D) pyd mutant clones in the larval wing disc, which were marked by the absence of GFP. Mutant cells, outlined by E-Cad, are smaller and more packed than in neighboring territories. (E) Ratio of cell numbers in wild-type (WT) versus pyd mutant territories. Cells were counted in a given area in pyd mutant clones and in adjacent wild-type tissues and expressed as a ratio (pyd147/WT). The ratio between two immediately adjacent wild-type tissues was scored as a control (WT/WT; P < 0.0001). n = 10. (F) Removing one copy of Notch does not modify the width/length ratio of the hypomorphic pydtamou/pydex147 combination (P = 0.25). n = 15–20. (B, C, E, and F) Error bars represent SD. (G) Expression of Pyd in the GSC niche. Pyd is localized around the cortex of niche cap cells marked with Engrailed. The spectrosome is marked with Shot to identify GSCs where the spectrosome forms a dot or exclamation mark (arrowheads). (H) pyd mutant germaria have an increased number of cap cells marked by Engrailed and of GSCs marked by Shot (arrowheads). (I) Quantification of the number of GSCs and cap cells per germarium in wild-type and indicated genotypes (error bars are standard error of the mean; n = 20–40; P < 0.0001). (J and K) E(spl)m7–lacZ expression (red) in the cap cells of wild-type ovarioles (J, arrowhead) becomes expanded in the pydex147/+ heterozygous ovarioles (K, arrowhead) where extra cap cells form. Ovarioles were also stained with Pyd (green) to mark the cap cells and Shot (blue) to mark the spectrosome and highlight the GSCs. Bars, 10 µm.
Third, we uncovered a novel function for Pyd in controlling the size of the ovary germline stem cell (GSC) niche, the cap cells (Fig. 3, G and H). Pyd is highly expressed in the cap cells, where it localizes at the cortex (Fig. 3 G). In heterozygous and homozygous pyd mutants, the numbers of niche cap cells and of stem cells per ovariole were increased (Fig. 3, G–I). Interestingly, this phenotype, observed in independently derived pyd alleles, is characteristic of increased Notch signaling (Ward et al., 2006), suggesting that Pyd has the opposite effect on the outcome of the Notch signaling in the niche cells and in the SOPs. In support of this, we found that there were fewer cap cells and germ cells in Notch heterozygous ovarioles and that Notch alleles suppressed the increased cap cell numbers of pyd (Fig. 3 I). Furthermore, there was a strong expression of E(spl)m7–lacZ, a reporter of Notch activity, associated with the extra cap cells in pydex147/+ heterozygous ovarioles (Fig. 3, J and K), which is consistent with an increase in Notch signaling when pyd function is impaired. These genetic interactions suggest that the relationship between Pyd and Notch is context dependent and that Pyd is therefore likely to influence Notch in conjunction with additional factors.
Domains required for Pyd function
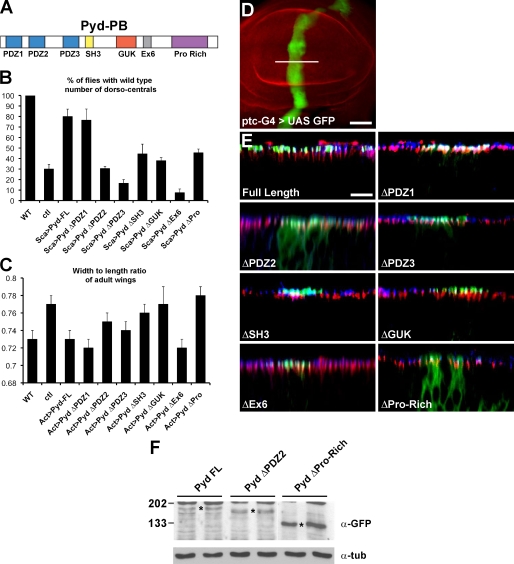
Pyd is a membrane-associated guanylate kinase protein containing three PDZ domains, one SH3 domain, one guanylate pseudokinase domain, an acidic domain encoded by the alternatively spliced sixth exon (Ex6), and a C-terminal Pro-Rich domain (Fig. 4 A). As a first step to identify other factors acting in conjunction with Pyd, we tested the requirements for the different domains in Pyd functions in the sensory organs and wing shape development.
Figure 4.
Domains of Pyd required for function and localization. (A) Schematic representation of Pyd/ZO-1; PDZ, SH3, Pro-Rich protein interaction domains, guanylate kinase domain (GUK), and alternatively spliced acidic Ex6 domain are indicated. (B) Pyd domains required for normal sensory organ specification. Graph showing the extent of rescue provided by expressing different GFP-Pyd variants (using scabrous (Sca)-Gal4) in pydtamou/pydC5. Full-length pyd (Pyd-FL) and PydΔPDZ1 both restored wild-type (WT) DC numbers to >80%. Other variants gave partial (PydΔSH3 and PydΔPro-Rich) or no rescue (PydΔPDZ2, PydΔPDZ3, PydΔGUK, and PydΔEx6). n = 4. ctl, control. (C) Pyd domains required for normal wing shape. Graph showing the extent of rescue provided by expressing different GFP-Pyd variants (using actin (Act)-Gal4) in pydtamou/pydex147. Full-length pyd, PydΔPDZ1, and PydΔEx6 restored the width/length ratio to wild type. Other variants gave partial (PydΔPDZ3) or no rescue (PydΔPDZ2, PydΔSH3, PydΔGUK, and PydΔPro-Rich). n = 15–20. (B and C) Error bars represent SD. (D) Expression domain of patched (ptc)-Gal4 (visualized with UAS-GFP) used to express N-terminal GFP-tagged Pyd variants in the wing disc in E. Bar, 50 µm. (E) Pyd domains required for localization. High power Z sections (corresponding to the position indicated by the white line in D) from discs expressing the GFP-Pyd variants indicated (green). Most colocalize with E-Cad (blue) at the AJ, apical to SJ (Dlg; red), except ΔPDZ2 and ΔPro-Rich, which are detected throughout the cytoplasm. Bar, 10 µm. (F) Expression levels of Pyd variants. Western blot of protein lysates from larval brain and discs. Two independent lines per GFP-Pyd construct are shown for the GFP-Pyd full length, GFP-Pyd ΔPDZ2, and GFP-Pyd ΔPro-Rich. (top) Western blot of extracts from imaginal discs expressing the GFP-Pyd variants indicated (duplicate lanes are shown for each), which were probed with anti-GFP (Asterisks mark the positions of GFP-Pyd bands). GFP-Pyd ΔPro-Rich is present at much higher levels than GFP-Pyd and GFP-PydΔPDZ2 (and other deletion mutants; not depicted). (bottom) Western blot probed with antitubulin (α-tub) as a loading control. Molecular masses are given in kilodaltons.
When expressed in the proneural clusters, full-length Pyd and PydΔPDZ1 restored the percentage of flies with the wild-type number of DC bristles from 30 to ∼80%, which was indicative of a significant rescue of the mutant phenotype (Fig. 4 B). In contrast, deletion of any of the four other domains, ΔPDZ2, ΔPDZ3, ΔGUK, and ΔEx6, generated proteins that failed to provide any rescue of bristle numbers. The effects of PydΔSH3 and PydΔPro-Rich were intermediate, with a low level of rescue detected (Fig. 4 B). The effects of ΔEx6 were the most striking because they produced more severe phenotypes, suggesting that PydΔEx6 behaves as a dominant negative. These results highlight a critical role for the Ex6 domain in Pyd function, which is consistent with previous observations (Wei and Ellis, 2001).
When expressed at low levels in the wing, PydΔPDZ2, ΔGUK, ΔSH3, and ΔPro-Rich all failed to rescue, suggesting that these domains have a role in the wing shape function (Fig. 4 C). Strikingly, Ex6 is totally dispensable in this process because the PydΔEx6 was able to fully rescue the phenotype (Fig. 4 C). These results suggest that Pyd controls the overall shape of the wing and the number of sensory bristles via at least one different mechanism.
One explanation for the failure of specific Pyd variants to rescue the SOP and wing mutant phenotype is that they no longer localize to the AJ. We therefore expressed the GFP-tagged Pyd variants in the disc epithelium and compared their localization with cadherin (E-Cad). The majority of Pyd variants localized to the AJ region in the same manner as full-length Pyd (Fig. 4 E) and were expressed at similar levels (Fig. 4 F and not depicted). However, ΔPDZ2 and ΔPro-Rich both failed to localize properly, and the mutant protein was detected throughout the cytoplasm (Fig. 4 E). PydΔPro-Rich was also present at significantly higher levels (Fig. 4 F and not depicted), suggesting that the Pro-Rich domain is also important in regulating Pyd stability. Similar results were obtained when the Pyd variants were expressed in clones of cells mutant for pyd, confirming that only the PDZ2 and Pro-Rich domains are important for localization even in the absence of any wild-type protein (unpublished data). The increased stability of PydΔPro-Rich may partially compensate for its mislocalization and/or weaker activity because this form of Pyd resulted in some rescue in the SOP assay.
A physical interaction between Pyd and Drosophila HECT domain ligases
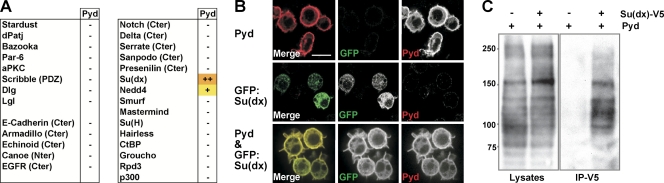
To gain insights into the mechanisms by which Pyd acts and given Pyd localization and effect on Notch signaling, we tested systematically whether Pyd could interact with components of the different epithelial polarity complexes and of the Notch pathway in a yeast two-hybrid assay. Of the fragments tested (note that not all parts of the proteins could be tested in some cases), we found that full-length Pyd interacted specifically with Su(dx) and Nedd4 (Fig. 5 A), E3 ubiquitin ligases that have been shown to regulate the endocytic trafficking of Notch and to influence the outcome of signaling (Mazaleyrat et al., 2003; Sakata et al., 2004; Wilkin et al., 2004; Chastagner et al., 2008).
Figure 5.
Su(dx) binds to Pyd/ZO-1. (A) Indicated proteins were tested for binding to full-length Pyd in yeast two-hybrid assays; − indicates no interaction. Orange indicates strong growth on selective media (++). Yellow indicates slow growth on selective media (+). Cter, C terminal. Nter, N terminal. (B) Pyd recruits Su(dx) to the cell cortex in transfected cells. GFP-Su(dx) is primarily in the cytoplasm of transfected S2 cells. When cotransfected with Pyd, GFP-Su(dx) (green) is relocalized to the cell cortex, colocalizing with Pyd (red). Bar, 6 µm. (C) Su(dx) coimmunoprecipitates with Pyd. (left) Western blot of whole lysates from cells expressing Pyd with or without Su(dx)-V5. (right) Anti-Pyd Western blot after immunoprecipitation (IP) with anti-V5. Molecular masses are given in kilodaltons.
We further investigated the interaction between Pyd and Su(dx) using a recruitment assay in S2 cells. Su(dx), predominantly detected in the cytoplasm when transfected alone, became recruited at the cell cortex and colocalized with Pyd when the two were cotransfected (similar interactions were detected with Nedd4; Fig. 5 B and not depicted). In addition, Pyd could be coimmunoprecipitated with Su(dx) from cells cotransfected with Pyd and a V5-tagged Su(dx), suggesting that Su(dx) and Pyd are present in a complex (Fig. 5 C). Together, these results suggest a direct interaction between Pyd and Su(dx), which is sufficient to cause Su(dx) relocalization in cultured cells.
Interaction between Pyd and Su(dx) requires Ex6 and Pro-Rich domains
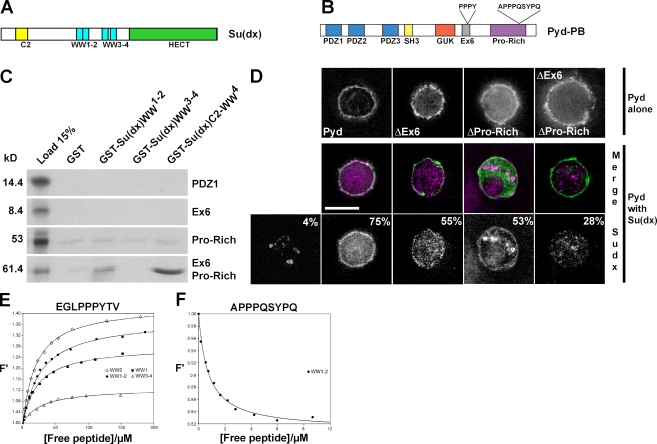
To investigate which domains are required for interactions with Su(dx) and whether Ex6 is important, different fragments of Pyd were tested in yeast two-hybrid and GST pull-down experiments (Fig. 6 C and not depicted). Robust interactions with Su(dx) GST fusion proteins and, in particular, with the Su(dx) WW domains were detected when Ex6 and Pro-Rich domains were both present (Fig. 6 C). This suggests that both regions of Pyd contribute to the interaction with Su(dx) and that this interaction is mediated by the WW domains of Su(dx). Furthermore, a peptide from Pyd Ex6 was independently isolated in an unbiased screen for proteins binding to WW3–4 of Su(dx) (Fig. S2), suggesting that this region is important for the interaction.
Figure 6.
The Ex6 and Pro-Rich domains of Pyd mediate the interaction with Su(dx). (A) Schematic representation of the E3 ubiquitin ligase Su(dx). C2 phospholipid-binding domain, WW protein interaction domains, and the catalytic HECT domain are indicated. (B) Schematic representation of Pyd/ZO-1 indicating candidate WW domain–interacting motifs. (C) Fragments of Pyd, as indicated, were tested for binding to GST fusion proteins containing the WW domains of Su(dx). Autoradiograph of 35S-labeled input proteins (input, left) and of eluted bound fractions from the indicated combinations. Only fragments containing both Ex6 and Pro-Rich domains were retained by GST-Su(dx)-WW. Weak binding was detected with the Pro-Rich domain alone, and no specific binding was detected with other domains (PDZ2, PDZ3, SH3, and GUK; not depicted). (D) Pyd domains required for Su(dx) recruitment. Su(dx) (HA-tag: purple, middle; white, bottom) is recruited by full-length and, to a lesser extent, by ΔPro-Rich or by ΔEx6 Pyd variants but not by ΔEx6ΔPro-Rich (GFP tags: green, middle). (top) S2 cells transfected with the Pyd constructs only and stained for GFP (white). Percentages in the bottom images indicate percentage of transfected cells with some HA-Su(dx) detected at the cell cortex as the mean of more than three experiments. Bar, 7 µm. (E and F) Tryptophan fluorescence change upon titration of Su(dx) WW domains with synthetic Pyd-derived peptides. Single (WW1 and WW2) and paired (WW1–2 and WW3–4) WW domains were tested with peptide sequences EGLPPPYTV (E) and APPPQSYPQ (F). F′ are normalized fluorescence values. The data were fitted to a single-site binding model, as described in Materials and methods, to obtain Ka (association constant) and converted to Kd (dissociation constant). Errors of fit are smaller than SDs from multiple experiments; SD and mean Kd values are reported in Table S1 for each WW domain–peptide combination.
Using the cell cortex recruitment assay reported earlier in this paper, we subsequently tested whether deleting the Ex6 and Pro-Rich domains of Pyd affected its ability to recruit Su(dx) (Fig. 6 D). Deletion of either domain alone reduced the ability to relocalize Su(dx). This was further compromised when the two domains were deleted together (PydΔEx6ΔPro-Rich). These results suggest that efficient relocalization of Su(dx) by Pyd requires both Ex6 and Pro-Rich domains.
WW domains bind to peptides with the consensus sequence PPxY (Chen and Sudol, 1995). To confirm that the Ex6 and Pro-Rich domains of Pyd contain Su(dx) binding sites, we used a solution assay in which the binding is detected by a change in fluorescence of a conserved tryptophan residue within the E3 ligase WW domains (Fig. 6, E and F). The Ex6 domain of Pyd contains a classical WW recognition motif, PPPY, that binds to individual Su(dx) WW repeats, WW1 and WW2, to the WW1–2 pair, and, less strongly, to the WW3–4 pair (Fig. 6 E). Although the Pro-Rich domain of Pyd does not contain any canonical PPxY motif, it does contain a related APPPQSYPQ peptide. Using the same assay, only the WW1–2 pair showed any binding to this peptide (Fig. 6 F). Significantly, the direction of the fluorescence change was reversed, suggesting an atypical mode of interaction. Calculations of the dissociation constant (Kd) indicate that this noncanonical peptide binds with a higher affinity than the classical PPPY motif from Ex6 (Kd = 0.7 ± 0.06 µM compared with Kd = 28.0 ± 4.1 µM; Table S1).
Functional interactions between Pyd and Su(dx)
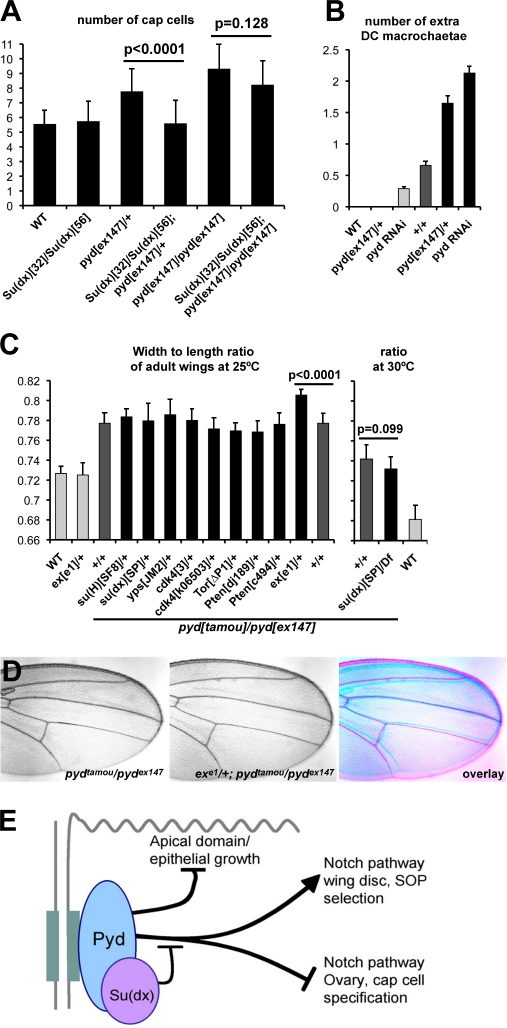
To explore the functional relationship between Su(dx) and pyd, we investigated the consequence of altering the dosage of the two genes. This approach required that we could identify developmental processes in which mutations produce distinct phenotypes that are modifiable. One such scenario is in the niche cells of the ovary (cap cells), where the pyd heterozygous combinations give rise to an increase in cap cells associated with more cells expressing the Notch activity reporter E(spl)m7–lacZ (Fig. 3, J and K). Here, the dominant effect of heterozygous pyd alleles on cap cell number was suppressed in combinations with alleles of Su(dx) (Fig. 7 A), arguing that Pyd and Su(dx) have opposing functions in cap cell regulation. In addition, expression of E(spl)m7–lacZ was reduced in Su(dx) homozygotes (Fig. S2), which was consistent with the hypothesis that effects of Su(dx) on Notch activity in this context are also opposite to those of Pyd. However, as mutations in Su(dx) were unable to suppress the elevated cap cell numbers in homozygous null pyd mutants, this suggests that Pyd and Su(dx) act together rather than independently to regulate cap cell number (Fig. 7 A). It further shows that the expanded niche phenotype resulting from complete loss of Pyd does not depend on the presence of functional Su(dx).
Figure 7.
Pyd acts through Su(dx) to control macrochaetae and cap cell numbers. (A) Number of cap cells in the female germaria of the stated genotypes. Su(dx) mutations suppress the extra cap cell phenotype of pyd mutants. n ≥ 40. (B) Number of extra DC macrochaetae from the different stated genotypes. dppG4 drives expression in a stripe that encompasses the scutellar and DC bristles. Light gray bars indicate the wild type (WT), pydex147/+, and dppG4>pyd RNAi controls; dark gray bars show the dppG4>Su(dx) baseline (+/+); and black bars represent experiments in which dpp-Gal4 drives the expression of Su(dx) in combination with altered pyd dosage as indicated (P < 0.001). n = 38–114. (C) Width/length ratio of adult female wings. Light gray bars indicate the wild-type and exe1/+ controls, dark gray bars show the pydtamou/pydex147 baseline (+/+), and black bars represent genetic combinations with the pydtamou/pydex147 hypomorphic background. exe1/+ showed an enhancement (P < 0.0001). None of the other tested genotypes, in particular the Su(dx) strong loss-of-function combination Su(dx)SP/Df(2L)Exel6006 at 30°C, modified the pydtamou/pydex147 broad-wing phenotype (all P ≥ 0.05). n = 15–20. (A–C) Error bars represent SD. (D) Removing one copy of ex enhances the broad-wing phenotype of hypomorphic pydtamou/pydex147. An overlay is shown on the right. (E) Summary of Pyd/ZO-1 function in Drosophila epithelia. Pyd is localized to AJs (gray) and regulates apical domain/epithelial growth, sensory organ precursor (SOP) numbers, and female GSC niche cap cells. The last two involve different effects on final output of the Notch pathway and are inhibited by recruitment of Su(dx) E3 ubiquitin ligase (pink).
We then looked into a possible modification of the phenotypes produced by Su(dx) overexpression in the imaginal discs. When overexpressed using a dpp-Gal4 driver, Su(dx) consistently caused a gain of macrochaetae, which was similar to pyd loss of function. This phenotype was enhanced by reducing pyd levels, suggesting that Pyd and Su(dx) can have antagonistic effects on bristle development (Fig. 7 B). Thus, Pyd and Su(dx) appear to have consistent antagonistic interactions in cap cells and macrochaetae, despite the fact that the outcome for Notch signaling differs.
To test the interaction between Pyd and Su(dx) in the control of wing shape, we investigated whether removing Su(dx) function could rescue a dominant phenotype produced in pydex147/+ heterozygous flies, measuring the number of cells between the L3 and L4 wing veins. In contrast with the niche phenotype, Su(dx) alleles did not rescue the wing shape phenotype in pyd heterozygotes (Table S2 and Table S3). We note also that neither Su(dx) mutant combinations (Fig. 7 C) nor changes in the dosage of Notch were able to modify the pyd wing shape phenotype (Fig. 3 F), further supporting a different effector in this process.
As ZO-1 is proposed to influence proliferation by trapping the Y-box factor ZONAB (ZO-1–associated nucleic acid binding protein) at junctions and, consequently, inhibiting CDK4 activity (Balda and Matter, 2000; Balda et al., 2003), we tested whether the wing phenotype could be modified by alleles affecting growth pathways. These included alleles of the Drosophila homologues of ZONAB (yps) and CDK4, of expanded (ex) involved in the control of epithelial growth through modulation of the Hippo pathway, and of members of the Tor and Pten growth pathways. Of those tested, only the combination with ex alleles significantly modified the pyd phenotype, resulting in an enhanced broad-wing ratio (ex/+ alone have normal-shaped wings; Fig. 7, C and D). Thus, the effects of pyd on wing shape appear to involve a pathway related to ex. Interestingly, ex mutant cells also show accumulation of the AJ proteins Notch, Cad, and Fat, albeit to higher levels than observed here in pyd mutant cells, in support of these results linking the functions of Pyd and Expanded (Maitra et al., 2006).
Discussion
ZO proteins are thought to be adaptors that link junction proteins with the cytoskeleton and with cytoplasmic factors (Fanning et al., 2007; Hartsock and Nelson, 2008). Here, we uncover a novel interaction between the Drosophila ZO-1 homologue Pyd and the cytosolic E3 ligase Su(dx). Su(dx) acts antagonistically on Pyd function, with tissue-specific consequences on Notch signaling.
Physical interactions between the E3 ubiquitin ligase Su(dx) and Pyd were identified by two complementary screening strategies and validated using a wide range of biochemical assays. Binding of Su(dx) WW domains to Pyd requires two motifs in the Ex6 and Pro-Rich domains of Pyd. The motif found in the Pro-Rich is of a novel type, PPxxY, which diverges from the canonical PPxY motif by an increased spacing between the tandem prolines and the tyrosine. A further novel feature is that this site only interacts with a pair of adjacent WW domains, specifically WW1–2, in contrast to conventional PPxY motifs, which interact with single WW domains. Both motifs together contribute to the Su(dx) recruitment in cells, and both are necessary for Pyd function during sensory organ number restriction.
Several data indicate that Su(dx) and Pyd interact antagonistically in two of the contexts we have analyzed. An antagonistic relationship is suggested by genetic interactions observed in the macrochaetae between overexpressed Su(dx) and pyd loss of function and by the suppression of pyd mutant phenotypes in the GSC niche by Su(dx) alleles. Although we were not able to carry out a detailed epistasis analysis in all tissues investigated, the phenotypes in cap cells are most consistent with a model in which Su(dx) antagonizes Pyd. This is because loss of Su(dx) suppressed the phenotypes caused by a reduction in Pyd but not those resulting from complete removal of Pyd, although the presence of additional Su(dx)-related E3 ligases (e.g., Nedd4) may complicate interpretations. Su(dx) may thus act by physically blocking the access to Pyd of another effector (e.g., Notch) or may modify either the effector or Pyd by ubiquitinylation. Ultimately, this may translate to Pyd having a role similar to the adaptor 14-3-3, which regulates the location and interaction of E3 ligases with their targets (Ichimura et al., 2005; Sato et al., 2006).
However, there are several points of complexity. One is on the outcome for Notch activity in different tissues. Our analysis suggests that Pyd normally limits the amount of Notch at the apical surface, most likely through its effects on the apical domain. However, the pyd phenotypes in the GSC niche and in the macrochaetae correlate with increased and decreased Notch signaling, respectively (Ward et al., 2006). As the core antagonistic interaction between Pyd and Su(dx) remains the same in all tissues, one possibility is that the consequences of the Pyd–Su(dx) interaction on Notch, for example ubiquitinylation, have different outcomes caused by tissue-specific differences in the downstream endosomal sorting of the Notch receptor brought about by effectors such as Deltex (Wilkin et al., 2008). Another possibility is that the differing effects on the output of the Notch signaling are related to the response window associated with each process. For example, if the accumulation of Notch at the membrane in pyd mutant cells is associated with slower endocytosis and cleavage/activation of Notch, this may impede those processes in which there are rapid transitions in activity, as in Notch-mediated lateral inhibition. On the other hand, where signaling is more extended, as in the ovarioles where the cap cells will persist for the life span of the adult female, the slower activation may not have a significant impact. Instead, the reduced turnover of an unliganded receptor could increase its availability for signaling, so leading to an elevated Notch response overall.
A second complexity is that Ex6 is required for bristle number but not for wing shape control. This implies differences in the mechanisms of Pyd action in these two contexts, suggesting that different Pyd partners are important for these two processes. Because Ex6 participates in the interaction between Pyd and Su(dx), Pyd could exert effects on wing shape independently of Su(dx). This conclusion is supported by the lack of detectable modifications to wing shape by combinations between pyd and Su(dx) loss-of-function mutations.
Mammalian ZO-1 has been reported to regulate cell proliferation by sequestering the Y-box transcription factor ZONAB and the cyclin regulator CDK4 outside of the nucleus (Balda and Matter, 2000; Balda et al., 2003). This is an unlikely explanation for the wing shape effects of Drosophila Pyd, as we were unable to detect any genetic interaction between pyd and yps, the unique fly ZONAB homologue. Furthermore, yps-null mutants are viable and have a similar size and shape of wing to wild type (unpublished data). A more likely candidate is the FERM domain protein Expanded, based on the genetic interactions between pyd and ex alleles. Expanded is proposed to cooperate with other cortically localized proteins to relay information about epithelial architecture to the Salvador–Warts–Hippo network and, hence, to control tissue growth (Hamaratoglu et al., 2006; Badouel et al., 2009), and the cortically localized Pyd may contribute to this relay of information. Like pyd, ex mutations also result in the accumulation of receptors at apical membranes (Maitra et al., 2006), and mutations in the Hippo pathway also result in increased apical domain (Genevet et al., 2009; Hamaratoglu et al., 2009), further strengthening a functional link between the two.
Our results suggest that whereas Pyd is not required for epithelial viability or polarization, it has effects on the regulation of Notch signaling and epithelial cell numbers, which are likely controlled by the endocytic regulators Su(dx) and Expanded. As mammalian ZO proteins have also been proposed to regulate membrane trafficking (Köhler and Zahraoui, 2005), it is possible that a conserved role of ZO family proteins may be to coordinate trafficking, apical domain structure, and cell signaling with the junctional organization of epithelial cells.
Materials and methods
Drosophila genetic experiments
New pyd alleles, pydex79, pydex147, and pydex180, were generated by imprecise excision of the P element NP4400. Other pyd alleles were pydtamou and pydC5 (gifts from R. Ueda, National Institute of Genetics, Mishima-shi, Japan) and the deletion Df(3R)p-XT103. Genetic interactions were performed at 25°C, assaying for modification of the extra DC macrochaetes of pydtamou/pydC5 or of the broad wing of pydtamou/pydex147 using the alleles stated. Details are available at Flybase. For the broad-wing phenotype, width and length were measured using fixed wing landmarks on 15–20 wings from independent females. scabrous[537.4]-Gal4 (proneural clusters), patched[559.1]-Gal4 (stripe in the wing disc), and actin-Gal4 (ubiquitous) were used to drive the expression of different pUAST transgenes coding for N-terminally GFP-tagged variants of Pyd. These were generated by PCR using a cDNA-encoding full-length Pyd-RB isoform (gift from R. Ueda) as a template and are as described in Expression constructs for Pyd and other proteins. pUAST Su(dx) was generated as previously described (Cornell et al., 1999). Mutant clones were generated at high frequency in the wing using abxUbxFLPase in combination with FRT82B pydtamou or FRT82B pydex147.
Immunocytochemistry
Antibody staining of wing imaginal discs or female germaria was performed using standard protocols. The primary antibodies used were rabbit anti-Pyd (polyclonal directed against Pyd aa 1–592 generated for this study; 1:2,000), rat anti–E-Cad (DCAD2, 1:25; Developmental Studies Hybridoma Bank), mouse anti-Notch intra (C17.9C6, 1:25; Developmental Studies Hybridoma Bank), mouse anti-Notch extra (C458.2H, 1:25; Developmental Studies Hybridoma Bank), mouse anti-Arm (N2 7A1, 1:25; Developmental Studies Hybridoma Bank), mouse anti-Dlg (4F3, 1:25; Developmental Studies Hybridoma Bank), guinea pig anti-Scribble (1:2,000; gift from D. Bilder, University of California, Berkeley, Berkeley, CA), rabbit anti-aPKC (anti-PKCz C-20, 1:1,000; Santa Cruz Biotechnology, Inc.), guinea pig anti-Senseless (1:5,000; gift from H. Bellen, Baylor College of Medicine, Houston, TX), rat anti-Fat (1:2,000; gift from D. Strutt, University of Sheffield, Sheffield, England, UK), mouse anti-Engrailed (4D9, 1:25; Developmental Studies Hybridoma Bank), guinea pig anti-Shot (1:2,000; gift from K. Röper, University of Cambridge, Cambridge, England, UK), mouse anti-GFP (A-11120, 1:250; Invitrogen), rabbit anti-HA (1:300; Takara Bio Inc.), and mouse anti-V5 (1:500; Invitrogen). Images were acquired with a scanning confocal microscope (SP2-405; Leica) and processed using Photoshop (Adobe).
X-gal staining
3–4-d-old female germaria were dissected in PBS, fixed for 20 min in 1.7% glutaraldehyde in PBS, and then washed in PBS. They were stained overnight at 37°C in 1 mM MgCl2, 6 mM K4FeIICN6, 6 mM K3FeIICN6, and 0.2% X-gal. After washing in PBS, they were then mounted in vectamount. Images were acquired with a microscope (Axioskop 2 plus; Carl Zeiss, Inc.) using a 40× Plan-Neofluar and a color camera (AxioCam 412–312; Carl Zeiss, Inc.) and processed in Photoshop.
Transmission electron microscopy
Wild-type and pydex147/Df(3R)p-XT103 fully pyd mutant third instar larval epithelial wing discs were dissected in cold PBS, rinsed in cold 0.9% NaCl solution, and then fixed in 3% glutaraldehyde in 0.1-M Pipes buffer for 2 h at 4°C. After rinsing, they were treated with 1% osmium tetroxide containing 0.155-M potassium ferricyanide for 1 h, rinsed in water, and stained with 2% uranyl acetate in 0.05-M maleate buffer. They were dehydrated in an ascending series of ethanol solutions from 70 to 100%, washed twice in acetonitrile, and infiltrated with epoxy resin (Quetol). The resin was cured at 60°C for 48 h. Thin sections were cut using an ultramicrotome (Ultracut UCT; Leica) mounted on 300 mesh copper grids and stained with uranyl acetate and lead citrate. The sections were viewed in an electron microscope (Tecnai G2; FEI) operated at 120 kV, and images were recorded with a digital camera (AMT XR80B; Deben).
Expression constructs for Pyd and other proteins
All Pyd constructs were derived from Pyd-PB (aa 1–1,367) to produce proteins with the amino acids deleted as indicated: PydΔEx6, Δ791–869; PydΔPro-Rich, Δ870–1,367; PydΔEx6ΔPro-Rich, Δ791–1,367; PydΔPDZ1, Δ1–134; PydΔPDZ2, Δ135–343; PydΔPDZ3, Δ344–492; PydΔSH3, Δ493–592; PydΔGUK, Δ593–790; PydΔEx6, Δ791–869; PydΔPro-Rich, Δ870–1,367; and PydΔEx6ΔPro-Rich, Δ791–1,367. These were subcloned into the appropriate plasmids for transgenic Drosophila (pUAST), yeast expression (pAS and pACT; Takara Bio Inc.), and GST pull-downs (pGEX 4T1; GE Healthcare). In cases in which GFP fusions were generated, the coding sequence for EGFP was fused in frame at the N terminus. Su(dx)-, Nedd4-, and Smurf-expressing plasmids were created in a similar manner. Su(dx) was also subcloned into pMT (Invitrogen) ± C-terminal V5 tag for the cell culture experiments. Fragments encompassing other proteins, as indicated, were subcloned into pAS and pACT (Takara Bio Inc.) for yeast two-hybrid experiments.
Biochemistry
To identify Pyd binding partners, a systematic two-hybrid screen was performed using full-length Pyd protein as bait and different A/B determinants, AJ-associated proteins, and Notch pathway–related proteins as preys (similar to Djiane et al., 2005). Constructs were generated by PCR from genomic DNA, ESTs, or cDNA obtained from S2 cells.
For coimmunoprecipitation, S2 cells were grown in 90-mm dishes in Schneider’s medium (Invitrogen) at 25°C and transfected with pMT-Gal4 and pUAST-Pyd with or without pMT-HA-Su(dx)-V5. The cells were further grown for 24 h with the addition of CuSO4 (1-mM final concentration) and then homogenized in lysis buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 2 mM EGTA, protease inhibitor cocktail [Complete; Roche], and 50 mM MG132 [Enzo Life Sciences, Inc.]), and the resulting lysates were incubated with 1 mg mouse anti-V5 (Invitrogen) for 3 h at 4°C followed by incubation with protein G–Sepharose (Sigma-Aldrich) for 1 h at 4°C. After several washes in lysis buffer, bound proteins were eluted, and Western blotting was performed with rabbit anti-Pyd (see Immunocytochemistry; 1:10,000).
To map the domains of Pyd interacting with the WW domains of Su(dx) and Nedd4, GST pull-down experiments were performed as described previously (Djiane et al., 2005) by incubating GST fusions for the WW domains (GST-Su(dx)-C2-WW4 [aa 1–570], GST-Su(dx)-WW1–2 [aa 350–453], and GST-Su(dx)-WW3–4 [aa 453–570]) with 35S-labeled Pyd domains as indicated.
Su(dx) cortex recruitment assay
S2 cells were grown in Schneider’s medium at 25°C and transfected with pMT-Gal4 and pUAST HA-Su(dx) with or without different pUAST GFP-Pyd mutants. The cells were allowed to grow for another 24 h, fixed, and then stained for HA and Pyd (see Immunocytochemistry).
Peptide binding
Individual and paired Su(dx) WW domains were expressed and purified as GST fusion proteins, and the WW domain fragments were released by cleavage with protease (PreScission; Fedoroff et al., 2004; Jennings et al., 2007). Synthetic substrate peptides (Pepceuticals Ltd.) were EGLPPPYTV (residues 861–870 of Pyd) and APPPQSYPQ (residues 1,287–1,295 of Pyd). WW domain fragments were diluted in PBS to give ∼50 arbitrary units of intrinsic fluorescence (final concentrations were 4 µM WW1, 5 µM WW2, 15 µM WW1–2, and 2 µM WW3–4). Fluorescence experiments were performed using a Cary Eclipse Fluorimeter (Varian) at 20°C with a 5-mm emission and excitation slit width and excitation and emission wavelengths of 295 and 340 nm, respectively. Peptide ligands were titrated into WW domain solutions in 10-µl increments over a concentration range suitable for accurate Kd determination. Fluorescence values corrected for WW domain dilution were normalized (to give F′) relative to the fluorescence of the apoprotein (F0). Calculations of free ligand concentrations were performed (ligand is not in great excess of WW domain in these experiments), and the data were fitted to a single-site binding model according to the equation F′ = (1 + F′MLKa[L])/(1 + Ka[L]), using WinCurveFit (Kevin Raner Software). F′ML is the normalized maximum fluorescence of the complexed macromolecule, [L] is the free ligand concentration, and Ka is the association constant. Kd was derived by taking the reciprocal of the Ka values obtained from the single-site binding curves. The single-site binding equation was also used for paired domains (a two-site model proved inaccurate as expected where binding affinities of individual domains within a pair are not orders of magnitude apart). Each WW domain/peptide combination was analyzed three times.
Online supplemental material
Fig. S1 provides more details of the effects of pyd mutations on the architecture of larval wing disc epithelial cells. Fig. S2 shows results from a phage expression library screen that identified Pyd binding to the WW3–4 domains of Su(dx) and demonstrates that the expression of the Notch reporter E(spl)m7–lacZ in the ovary is strongly reduced in Su(dx) mutant combinations. Table S1 summarizes dissociation constants for interactions between two Pyd peptides and the different Su(dx) WW domains. Results in Tables S2 and S3 show that Su(dx) and pyd do not interact genetically in the control of cell numbers in the wing. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007023/DC1.
Acknowledgments
We are grateful to M. Affolter (Biozentrum, Basel, Switzerland), D. Bilder, D. St Johnston (University of Cambridge, Cambridge, England, UK), I. Palacios (University of Cambridge, Cambridge, England, UK), K. Röper, J. Silber (University of Paris 7, Paris, France), D. Strutt, R. Ueda, the Drosophila Genetic Resource Center Kyoto and Bloomington Drosophila Stock Centers, the Drosophila Genomics Resource Center, and the Developmental Studies Hybridoma Bank for flies, cDNAs, and antibodies. We thank D. Strutt for sharing reagents before publication. We thank K. Röper and I. Torres for constructive comments on the manuscript.
This work was supported by a Medical Research Council program grant to S. Bray, a Biotechnology and Biological Sciences Research Council project grant to M. Baron, and a Wellcome Trust project grant to J. Avis and M. Baron. A. Djiane was supported by an FP6 Marie Curie Intra-European Fellowship postdoctoral fellowship.
Footnotes
Abbreviations used in this paper:
- AJ
- adherens junction
- aPKC
- atypical PKC
- DC
- dorsocentral
- E-Cad
- E-cadherin
- GSC
- germline stem cell
- Pyd
- Polychaetoid
- SJ
- septate junction
- SOP
- sensory organ precursor
- TJ
- tight junction
- ZO
- zona occludens
References
- Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., McNeill H. 2009. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 16:411–420 10.1016/j.devcel.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Balda M.S., Matter K. 2000. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19:2024–2033 10.1093/emboj/19.9.2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S., Garrett M.D., Matter K. 2003. The ZO-1–associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160:423–432 10.1083/jcb.200210020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner P., Israël A., Brou C. 2008. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 3:e2735 10.1371/journal.pone.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M., Freedman J.A., Bettler D.R., Jr, Manning S.D., Giep S.N., Steiner J., Ellis H.M. 1996. Polychaetoid is required to restrict segregation of sensory organ precursors from proneural clusters in Drosophila. Mech. Dev. 57:215–227 10.1016/0925-4773(96)00548-5 [DOI] [PubMed] [Google Scholar]
- Chen H.I., Sudol M. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA. 92:7819–7823 10.1073/pnas.92.17.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell M., Evans D.A., Mann R., Fostier M., Flasza M., Monthatong M., Artavanis-Tsakonas S., Baron M. 1999. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics. 152:567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A., Yogev S., Mlodzik M. 2005. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell. 121:621–631 10.1016/j.cell.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Fanning A.S., Little B.P., Rahner C., Utepbergenov D., Walther Z., Anderson J.M. 2007. The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol. Biol. Cell. 18:721–731 10.1091/mbc.E06-08-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff O.Y., Townson S.A., Golovanov A.P., Baron M., Avis J.M. 2004. The structure and dynamics of tandem WW domains in a negative regulator of notch signaling, Suppressor of deltex. J. Biol. Chem. 279:34991–35000 10.1074/jbc.M404987200 [DOI] [PubMed] [Google Scholar]
- Genevet A., Polesello C., Blight K., Robertson F., Collinson L.M., Pichaud F., Tapon N. 2009. The Hippo pathway regulates apical-domain size independently of its growth-control function. J. Cell Sci. 122:2360–2370 10.1242/jcs.041806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8:27–36 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Gajewski K., Sansores-Garcia L., Morrison C., Tao C., Halder G. 2009. The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J. Cell Sci. 122:2351–2359 10.1242/jcs.046482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock A., Nelson W.J. 2008. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta. 1778:660–669 10.1016/j.bbamem.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., Isobe T. 2005. 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 280:13187–13194 10.1074/jbc.M412884200 [DOI] [PubMed] [Google Scholar]
- Ikenouchi J., Umeda K., Tsukita S., Furuse M., Tsukita S. 2007. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 176:779–786 10.1083/jcb.200612080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M.D., Blankley R.T., Baron M., Golovanov A.P., Avis J.M. 2007. Specificity and autoregulation of Notch binding by tandem WW domains in suppressor of Deltex. J. Biol. Chem. 282:29032–29042 10.1074/jbc.M703453200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A.C., Ribeiro C., Michaut L., Certa U., Affolter M. 2006. Polychaetoid/ZO-1 is required for cell specification and rearrangement during Drosophila tracheal morphogenesis. Curr. Biol. 16:1224–1231 10.1016/j.cub.2006.04.048 [DOI] [PubMed] [Google Scholar]
- Köhler K., Zahraoui A. 2005. Tight junction: a co-ordinator of cell signalling and membrane trafficking. Biol. Cell. 97:659–665 10.1042/BC20040147 [DOI] [PubMed] [Google Scholar]
- Maitra S., Kulikauskas R.M., Gavilan H., Fehon R.G. 2006. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16:702–709 10.1016/j.cub.2006.02.063 [DOI] [PubMed] [Google Scholar]
- Mazaleyrat S.L., Fostier M., Wilkin M.B., Aslam H., Evans D.A., Cornell M., Baron M. 2003. Down-regulation of Notch target gene expression by Suppressor of deltex. Dev. Biol. 255:363–372 10.1016/S0012-1606(02)00086-6 [DOI] [PubMed] [Google Scholar]
- Sakata T., Sakaguchi H., Tsuda L., Higashitani A., Aigaki T., Matsuno K., Hayashi S. 2004. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 14:2228–2236 10.1016/j.cub.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Sato S., Chiba T., Sakata E., Kato K., Mizuno Y., Hattori N., Tanaka K. 2006. 14-3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 25:211–221 10.1038/sj.emboj.7600774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppa M.J., Johnson R.I., Bao S., Cagan R.L. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 318:1–16 10.1016/j.ydbio.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K., Fogg V.C., Margolis B. 2006. Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 22:207–235 10.1146/annurev.cellbio.22.010305.104219 [DOI] [PubMed] [Google Scholar]
- Takahisa M., Togashi S., Suzuki T., Kobayashi M., Murayama A., Kondo K., Miyake T., Ueda R. 1996. The Drosophila tamou gene, a component of the activating pathway of extramacrochaetae expression, encodes a protein homologous to mammalian cell-cell junction-associated protein ZO-1. Genes Dev. 10:1783–1795 10.1101/gad.10.14.1783 [DOI] [PubMed] [Google Scholar]
- Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., Tsukita S. 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 126:741–754 10.1016/j.cell.2006.06.043 [DOI] [PubMed] [Google Scholar]
- Ward E.J., Shcherbata H.R., Reynolds S.H., Fischer K.A., Hatfield S.D., Ruohola-Baker H. 2006. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr. Biol. 16:2352–2358 10.1016/j.cub.2006.10.022 [DOI] [PubMed] [Google Scholar]
- Wei X., Ellis H.M. 2001. Localization of the Drosophila MAGUK protein Polychaetoid is controlled by alternative splicing. Mech. Dev. 100:217–231 10.1016/S0925-4773(00)00550-5 [DOI] [PubMed] [Google Scholar]
- Wilkin M.B., Carbery A.M., Fostier M., Aslam H., Mazaleyrat S.L., Higgs J., Myat A., Evans D.A., Cornell M., Baron M. 2004. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 14:2237–2244 10.1016/j.cub.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Wilkin M., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K., Baron M. 2008. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell. 15:762–772 10.1016/j.devcel.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Wittchen E.S., Haskins J., Stevenson B.R. 1999. Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. J. Biol. Chem. 274:35179–35185 10.1074/jbc.274.49.35179 [DOI] [PubMed] [Google Scholar]