Abstract
The 37/67 kDa laminin receptor (LAMR) is a multifunctional protein, acting as an extracellular receptor, localizing to the nucleus, and playing roles in rRNA processing and ribosome assembly. LAMR is important for cell viability; however, it is unclear which of its functions are essential. We developed a silent mutant LAMR construct, resistant to siRNA, to rescue the phenotypic effects of knocking down endogenous LAMR, which include inhibition of protein synthesis, cell cycle arrest, and apoptosis. In addition, we generated a C-terminal-truncated silent mutant LAMR construct structurally homologous to the Archaeoglobus fulgidus S2 ribosomal protein and missing the C-terminal 75 residues of LAMR, which displays more sequence divergence. We found that HT1080 cells stably expressing either silent mutant LAMR construct still undergo arrest in the G1 phase of the cell cycle when treated with siRNA. However, the expression of full-length silent mutant LAMR rescues cell viability, whereas the expression of the C-terminal-truncated LAMR does not. Interestingly, we also found that both silent mutant constructs restore protein translation and localize to the nucleus. Our findings indicate that the ability of LAMR to regulate viability is associated with its C-terminal 75 residues. Furthermore, this function is distinct from its role in cell proliferation, independent of its ribosomal functions, and may be regulated by a nonnuclear localization.
Keywords: laminin receptor, extraribosomal, viability, translation, silent mutant
The 37/67 kDa laminin receptor (LAMR), as a cell surface receptor, has roles in cell migration,1 invasion,2 angiogenesis,3 and extracellular matrix remodeling.4, 5 LAMR also serves as a cellular receptor for many pathogens, including prion protein,6 bacteria,7 and numerous viruses.8, 9, 10, 11 Since its initial discovery, however, LAMR was determined to be a highly conserved ribosomal protein that acquired its extracellular functions during evolution.12 The human sequence of LAMR has homologs in many species, including prokaryotes; Archaea encode a protein that is 40% identical to LAMR, which places it as a member of the S2 family of ribosomal proteins.13 The crystal structure of recombinant human LAMR from residues 1 to 220, generated by our laboratory, shows structural homology with the Archaeoglobus fulgidus S2 ribosomal protein.14 Such sequence and structural conservation implies that LAMR is important for basic cellular functioning. Indeed, yeast homologs of LAMR are essential for cell viability,15 having roles in 20s–18s rRNA processing and ribosome assembly.16 LAMR homologs are ribosome associated in higher organisms as well, including plant17 and mouse,18 although, whether or not the ribosomal functions of LAMR are important for cell viability in higher organisms remains unclear. Reduction of LAMR expression in HeLa cells results in apoptosis;19 however, translation was reportedly not affected. Apoptosis was observed in Hep3b cells upon reduction of LAMR, but the effect on translation was not reported.20 LAMR can also localize to the nucleus and interact with histones;21 therefore, nuclear functions of LAMR may have a role in cell viability. LAMR has been implicated in cell signaling pathways that are important for cell survival as well.22
Interestingly, sequence homology between human LAMR and homologs in invertebrates resides only within the first two-thirds of the molecule, whereas the C-terminal sequence is more divergent. The human LAMR sequence is 73.5% homologous to that of hydra from residues 1 to 218, but only 20% homologus in the C-terminal 76 residues.23 In contrast, the entire human LAMR sequence is highly conserved among vertebrates as the human LAMR sequence displays >98% homology with the mouse, bovine, and rat sequences.24 C-terminal divergence among higher organisms is considered to be the process by which LAMR acquired its extracellular functions. It may have also enabled LAMR to become a dimeric molecule, as the 37 kDa LAMR has been shown to serve as a precursor to the 67 kDa LAMR,25 which has only been observed in vertebrates and is dependent on posttranslational modifications.26, 27 Extraribosomal functions associated with the C terminus of LAMR may also have enabled alternative mechanisms to regulate cell viability in higher organisms, including nuclear localization.
The elucidation of how LAMR regulates cell survival is important from a therapeutic standpoint. LAMR is overexpressed in many different types of cancers and is considered a prognostic factor for determining the severity of tumors.28 Transplanted tumor cells with reduced expression of LAMR grow slower in vivo, resulting in prolonged survival of recipient mice.29 Furthermore, the green tea polyphenol (−)-epigallocatechin-3-gallate (EGCG) targets tumors using LAMR as a cellular receptor30 and induces G1 arrest and apoptosis in multiple myeloma cells.31 Previously, we have shown that the reduction of LAMR expression in HT1080 human fibrosarcoma cells results in G1-phase cell-cycle arrest.32 We found cell cycle- and survival-related genes to be altered at the mRNA and protein level in a manner consistent with these observations. We also found protein translation to be dramatically reduced as knockdown of LAMR in HT1080 cells leads to fewer 40 s subunits and 80 s monosomes, as well as an increase in unassociated 60 s subunits. These results were consistent with yeast studies and show that LAMR can serve as a critical component of the translational machinery in mammalian cells. In turn, HT1080 cells with reduced LAMR expression have an impaired ability to grow tumors in vivo.
In this study, we used a silent mutant LAMR, construct resistant to siRNA, to rescue cell viability and translation in siRNA-treated HT1080 cells. Furthermore, using a silent mutant 1–220 truncated LAMR construct, homologous to the A. fulgidus S2 ribosomal protein and devoid of the C-terminal 75 residues, we clarified whether cell viability in HT1080 cells is dependent on the highly conserved ribosomal functions of LAMR.
Results
Silent mutant LAMR constructs are resistant to siRNA and rescue the phenotypic effect of knocking down endogenous LAMR
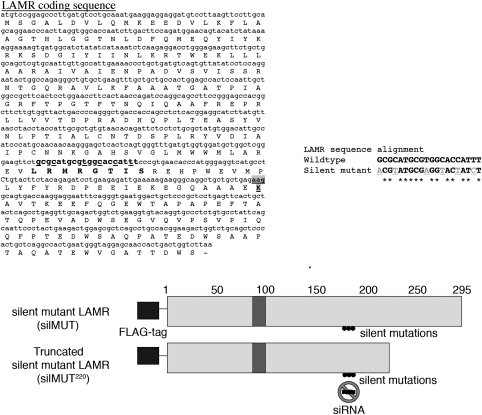
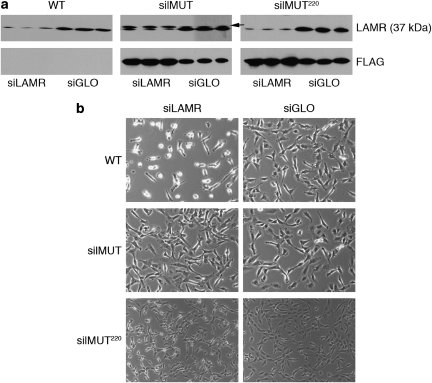
We mutated the third base of 6 codons within the coding region of LAMR targeted by siRNA (siLAMR) to generate a silent mutant cDNA/mRNA sequence that still encodes a wild-type protein but is resistant to siLAMR (Figure 1). To distinguish from endogenous LAMR, silent mutant LAMR (silMUT) was cloned with an N-terminal flag tag and introduced into the human fibrosarcoma HT1080 cell line. In addition, we introduced silent mutations and an N-terminal flag tag into a 1–220 residue LAMR construct previously used to generate a crystal structure. This silent mutant construct (silMUT220), missing the C-terminal 75 residues of LAMR, is homologous to the A. fulgidus S2 ribosomal protein. We selected several clonally derived HT1080 cell lines that stably express silMUT and silMUT220. Although we performed experiments with all selected cell lines, yielding similar results, we report herein our findings with one of each. silMUT, silMUT220, and parental HT1080 (WT) cells were transfected with either siLAMR or a fluorescently labeled nontargeting control siRNA (siGLO) and analyzed for total LAMR protein expression. Four days after transfection, endogenous LAMR is reduced in all cells treated with siLAMR (Figure 2a, top panel). Exogenous silMUT LAMR, detected by both an antibody for LAMR (Figure 2a, arrow) and an antibody for the FLAG tag (Figure 2b, middle), is not reduced by siLAMR treatment but rather appears to be increased in expression compared with siGLO-treated silMUT cells. silMUT220 is not readily detected by the antibody we used for LAMR as it is missing a part of its C-terminal epitope. However, using an antibody for the FLAG tag, we observed that silMUT220 LAMR protein expression (Figure 2b, right), is not reduced by siLAMR. We also observed that siLAMR-treated silMUT and silMUT220 cells display higher LAMR mRNA expression levels than do siLAMR-treated WT cells (Supplementary Figure 1).
Figure 1.
Constructing silent mutant LAMR. The coding sequence of LAMR (accession no. NM_001012321.1) is shown along with the sequence that siLAMR targets (bold). The third base pair of each codon (underlined) within this region was changed as displayed by sequence alignment. The last residue of silMUT220 is bold and shaded gray. A schematic of FLAG-tagged silMUT and silMUT220 and the region that siLAMR targets is shown below the coding sequence. Numbers represent every 50 residues of LAMR starting with 1 at the N terminus (N), and ending with 295 at the C terminus (C). Shaded area represents the proposed transmembrane region of LAMR
Figure 2.
Silent mutant LAMR protein expression and cell phenotype. (a) Western blot analysis of LAMR protein expression in WT, silMUT, and silMUT220 cells 4 days after transfection with siLAMR and siGLO. Antibodies against LAMR (top panel) and FLAG (bottom panel) were used to detect expression of endogenous and exogenous LAMR protein expression levels, respectively. Full-length silMUT LAMR runs slightly higher than endogenous LAMR (37 kDa) and can be detected with antibodies for LAMR (arrow). silMUT220 is not detected by the antibody used for LAMR and runs at ∼25 kDa. Blots represent three separate transfections. (b) Images of WT, silMUT, and silMUT220 cells treated with siLAMR and siGLO 4 days after transfection. Images are indicative of at least three separate transfections
As visualized by microscopy, WT cells treated with siLAMR are reduced in number and cell size with many rounded cells (Figure 2b, top). In contrast, silMUT cells treated with siLAMR have normal morphology with a phenotype similar to cells treated with siGLO (Figure 2b, middle). It should be noted that similar results were obtained with several additional FLAG-tagged silMUT stable clones (data not shown) and stable cell lines that express either untagged or C-terminal v5/his-tagged silMUT LAMR constructs (Supplementary Figure 2). Conversely, cell lines that stably express nonmutated exogenous LAMR do not rescue the phenotypic effect of siLAMR (data not shown). The phenotype of silMUT220 cells treated with siLAMR does not appear to be as severe as in WT cells (Figure 2b, bottom), although siLAMR-treated silMUT220 cells are still smaller in size (including rounded cells) than their siGLO-treated counterparts. This suggests that unlike full-length silMUT, silMUT220 LAMR is unable to fully restore the phenotypic effect of siLAMR.
Silent mutant LAMR cells still undergo G1-phase cell-cycle growth arrest
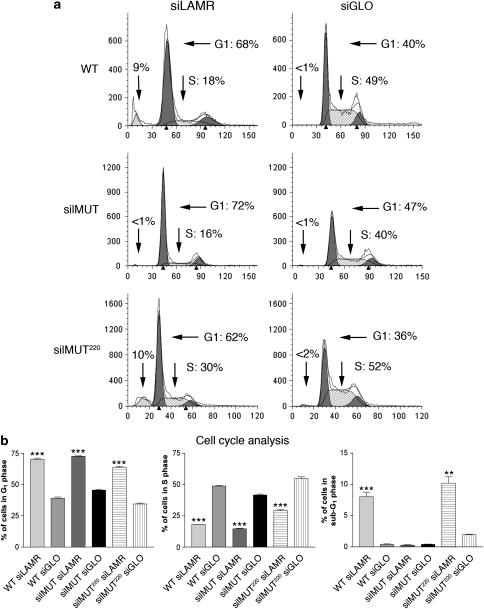
Analyzing the cell-cycle profile of WT, silMUT, and silMUT220 cells, we found that all cell lines are arrested in the G1 phase of the cell cycle 4 days after treatment with siLAMR. Overall, 68% of WT, 72% of silMUT, and 62% of silMUT220 siLAMR-treated cells are in the G1 phase of the cell cycle compared with 40, 47, and 36% for their siGLO-treated controls, respectively (Figure 3a). Furthermore, only 18% of WT, 16% of silMUT, and 30% of silMUT220 siLAMR-treated cells are in the S phase compared with 49, 40, and 52% of siGLO-treated cells, respectively.
Figure 3.
silMUT and silMUT220 cells still undergo G1-phase cell-cycle growth arrest. (a) Cell cycle profiles of WT (top), silMUT (middle), and silMUT220 (bottom) cells treated with siLAMR (left) and siGLO (right) 4 days after transfection. Percentages of cells in the G1, S, and sub-G1 phases are indicated (arrows). Plots are representative of at least three separate transfections. (b) The percentages of cells in the G1 (left), S (middle), and sub-G1 phases (right) 4 days after transfection are summarized. Bar=S.E.M. Statistical analysis was performed using a standard Student's t-test to generate P-values. All P-values are two-tailed. ***P<0.0005. **P<0.005
While analyzing cell-cycle profiles, we noticed that ∼9% of siLAMR-treated WT cells are in the sub-G1 phase 4 days after transfection, indicative of cells undergoing apoptosis (Figure 2a). Interestingly, we found no increase in a sub-G1 population for silMUT cells treated with siLAMR; however, silMUT220 cells treated with siLAMR display an increase in the sub-G1 population similar to WT cells. The percentages of cells in G1, S, and sub-G1 for each cell line are shown in Figure 2b (left, middle, and right panel, respectively). These results indicate that although all siLAMR-treated cells are still arrested in the G1 phase of the cell cycle, full-length silMUT LAMR rescues the apoptotic effects of reducing endogenous LAMR, whereas the C-terminal-truncated silMUT220 LAMR does not.
The C-terminal 75 residues of LAMR are required for maintaining cell viability
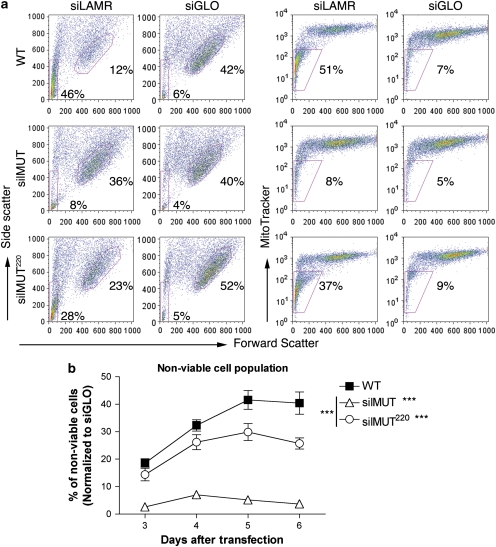
To further analyze cell viability, we stained siLAMR- and siGLO-treated WT, silMUT, and silMUT220 cells 3–6 days after transfection with MitoTracker Deep-Red (Invitrogen, Carlsbad, CA, USA), a fluorescent dye that stains the mitochondria only in viable cells. Cell viability was accessed by fluorescence-activated cell sorting (FACS) with forward and side scatter (Figure 4a, left), as well as MitoTracker (Figure 4a, right). Viewing side scatter 5 days after transfection, we observed that WT and silMUT220 cells treated with siLAMR develop a large population of nonviable cells (red rectangle), whereas silMUT cells do not. Overall, 46% of siLAMR-treated WT and 28% of silMUT220 cells are in this nonviable population, compared with only 8% of siLAMR-treated silMUT cells. Similar results were obtained with MitoTracker (Figure 3a, right). Overall, 51% of siLAMR-treated WT and 37% of silMUT220 cells are negative for MitoTracker compared with only 8% of siLAMR-treated silMUT cells. The percentage of nonviable cells for the entire time course, determined by MitoTracker, is shown in Figure 4b.
Figure 4.
The C-terminal 75 residues of LAMR are required for maintaining cell viability. (a) FACS analysis of siLAMR- and siGLO-treated WT (top), silMUT (middle), and silMUT220 (bottom) cells 5 days after transfection. Cell viability was assessed through forward and side scatter (left panel), and the MitoTracker Deep-Red stain (right panel). Nonviable cell populations were gated in siLAMR-treated WT cells (red rectangle) and applied to all plots to determine the percentage of nonviable cells, as indicated. The most dense viable cell population was gated in each siGLO-treated plot (red circle, left panels) and applied to siLAMR plots of each respective cell line to determine changes in cell size/morphology. Each plot is indicative of at least three separate transfections. (b) The percentage of nonviable cells 3–6 days after transfection, according to MitoTracker Deep Red and normalized to siGLO-treated cells, are summarized. Bar=S.E.M. Statistical analysis was performed using a two-way ANOVA to generate P-values. ***P<0.0005
Furthermore, using side scatter, we gated the most dense population of viable siGLO-treated WT, silMUT, and silMUT220 cells (Figure 4a left panel, right plots, 42, 40, and 52%, respectively). Each gate was applied to its respective cell line treated with siLAMR. We found that viable WT cells treated with siLAMR become smaller and more granular, as determined by their shift in forward and side scatter, respectively (12% of cells remain in the gate applied for siGLO viable cells). silMUT220 treated with siLAMR also become smaller (23% of cells remain in the gate applied for siGLO viable cells). In contrast, viable siLAMR-treated silMUT cells have the same size and morphology as siGLO-treated silMUT cells (36% of cells remain in the gate applied for siGLO viable cells), indicating that morphological functions are also restored by silMUT LAMR. In all, the viability/morphology results from our FACS analysis are consistent with the rescue of phenotypic effects we observed by microscopy (Figure 1), and indicate that the C-terminal residues of LAMR are required for maintaining cell viability.
The C-terminal 75 residues of LAMR are not required for ribosomal functions
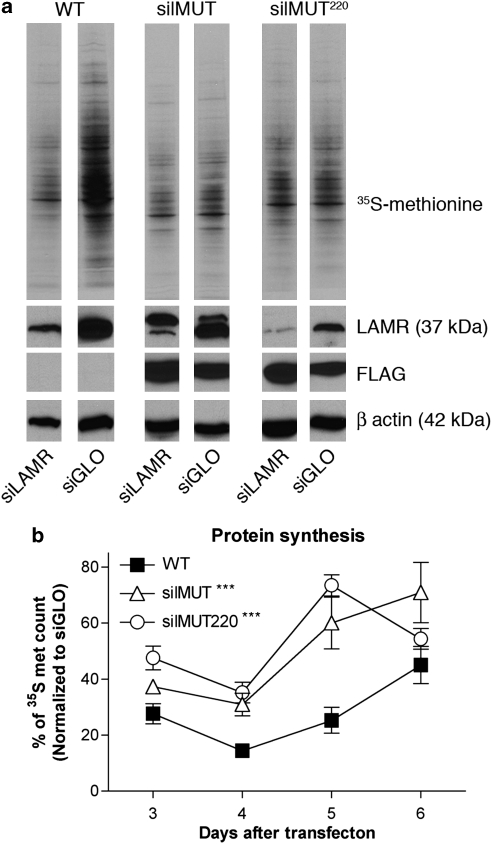
As silMUT220 is structurally homologous to the A. fulgidus S2 ribosomal protein, we reasoned that it should maintain all ribosomal functions of LAMR, and therefore its inability to rescue viability must be due to an extraribosomal function. To clarify whether the ribosomal functions of LAMR are required for cell viability, we measured protein synthesis in WT, silMUT, and silMUT220 cells treated with siLAMR and siGLO 3–6 days after transfection. Five days after transfection, translation in siLAMR-treated WT cells is dramatically reduced, whereas that in siLAMR-treated silMUT and silMUT220 cells appears normal (Figure 5a). It should be noted that translation appears to be higher in control siGLO-treated WT cells than in siGLO-treated silMUT and silMUT220 cells, which may be due the clonal selection of both cell lines. Therefore, protein synthesis for the entire time course was quantified by scintillation counting (Figure 5b). The percentage of translation for WT, silMUT, and silMUT220 siLAMR-treated cells, compared with that of their respective siGLO-treated cells, is shown. Surprisingly, the trend of translation inhibition is similar for all cell lines 3 and 4 days after transfection, although siLAMR-treated silMUT and silMUT220 do have higher protein synthesis levels than do WT siLAMR cells 4 days after transfection (31 and 37% compared with 14%, respectively). Five days after transfection, siLAMR-treated WT cells have a modest increase in protein synthesis to 25% of siGLO-treated WT cells. However, siLAMR-treated silMUT and silMUT220 cells dramatically increase their protein synthesis up to 60% and 74, respectively. These results indicate that both silMUT and silMUT220 LAMR restore protein translation in siLAMR-treated cells, thus showing that the C-terminal 75 residues of LAMR are not required for ribosomal functions.
Figure 5.
The C-terminal 75 residues of LAMR are not required for ribosomal functions. (a) In all, 20 μg of protein from 35S-methionine-labeled lysates was loaded for SDS-PAGE to detect newly synthesized proteins (top panel). Lysates were also used for western blot analysis to detect endogenous LAMR (second panel), exogenous silMUT and silMUT220 LAMR (third panel), and β-actin (fourth panel). Blots represent protein synthesis 5 days after transfection and are indicative of at least three separate transfections. (b) Newly synthesized protein was quantified by scintillation counting 3–6 days after transfection. Equal volumes of siLAMR and siGLO lysates were used for counting and normalized according to protein concentration. 35S-methionine scintillation counts for siLAMR-treated cells are summarized as a percentage of siGLO-treated cells for each respective cell line. Each percentage is representative of at least three separate transfections. Bar=S.E.M. Statistical analysis was performed using a two-way ANOVA to generate P-values. ***P<0.0005
The C-terminal 75 residues of LAMR are not required for nuclear localization
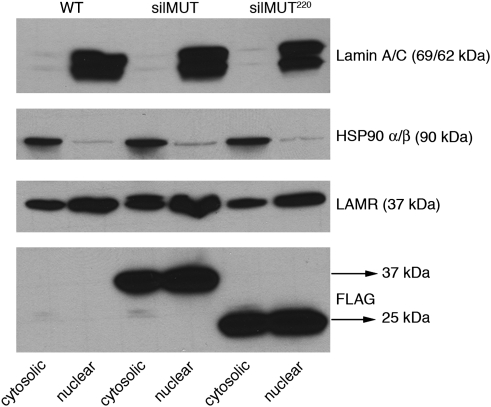
As LAMR was previously shown to interact with histones and localize to the nucleus, we decided to see whether endogenous, silMUT, and silMUT220 LAMR can be detected in HT1080 nuclear extracts by western blot. The nuclear marker Lamin A/C and the cytosolic marker HSP 90-α/β show that we successfully separated both fractions (Figure 5, top and second panel, respectively). We found endogenous LAMR to be both cytosolic and nuclear in all cell lines (Figure 5, third panel, LAMR). Furthermore, we found that both silMUT and silMUT220 are cytosolic and nuclear as well. Therefore, our results show that LAMR localizes to the nucleus in HT1080 cells and that the C-terminal 75 residues are not required for this process.
Discussion
Reduction of LAMR expression by siRNA results in translation inhibition, G1-phase cell-cycle arrest, and apoptosis. Therefore, we had to use a transient siRNA system to study LAMR. This complicates determining the kinetics of our observations, as well as identifying which functions/localizations of LAMR (namely ribosomal, nuclear, extracellular) are important for proliferation and cell viability. Our silent mutant LAMR system addresses these issues by providing a novel way in which to study the multiple functions of LAMR in a cellular environment. Interestingly, silMUT LAMR rescues different functions of LAMR separately. We observe that siLAMR-treated silMUT cells still undergo cell cycle arrest (Figure 3) and that restoration of protein synthesis is delayed (Figure 5). This contrasts with the rescue of morphology and cell viability, which happens during times of cell-cycle arrest and translational inhibition (Figure 4). The inability of exogenous LAMR to rescue all phenotypes resulting from inhibition of endogenous LAMR has previously been observed in another system as well.33 It is possible that there are threshold levels of LAMR expression required for each cellular function it regulates and thus with limited expression of exogenous LAMR, only certain functions can be restored. In this case, functions required for cell viability would be restored before those for translation and proliferation as they are more essential. This is suggested by studies in yeast, which have two copies of the lamr gene. Disruption of one gene leads to inhibition of cell growth, whereas knockout of both gene copies is lethal.15
LAMR cDNA cloned from yeast has been previously used to complement double mutant yeast strains lacking LAMR to restore cell viability.16, 34 In addition, exogenous LAMR has been expressed to rescue both lethal and viable phenotypes in Drosophila due to mutations of the endogenous gene.33 We are the first to achieve such rescue in mammalian cells. Although studies conducted in yeast and Drosophila concluded that the rescue of cell viability is most likely attributed to the ribosomal functions of LAMR, our results with silMUT220 cells indicate that extraribosomal functions of LAMR are important for this process. We found that silMUT220 LAMR can restore protein synthesis in a delayed manner similar to full-length silMUT LAMR (Figure 5). This is consistent with sequence analysis showing that, as a ribosomal protein, the first two-thirds of LAMR are highly conserved, whereas the C terminus is more divergent among higher organisms (most likely to acquire extracellular functions). This also supports our structural analysis showing the homology between the 1–220 LAMR and the A. fulgidus S2 ribosomal protein.14 However, despite retaining ribosomal functions, silMUT220 is unable to rescue cell viability (Figure 4), indicating that extraribosomal functions of the C-terminal 75 residues are essential for this process.
Furthermore, we have shown that the C-terminal 75 residues of LAMR are not required for nuclear localization (Figure 6), as silMUT220 localizes to the nucleus in a similar capacity as full-length silMUT and endogenous LAMR. Future studies are required to determine whether silMUT220 is functional in a nuclear context. Although silMUT220 may localize to the nucleus, its inability to rescue cell viability may be due to a lack of the necessary residues/domains required for nuclear interactions. Still, these findings suggest that the functions of LAMR associated with viability may be dependent on a nonnuclear localization.
Figure 6.
The C-terminal 75 residues of LAMR are not required for nuclear localization. In all, 20 μg of protein was loaded for western blot from both cytosolic and nuclear fractions. Blots were probed with Lamin A/C (panel 1) and HSP 90-α/β (panel 2) for nuclear and cytosolic markers, respectively. Fractions were also probed for endogenous LAMR (panel 3) and exogenous FLAG-tagged silMUT and silMUT220 LAMR (panel 4)
As mentioned earlier, LAMR was initially discovered as a cell surface receptor for laminin. Surface localization of LAMR may therefore be important for maintaining cell viability. Others have shown that the 37 kDa LAMR is expressed on the cell surface of numerous cell types,6 including HT1080 cells.35 However, we were unable to detect 37 kDa LAMR at the cell surface of our HT1080 cells (data not shown). Surface localization may instead be reserved for a different form of LAMR, in particular the 67 kDa dimer. As this molecule is only observed in vertebrates, it is believed that C-terminal sequence divergence has a role in its formation. Therefore, silMUT220 may be missing the required residues/domain to form a dimer. Alternatively, silMUT220 may still be able to form a dimer, but one that is missing residues important for essential contacts with the extracellular matrix. It should be noted that three binding sites for laminin have been discovered on LAMR thus far: residues 161–181 (peptide G),36 205–229,37 and the C-terminal 53 residues.38 Although silMUT220 contains the peptide G binding region and is functional in laminin-1 binding,14 it is missing portions or all of the other two laminin-binding sites. The laminin-binding regions missing in silMUT220 may be required for interactions important for cell viability. Interestingly, cleavage of LAMR by stromelysin-3, which separates the laminin-binding regions of LAMR from the cell surface, may have a role in promoting apoptosis.39 Therefore, the expression of silMUT220, missing these binding sites, may have a similar effect.
In conclusion, our silent mutant rescue system has determined that translation and cell viability are regulated in distinct manners by LAMR and that the extraribosomal functions of LAMR are required for maintaining cell viability. In addition, we used truncated LAMR to examine and distinguish its role in proliferation, translation, morphology, and viability. Further use of silMUT220 as well as additional truncated/mutated silent mutant LAMR constructs, will ultimately help define the multiple functions/localizations of LAMR.
Materials and Methods
Cloning
The LAMR coding sequence was cloned into the pcDNA 3.1/c N-terminal flag vector (Invitrogen) using BamH1 and Xho1 restriction sites. Primers used were as follows:
Forward primer: 5′-GGATCCTCCGGAGCCCTTGATGT-3′ and reverse primer: 5′-CTCGAGTTAAGACCGGTCAGTGGTTGCTCC-3′.
This vector served as a backbone for generating six silent mutations in the LAMR coding sequence using the Quick change II XL site-directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). Primers used for silent mutant cloning were as follows:
Forward primer: 5′-GGGAAGTTCTACGTATGCGAGGTACTATCTCCCGTGAACACCC-3′ and reverse primer: 5′-GGGTGTTCACGGGAGATAGTACCTCGCATACGTAGAACTTCCC-3′.
To generate the C-terminal-truncated silent mutant LAMR construct, the above forward primer was used with a reverse primer starting at the codon for the 220th residue of LAMR (out of 295).
Reverse truncated primer: 5′-CTCGAGTTACTTCTCAGCAGCAGCCTGCCC-3′.
Gene analysis
Extraction of total cellular RNA was performed using the RNA easy mini kit (Qiagen, Valencia, CA, USA). In all, 0.5 μg of RNA was used for the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Overall, 0.5 μl of cDNA was used to determine LAMR expression levels by quantitative real-time PCR using a Bio-Rad iCycler. Human GAPDH was chosen as the housekeeping gene for comparative analysis between samples. Human LAMR and GAPDH primers were designed and used as described previously.40 The fold change in laminin receptor relative to the GAPDH endogenous control was determined by fold change=2−(CT), where CT=CT(laminin receptor)−CT(GAPDH) and (CT)=CT(silMUT)−CT(WT). CT is the threshold cycle determined by fluorescence data collection.
Cell culture
The HT1080 cell line was obtained from the ATCC (Manassas, VA, USA). Silent mutant LAMR cell lines were generated by transfecting HT1080 cells with pCDNA 3.1/c N-terminal flag vectors containing either full-length or 1–220 C-terminal-truncated silent mutant LAMR. Lipofectamine 2000 (Invitrogen) was used for transfection and clonal stable cell lines were selected with neomycin.
Endogenous LAMR expression was inhibited using the predesigned siGENOME individual oligonucleotide no. 3 targeting LAMR (Dharmacon, Lafayette, CO, USA). Control nontargeting siGLO RISC-free siRNA and DharmaFECT 4 transfection reagent were also purchased from Dharmacon. A final concentration of 100 nM of siRNA was used for each transfection. To analyze phenotypic effects of siRNA, cells were imaged using a Nikon Eclipse TE200-E microscope (Tokyo, Japan) using the NIS-Elements BR-2.30 program (Nikon).
Cell proliferation/viability
For cell-cycle profile analysis, cells were permeabilized overnight in 100% ethanol at 4°C then stained with final concentrations of 50 μg/ml of propidium Iodide (Invitrogen) and 100 μg/ml of RNAse A (Sigma-Aldrich, St. Louis, MO, USA). Cells were analyzed using a BD FACScan machine (BD Biosciences, San Jose, CA, USA) and the percentages of cells in sub-G1, G1, and S phase were determined using the Modfit LT 3.2 program (Verity software house, Topsham, ME, USA).
Cell morphology/viability was further analyzed by forward/side scatter FACS analysis and using the Deep-Red MitoTracker stain (Invitrogen). Analysis was performed using a BD LSRII machine. Viable/nonviable cells were gated and quantified using the Flowjo 8.2 program (Tree Star, Ashland, OR, USA).
Protein analysis
For western blots, cells were harvested in Mammalian Protein Extraction Reagent (MPER) (Pierce, Rockford, IL, USA). A total of 20 μg of total protein was used for SDS-PAGE and transferred to PVDF membrane. LAMR protein expression was detected using the rabbit polyclonal antibody H-141, purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). This antibody recognizes epitopes in the C terminus of LAMR and predominantly detects the 37 kDa form. Flag-tagged silent mutant LAMR was detected using the Flag M-2 antibody from Stratagene.
To study translation, cells were metabolically labeled with 35S-methionine (20 μCi/ml) (Perkin-Elmer, Waltham, MA, USA) in DMEM lacking methionine (MP Biomedicals, Santa Ana, CA, USA) supplemented with 0.1% FCS for 2 h at 37°C (pulse). Normal growth medium was then added and cells were incubated for an additional 90 min at 37°C (chase). Cells were harvested in MPER and 20 μg of protein was loaded onto SDS-PAGE for analysis. To quantify protein synthesis, 4 μl of protein lysate (12–16 μg total protein) was precipitated with 10% TCA for 20 min at 4°C. Precipitated protein was vacuum filtered through Whatman glass microfiber filters (Whatman, Piscataway, NJ, USA) and washed once with 5% TCA then 95% ethanol. Filters were dried under a heating lamp then immersed in 3 ml of scintillation fluid and counted using a Beckman LS 3801 scintillation counter (Beckman, Brea, CA, USA).
Nuclear extraction
Nuclear extracts were collected from 2 × 106 WT, silMUT, and silMUT220 cells using the Affymetrix Nuclear Extraction Kit (Affymetrix, Fremont, CA, USA). The procedure was carried out according to the manufacturer's protocol. The number of cells used for extraction, as well as buffer volume, were scaled up twofold.
Statistical analysis
Statistical analysis for cell-cycle data was performed using a standard Student's t-test. The P-values generated are two-tailed. Statistical analysis for MitoTracker staining (viability), scintillation counting (translation), and mRNA expression was performed with two-way ANOVA. All statistics were generated using GraphPad Prism, version 3.0cx for Macintosh (GraphPad Software, San Diego, CA, USA).
Acknowledgments
We thank Dr. Christine Pampeno for the critical reading of this paper. US Public Health grants CA100687 from the National Cancer Institute, National Institutes of Health, and Department of Health and Human Services supported this study. Funding was also provided by a gift from the Litwin Foundation.
Glossary
- LAMR
37/67 kDa laminin receptor
- silMUT
silent mutant LAMR
- silMUT220
C-terminal-truncated silent mutant LAMR
In addition, funding for this study was also provided by a Research and License agreement between NYU and CynVec. In particular, the contents of this study may be used for a patent. According to the rules and regulations of the New York University School of Medicine, if this patent is licensed by a third party, Mr Scheiman, Ms Jamieson, Dr. Tseng, and Dr. Meruelo may receive benefits in the form of royalties or equity participation. Ms Ziello declares no potential conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Supplementary Material
References
- Wewer UM, Taraboletti G, Sobel ME, Albrechtsen R, Liotta LA. Role of laminin receptor in tumor cell migration. Cancer Res. 1987;47:5691–5698. [PubMed] [Google Scholar]
- Mafune K, Ravikumar TS. Anti-sense RNA of 32-kDa laminin-binding protein inhibits attachment and invasion of a human colon carcinoma cell line. J Surg Res. 1992;52:340–346. doi: 10.1016/0022-4804(92)90113-e. [DOI] [PubMed] [Google Scholar]
- Bernard A, Gao-Li J, Franco CA, Bouceba T, Huet A, Li Z. Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. J Biol Chem. 2009;284:10480–10490. doi: 10.1074/jbc.M809259200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E, Sporchia B, Pollegioni L, Modugno M, Ghirelli C, Castiglioni F, et al. Identification of a novel function for 67-kDa laminin receptor: increase in laminin degradation rate and release of motility fragments. Cancer Res. 2002;62:1321–1325. [PubMed] [Google Scholar]
- Berno V, Porrini D, Castiglioni F, Campiglio M, Casalini P, Pupa SM, et al. The 67 kDa laminin receptor increases tumor aggressiveness by remodeling laminin-1. Endocr Relat Cancer. 2005;12:393–406. doi: 10.1677/erc.1.00870. [DOI] [PubMed] [Google Scholar]
- Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, et al. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J Biol Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig GV, Kondig JP, Smith JF. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J Virol. 1996;70:5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J Virol. 2004;78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, et al. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- Ouzonis C, Kyrpides N, Sander C. Novel protein families in archaean genomes. Nucleic Acids Res. 1995;23:565–570. doi: 10.1093/nar/23.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson KV, Wu J, Hubbard SR, Meruelo D. Crystal structure of the human laminin receptor precursor. J Biol Chem. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- Demianova M, Formosa TG, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are essential components of the 40 S ribosomal subunit. J Biol Chem. 1996;271:11383–11391. doi: 10.1074/jbc.271.19.11383. [DOI] [PubMed] [Google Scholar]
- Ford CL, Randal-Whitis L, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 1999;59:704–710. [PubMed] [Google Scholar]
- Garcia-Hernandez M, Davies E, Staswick PE. Arabidopsis p40 homologue. A novel acidic protein associated with the 40 S subunit of ribosomes. J Biol Chem. 1994;269:20744–20749. [PubMed] [Google Scholar]
- Auth D, Brawerman G. A 33-kDa polypeptide with homology to the laminin receptor: component of translation machinery. Proc Natl Acad Sci USA. 1992;89:4368–4372. doi: 10.1073/pnas.89.10.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda Y, Kinoshita K, Sato M, Saeki Y, Yamada R, Wataya-Kaneda M, et al. The induction of apoptosis in HeLa cells by the loss of LBP-p40. Cell Death Differ. 1998;5:20–28. doi: 10.1038/sj.cdd.4400315. [DOI] [PubMed] [Google Scholar]
- Susantad T, Smith DR. siRNA-mediated silencing of the 37/67-kDa high affinity laminin receptor in Hep3B cells induces apoptosis. Cell Mol Biol Lett. 2008;13:452–464. doi: 10.2478/s11658-008-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Kaneda Y, Sato M, Saeki Y, Wataya-Kaneda M, Hoffmann A. LBP-p40 binds DNA tightly through associations with histones H2A, H2B, and H4. Biochem Biophys Res Commun. 1998;253:277–282. doi: 10.1006/bbrc.1998.9699. [DOI] [PubMed] [Google Scholar]
- Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells: the role of the M(r) 67,000 laminin receptor. Cancer Res. 2004;64:3572–3579. doi: 10.1158/0008-5472.CAN-03-3424. [DOI] [PubMed] [Google Scholar]
- Keppel E, Schaller HC. A 33 kDa protein with sequence homology to the ‘laminin binding protein' is associated with the cytoskeleton in hydra and in mammalian cells. J Cell Sci. 1991;100 (Part 4:789–797. doi: 10.1242/jcs.100.4.789. [DOI] [PubMed] [Google Scholar]
- Menard S, Castronovo V, Tagliabue E, Sobel ME. New insights into the metastasis-associated 67 kD laminin receptor. J Cell Biochem. 1997;67:155–165. [PubMed] [Google Scholar]
- Rao CN, Castronovo V, Schmitt MC, Wewer UM, Claysmith AP, Liotta LA, et al. Evidence for a precursor of the high-affinity metastasis-associated murine laminin receptor. Biochemistry. 1989;28:7476–7486. doi: 10.1021/bi00444a047. [DOI] [PubMed] [Google Scholar]
- Landowski TH, Dratz EA, Starkey JR. Studies of the structure of the metastasis-associated 67 kDa laminin binding protein: fatty acid acylation and evidence supporting dimerization of the 32 kDa gene product to form the mature protein. Biochemistry. 1995;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- Buto S, Tagliabue E, Ardini E, Magnifico A, Ghirelli C, van den Brule F, et al. Formation of the 67-kDa laminin receptor by acylation of the precursor. J Cell Biochem. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Menard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Res Treat. 1998;52:137–145. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- Satoh K, Narumi K, Abe T, Sakai T, Kikuchi T, Tanaka M, et al. Diminution of 37-kDa laminin binding protein expression reduces tumour formation of murine lung cancer cells. Br J Cancer. 1999;80:1115–1122. doi: 10.1038/sj.bjc.6690474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H, Koga K, Fujimura Y, Yamada K. A receptor for green tea polyphenol EGCG. Nat Struct Mol Biol. 2004;11:380–381. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- Shammas MA, Neri P, Koley H, Batchu RB, Bertheau RC, Munshi V, et al. Specific killing of multiple myeloma cells by (-)-epigallocatechin-3-gallate extracted from green tea: biologic activity and therapeutic implications. Blood. 2006;108:2804–2810. doi: 10.1182/blood-2006-05-022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiman J, Tseng JC, Zheng Y, Meruelo D. Multiple functions of the 37/67-kd laminin receptor make it a suitable target for novel cancer gene therapy. Mol Ther. 2010;18:63–74. doi: 10.1038/mt.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick MB, Noll E, Perrimon N. The Drosophila stubarista phenotype is associated with a dosage effect of the putative ribosome-associated protein D-p40 on spineless. Genetics. 1993;135:553–564. doi: 10.1093/genetics/135.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero M, Marcilla A, Sentandreu R, Valentin E. A Candida albicans 37 kDa polypeptide with homology to the laminin receptor is a component of the translational machinery. Microbiology. 1998;144 (Part 4:839–847. doi: 10.1099/00221287-144-4-839. [DOI] [PubMed] [Google Scholar]
- Zuber C, Knackmuss S, Zemora G, Reusch U, Vlasova E, Diehl D, et al. Invasion of tumorigenic HT1080 cells is impeded by blocking or downregulating the 37-kDa/67-kDa laminin receptor. J Mol Biol. 2008;378:530–539. doi: 10.1016/j.jmb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Castronovo V, Taraboletti G, Sobel ME. Functional domains of the 67-kDa laminin receptor precursor. J Biol Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- Landowski TH, Uthayakumar S, Starkey JR. Control pathways of the 67 kDa laminin binding protein: surface expression and activity of a new ligand binding domain. Clin Exp Metastasis. 1995;13:357–372. doi: 10.1007/BF00121912. [DOI] [PubMed] [Google Scholar]
- Kazmin DA, Hoyt TR, Taubner L, Teintze M, Starkey JR. Phage display mapping for peptide 11 sensitive sequences binding to laminin-1. J Mol Biol. 2000;298:431–445. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- Mathew S, Fu L, Fiorentino M, Matsuda H, Das B, Shi YB. Differential regulation of cell type-specific apoptosis by stromelysin-3: a potential mechanism via the cleavage of the laminin receptor during tail resorption in Xenopus laevis. J Biol Chem. 2009;284:18545–18556. doi: 10.1074/jbc.M109.017723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng JC, Hurtado A, Yee H, Levin B, Boivin C, Benet M, et al. Using sindbis viral vectors for specific detection and suppression of advanced ovarian cancer in animal models. Cancer Res. 2004;64:6684–6692. doi: 10.1158/0008-5472.CAN-04-1924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.