Abstract
The Positive Transcriptional Elongation Factor b (P-TEFb), a heterodimer of CDK9 and Cyclin T1, is widely implicated in control of basal gene expression. Here, P-TEFb is involved in transitioning paused RNA polymerase II to enter productive transcriptional elongation mode by phosphorylating negative elongation factors and Ser2 of the heptad repeat in the RNA Pol II COOH terminal domain (CTD). This perspective will examine recent work in two unrelated inducible signaling pathways that illustrate the central role of P-TEFb in mediating cytokine inducible transcription networks. Specifically, P-TEFb has been recently discovered to play a key role in TNF-inducible NF-κB activation and IL-6-inducible STAT3 signaling. In these signaling cascades, P-TEFb forms protein complexes with the activated nuclear RelA and STAT3 transcription factor in the cellular nucleoplasm, an association important for P-TEFb’s promoter targeting. Studies using siRNA-mediated knockdown and/or selective CDK inhibitors show that P-TEFb plays a functional role in activation of a subset of NF-κB-dependent targets and all STAT3-dependent genes studied to date. Interestingly, cytokine inducible genes that are sensitive to P-TEFb inhibition share an induction mechanism requiring inducible RNA Pol II recruitment. Chromatin immunoprecipitation studies have preliminarily indicated that this recruitment is dependent on CDK enzymatic activity. The potential of inhibiting P-TEFb as an anti-inflammatory therapy in innate immunity and systemic inflammation will be discussed.
Keywords: NF-κB, cyclin dependent kinases (CDK), PTEF-b, STAT3, inflammation
Enhanced transcription is a major mechanism underlying metazoan gene expression that allows the cell to rapidly respond to developmental, hormonal or inflammatory stimuli, and still produce phenotypic memory of the stimulus. Transcription is a coordinated response involving several distinct, inter-dependent processes, including preinitiation complex formation, chromatin enhanceosome formation/coactivator recruitment, de-repression and transcriptional elongation. Preinitiation complex formation refers to the process where a basal promoter is bound by a multiprotein complex of TATA box binding protein, kinases, and ubiquitin ligases.1 In some promoters, hypophosphorylated RNA polymerase II is also bound; in this form, RNA Pol II produces a series of short, “unproductive”, transcripts that are truncated and rapidly degraded. Enhanceosome formation is linked to inducible gene expression, and is a process initiated by binding activated sequence specific transcription factors and their associated coactivators, such as p300/CBP or the Mediator complex.1, 2 These coactivators contain histone acetyltransferase activity that modify repressive nucleosomes and allow the upstream coactivators to productively couple with the basal/pre-initiation complex. In highly inducible genes, RNA Pol II recruitment occurs coincident with inducible transcription factor binding.3 Subsequently, RNA Pol II is relieved of repression, and acquires its processive form, producing full-length, spliced polyadenylated transcripts that encode the target protein, a process referred to as transcriptional elongation. The relative contributions of these steps in any inducible genetic network is not fully understood.
Because of its link with many pathological conditions, inducible gene expression in response to cytokine cascades has been intensively studied. Cytokines are secreted proteins that function as hormonal mediators of inflammation, innate responses to cellular infection, hepatic acute phase response and adaptive immunity.4, 5 Upon binding cognate cell surface receptors, signaling cascades are activated that affect the responding cell to induce genes that encode homeostatic proteins, adhesion molecules important in leukocyte activation or recruitment, and further amplify the inflammatory signal by secreting downstream cytokine cascades. This perspective will focus on recent work that has illuminated the critical of the positive transcription elongation factor b (P-TEFb), in cytokine inducible expression in two distinct cytokine signaling pathways. Understanding this process at the molecular level may lead to potential new therapeutics that can be used to modify the host inflammation, augment immune responses, or permit more rapid adaptive immune responses.
P-TEFb and Transcriptional Elongation
It has been observed for some time that RNA Pol II is paused on the 5’ end of promoters.6 Here, RNA Pol II is complexed with the 5,6-dichloro-1-β-D ribofuranosyl-benzimidaole (DSB)-sensitive factor complex (DRIF) and negative elongation factor (NELF). In the process of gene activation, RNA Pol II is phosphorylated at Ser5 in the repeated YSDPTSPS motif (“heptad repeats”) in the COOH terminal domain (CTD) by the preinitiation complex subunit TFIIH, an event that signals its transition to mode sufficient for promoter clearance and transcript elongation 1. Studies have shown phosphorylation of SPT4 and 5 (components of the DRIF complex) and NELF releases these proteins from Pol II. Concomitantly, phosphorylation of the Ser2 residue of the heptad repeats allows Pol II to enter processive mode where full-length transcripts are produced.7
The homologous proteins CDKs -7, -8 and -9 all share RNA Pol II CTD kinase activity. Of these, CDK9 has emerged as one of the major CTD kinases that phosphorylates Ser2 in the heptad repeat. CDK9 is a 43 kDa protein first identified by PCR amplification of a mouse library using degenerate primers to amplify genes homologous with the cdc2 NH2 terminal kinase domain.8 Sequence analysis showed CDK9 had a highly homologous PITALRE sequence to the cdc2 PSTAIRE motif in the NH2 terminal β sheet. Like other CDKs, CDK9 associates with cyclins (Ccns)-T, -K and –E; this association is important for CDK9 kinase activity. Most of the CDK9 in the cell is associated with nuclear CcnT1, found distributed primarily in the nucleus and associated with nuclear speckles and the splicing factor, SC35.9, 10 The complex of CDK9-Ccn T1 is homologous to the Drosophila melanogaster positive transcriptional elongation factor (P-TEFb7). P-TEFb is a kinase whose activity is not detectably regulated through the cell cycle.
Biochemical fractionation studies have shown that nuclear P-TEFb exists in two functionally distinct complexes. Half of nuclear P-TEFb is found as an inactive complex associated with HEXIM1 and 7SK snRNA.11 Tandem affinity isolation and mass spectrometry have shown that P-TEFb also associates with the bromodomain containing protein, termed Brd4.12 Functional assays using siRNA mediated Brd4 knockdown have indicated that Brd4 is required for P-TEFb induced HIV LTR transcriptional elongation. Currently it is thought that the bromodomain of Brd4 may be required for P-TEFb activity through its ability to associate with acetylated lysines and thereby facilitates its chromatin access or binding. In addition, Brd4 may bind components of the Mediator complex, including TRAPs-230, -170, -100 and -80.12 The Mediator binding function of Brd4 in PTEFb function has not yet been addressed.
Mechanisms underlying P-TEFb Activation
Despite its name, the kinase activity of CDK9 is not regulated in a cell cycle-dependent manner, but rather via its association with specific proteins (Figure 1). Work on the mode of regulation of P-TEFb in activation of the HIV TAR sequence has shown that the activation occurs in several distinct steps. Activated P-TEFb can be produced by dissociation of HEXIM1 and 7SK snRNA.13 First, CDK undergoes autophosphorylation, an event that allows its associated CcnT1 to bind TAT via release of an intermolecular association.14 The concept that there are cellular kinases that activate CDK9 is intriguing, but their identities have not yet been described. Additionally CDK9 is inducibly acetylated, a phenomenon whose effects on P-TEFb activity are currently unclear or controversial. One group has reported that the histone acetyltransferase activity of p300/CBP acetylates CDK9 on Lys44 in the ATP binding domain, enhancing its kinase activity toward the RNA Pol II CTD.15 By contrast, the GNAT histone acetylases, P/CAF and GCN5, acetylate Lys44 and Lys48 in the ATP binding domain of CDK9 resulting in inhibition of kinase activity and altered nuclear distribution into insoluble nuclear matrix.16 One explanation for these discordant results may be that other sites on CDK9 whose modification that affect the response to CDK9 acetylation on Lys44.
Figure 1. P-TEFb states in the resting cell.
Shown is a schematic view of the nuclear states of activated P-TEFb. Approximately half of nuclear P-TEFb is inactive state associated with HEXIM1 and 7SK RNA. The component that is actively involved in RNA Pol II dependent transcription is associated with Brd4. A small fraction of CDK9 is undergoing active nuclear-cytoplasmic shuttling mediated by CRM1-dependent nuclear export. Abbreviations: Brd4, bromodomain 4; P-TEFb, positive transcriptional elongation factor-b.
Although the majority of CDK9 is complexed with CcnT1 in nuclear speckles, a small fraction of CDK9 is found in an apparently uncomplexed form in the cytoplasm. Inhibition of nuclear exportin, CRM1, using leptomycin B leads to the nuclear accumulation of CDK9, leading to the conclusion that a fraction of the “free” (non Ccn-complexed) CDK9 is dynamically undergoing cytoplasmic-nuclear shuttling.9 An important unresolved issue is what controls physiological P-TEFb activation and/or partitioning into activated Brd4, or inactive HEXIM1-7SK snRNA complexes. Previous work has shown that P-TEFb primarily plays a role in expression of genes containing a TATA box 17, however its mechanism for recruitment to these promoters is not fully understood. Recent work has indicated that P-TEFb association with nuclear inducible transcription factors play an important role in P-TEFb recruitment in cytokine cascades.18, 19 These include the TNF-induced NF-κB and the IL-6 induced STAT3 signaling pathways, discussed separately in detail below.
The TNF-induced NF-κB pathway
TNF is released by tissue resident macrophages upon encountering infecting viral or bacterial products and activates neighboring cells by binding a ubiquitously expressed receptor, the TNFRI. Employing systematic microarray analyses of the TNF-induced transcriptional response, our laboratory has shown the transcription factor nuclear factor –κB (NF-κB) is responsible for mediating a significant fraction of the genomic response.20–23 The transcriptional activator subunits of the NF-κB family are complexed and inactivated in the cellular cytoplasm by binding inhibitors of NF-κBs, IκBs, a class of ankryin repeat domain-containing proteins that block nuclear translocation and inhibit DNA binding.24 NF-κB activation involves RelA liberation from its cytoplasmic inhibitors, allowing it to translocate into the nucleus and activate target genes (Figure 2).
Figure 2. Role of P-TEFb NF-κB-dependent transcription.
Schematic view of the TNF signaling pathway. Two pathways are activated downstream of liganded TNF, each required for NF-κB dependent activation. IKK mediates RelA translocation by phosphorylation of the IκBα inhibitor; PKAc responds to ROS generation to induce Ser276 phosphorylation. Phospho-Ser276 RelA is competent for P-TEFb association, a complex that activates IL-8 and other cytokine genes. This process involves RNA Pol II recruitment. By contrast, unphosphorylated RelA activates IκBα expression. Abbreviations: IKK, IκB kinase; TNFRI, TNF receptor-I.
NF-κB is controlled by distinct regulatory pathways, termed the “canonical”, “noncanonical” and “cross-talk” pathways, that are activated by selective stimuli and involve liberating RelA from distinct cytoplasmic complexes.25, 26 Of these, the canonical pathway is the major pathway induced by cytokines;27 the noncanonical and cross-talk pathways are induced by RNA virus infection or lymphokines and, for brevity, are not further discussed here.28–30 Recent work seeking to understand the canonical pathway has shown that it consists of separable “modules” by which we refer to signaling subpathways whose action are to control either nuclear translocation or transcriptional activation. Specifically, TNF stimulation controls release of RelA sequestered by IκBα via an IκB kinase (IKK)-IκBα module, whereas licensing of transcriptional activation of RelA by Ser276 phosphorylation via an reactive oxygen species (ROS)-catalytic subunit of protein kinase A (PKAc) module.19, 31
TNFα binding to TNFRI transduces intracellular signals by inducing submembranous complex formation with adapter signaling proteins that interact with the death domain of the TNFRI cytoplasmic tail. These adaptor proteins include the TNF receptor associated factor (TRAFs) -2, -6 and receptor interacting protein [RIP32], proteins with ubiquitin ligase activity responsible for downstream activation of MAP3Ks, particularly the TGFβ associated kinase-1 (TAK1), a process that occurs via a Lys63-linked ubiquitination step.33 Subsequently, the activated submembranous complex recruits and activates IKK 34, the first committed step in NF-κB translocation, by TAK1-initiated phosphorylation.33
The IKK is a multi-subunit kinase complex 35, whose core is composed of two highly homologous serine-threonine kinases, IKK-α and β, associated with a regulatory subunit, IKKγ.36–38 Although IKKβ plays the major catalytic role, the activation and behavior of the IKK is determined by the associated regulatory IKKγ subunit. IKKγ plays multiple roles in IKK activation through its ability to organize the assembly of IKKs into the activated high molecular weight complex,34, 39 bind ubiquitinated signaling adapters,40, 41 recruit the IκBα inhibitor into the activated IKK complex where it becomes phosphorylated marking IκBα for degradation,39 and to serve as an adapter molecule to recruit upstream kinases that phosphorylate the catalytic subunits.40, 42, 43 Moreover, we have found that IKKγ is alternatively spliced in an exon that encodes for its self-association and oligomerization domains, a feature that allows distinct IKK complexes to selectively couple to distinct activating cascades.26, 44 Through these activities, IKKγ forms a molecular bridge between IKK, its upstream activators, and its substrate.
TNF induced IKK activation is mediated by phosphorylation at Ser residues in the conserved IKK α/β activation loop, producing autophosphorylation.45 Activated IKKβ then phosphorylates IκBα on Ser residues 32 and 36 in its NH2 terminal regulatory domain,27 making it a substrate for proteolysis through the 26S proteasome and calpain pathways.35, 46 Liberated from its IκBα inhibitor, RelA rapidly enters the nucleus.
Systematic microarray studies conducted by our laboratory have revealed significant complexity in the genomic response.22, 47 For example, time series analyses in TNF stimulated cells have found that expression of NF-κB-dependent genes is nonsynchronous, separable by the timing of peak gene expression, with each expression group encoding distinct biological functions 21, 22, 25. This characteristic allows coordinated evoluation of cellular biological responses to TNF activation.22, 25
RelA Ser276 phosphorylation is necessary for transcriptional competency via P-TEFb
Although NF-κB translocation is necessary for target gene expression, it is not sufficient 48. Recent work has shown that site-specific phosphorylation of NF-κB is required for transcriptional activation. RelA is inducibly phosphorylated on a number of Ser residues; Ser276 and Ser536 are thought to be the most important in regulating transcriptional activity.19, 31, 49, 50 In this regard, we have defined a TNF-induced regulatory pathway mediated by intracellular ROS as a second messenger that is required for NF-κB dependent gene expression. Here, inhibition of ROS formation blocks Ser276 phosphorylation and NF-κB dependent gene expression without affecting NF-κB translocation.31, 48 This pathway is dependent on the catalytic subunit of protein kinase C (PKAc), a kinase that co-purifies with IκBα49 and whose activity is induced by TNF.31 Because inhibition of ROS formation or siRNA mediated PKAc knockdown prevents TNF-induced RelA Ser276 phosophorylation without affecting its nuclear translocation, we have interpreted these findings to indicate that the ROS-PKAc transcriptional activating module is a separate pathway whose function is also necessary for NF-κB dependent gene expression (Figure 2).
Phosphorylation at Ser276 in the mid molecule of RelA is thought to reduce intermolecular NH2 and COOH terminal interactions, allowing phospho-Ser276 RelA to complex with p300/CBP coactivators,51 resulting in RelA acetylation52 and form a stable enhanceosome on endogenous gene targets in ChIP assays.31
We have recently discovered that RelA Ser276 phosphorylation also allows complex formation with P-TEFb, a process that results in phospho-RelA to associate with both transcriptional coactivators as well as bridge with the transcriptional elongation machinery.19 Nondenaturing co-immunoprecipitation experiments show that RelA inducibly associates with P-TEFb, in a manner that is absolutely dependent on RelA Ser276 phosphorylation. This association results in inducible CDK9 binding to the downstream genetic targets of NF-κB, including IκBα, Groβ and IL-8 genes. Because we could selectively disrupt Ser276 phosphorylation without affecting NF-κB translocation using antioxidants, we used these agents to test the effect of Ser276 phosphorylation on activation of downstream genes. These findings revealed the surprising result that a subset of genes, Groβ and IL-8, were absolutely dependent on phospho- Ser276 RelA, but others, IκBα, were not (Figure 2).
To more selectively test the role of CDK9 interaction, CDK9 was downregulated by siRNA mediated transfection. The same subset of genes, Groβ and IL-8 were significantly inhibited, whereas the phospho-Ser276 RelA-independent genes were not affected. Interestingly only the genes requiring inducible RNA Pol II recruitment as a mechanism in their promoter activation were inhibited by CDK9 downregulation. These data suggest that P-TEFb is involved in the activation of a subset of NF-κB-dependent genes whose mechanism of activation involves Pol II recruitment.
Role of CDK9 in IL-6 signaling via the Jak-STAT3 pathway
IL-6 is a ubiquitously expressed anti-inflammatory cytokine that plays a major role in hepatic acute phase response and vascular inflammation.5 IL-6 signaling in target cells is initiated by its binding to the IL-6Rα receptor, an event that triggers ligand-mediated oligomerization with the ubiquitously expressed transmembrane gp130 β-subunit, and subsequent formation of a hexameric IL-6·IL-Rα·gp130 high-affinity complex [Figure 3, and ref. (53)]. Receptor ligation induces conformational changes in the cytoplasmic domains of gp130 that bring Janus tyrosine kinases (JAKs) into close proximity. This molecular crowing results in trans-autophosphorylation, an event that initiates IL-6 signaling.54 JAK1, in turn, phosphorylates gp130 on the docking sites for the signal transducers and activators of transcription (STAT); STAT isoforms -1 and -3 are then recruited, where they, too, become phosphorylated.55
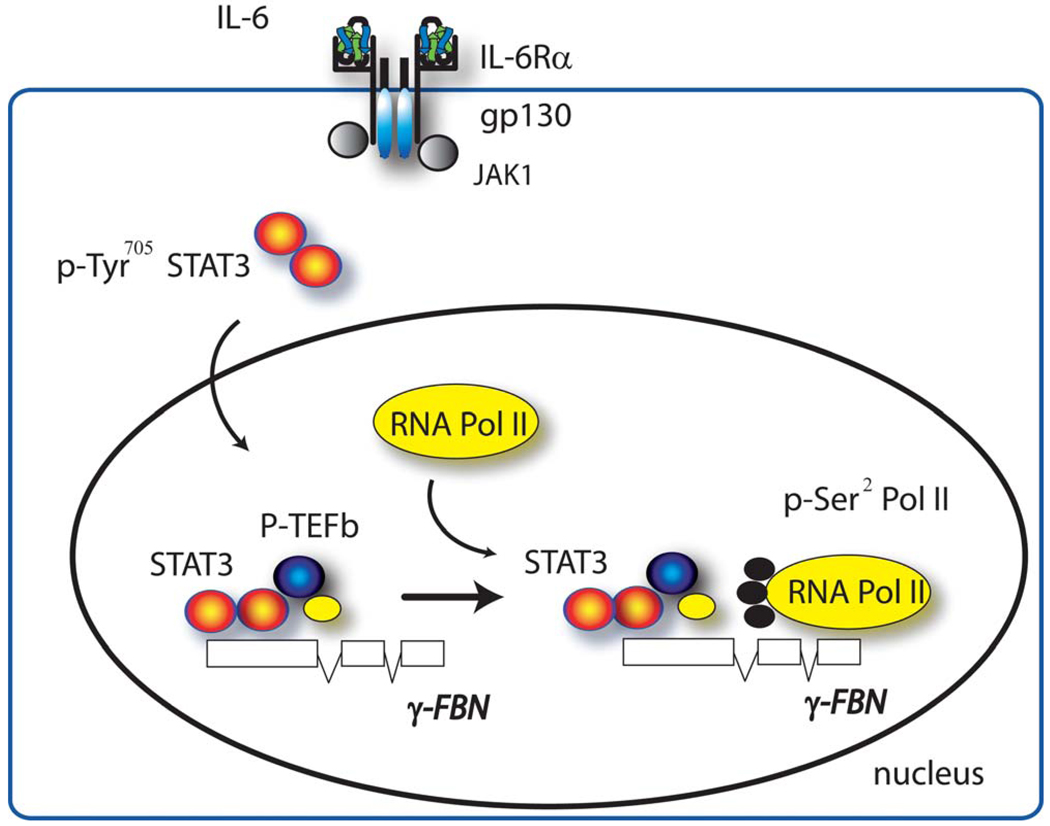
Figure 3. Role of P-TEFb in STAT3-dependent transcription.
Schematic view of IL-6 signaling pathway. Tyrosine phosphorylated STAT3 is induced to form a complex with P-TEFb via association with CDK-9. Abbreviations: IL-6Rα, IL-6 receptor-α; gp130, glycoprotein 130 (transducin); JAK, janus kinase.
Of the signaling pathways downstream of the IL-Rα·gp130 complex, STAT3 appears to play a major role. Tyr phosphorylated STATs-1 and -3 form intermolecular SH2–SH3 domain interactions, an event that is coupled to its nuclear translocation and transcriptional activation of target genes.54, 55 Analyses of complex formation with STAT3-dependent transcriptional enhancers have shown that STAT3 undergoes additional post-translational modifications that permits interactions with co-factors and co-activators.56
As with the NF-κB pathway, STAT3 associates with the p300/CBP coactivator, whose actions are to open chromatin structure, allowing other chromatin-modifying proteins to bind to DNA and activate transcription.57, 58 The p300/CBP association requires both the NH2-terminal modulatory domain and the COOH-terminal transactivation domain of STAT3.56, 57 Studies from our laboratory first described two novel acetylation sites on the STAT3 NH2 terminus at Lys49 and Lys87 that are required to stabilize the STAT3-p300/CBP complex through an additional interaction mediated by the modified STAT3 NH2 terminus.56
By several experimental techniques, we and others have found that activated nuclear STAT3 binds CDK9.18, 59 Importantly co-immunoprecipitation experiments from our laboratory indicate that both phospho-Tyr705 STAT3 and Ac Lys87 STAT3 are associated with P-TEFb.18 These observations indicate that the transcriptionally active form of STAT3 is complexed with P-TEFb, and that this complex forms in the nucleoplasm prior to DNA interaction. Interestingly, STAT3 appears to directly contact CDK9 in the P-TEFb complex via its NH2 and COOH terminal domains.18, 59 This interaction is apparently distinct from how NF-κB complexes with P-TEFb as prior mapping studies indicate that RelA binds the cyclin box of CcnT1.60 These findings indicate that multiple sites for transcription factor interaction exist on P-TEFb.
Chromatin immunoprecipitation (ChIP) assays using primers selective for the upstream and coding regions of STAT3-dependent γ-fibrinogen (FBN) show that P-TEFb is not uniformly concentrated on the 5’ regulatory site of the γ- FBN promoter. Interestingly a greater abundance of PTEFb is found in the coding region of the gene.18 These findings suggest additional roles of P-TEFb in inducible expression outside of initial Ser2 CTD phosphorylation, such as those described in the heat shock response in Drosophila where P-TEFb plays an important role in 3’RNA processing.61 Whether 3’ mRNA processing is mediated by P-TEFb in mammalian gene expression, and if so, if it plays a role in cytokine inducible gene expression will require further work.
siRNA mediated knockdown of CDK9 confirms the functional role of P-TEFb in STAT3-dependent gene expression of the endogenous γ- FBN gene52 and p21waf.59 As observed for the NF-κB-dependent genes, activation of the STAT3-dependent genes involves recruitment of Pol II. In this system, ChIP analysis of the effects of CDK9 inhibitor show that Pol II recruitment is itself dependent on functional CDK activity.18 These studies further indicate that the IL-6-STAT3 signaling pathway can be affected by modulating P-TEFb action. The question whether P-TEFb is a general transcriptional enhancer necessary for activating all STAT3-dependent genes, or whether only specific subsets of STAT3-dependent genes require P-TEFb has not been fully explored.
Implications for modulation of inflammation
Glucocorticoids are potent anti-inflammatory hormones that act to inhibit TNF-inducible gene expression. Work from the Yamamoto laboratory has discovered that the activated glucocorticoid receptor may antagonize NF-κB activation by blocking P-TEFb recruitment.62 This study also indicated that heterogenous mechanisms are involved in NF-κB-dependent gene expression with the IL-8 gene being sensitive to the inhibitory actions of glucocorticoids, whereas IκBα is not. These findings are remarkably similar to those we reported after antagonizing NF-κB·P-TEFb complex formation using antioxidants to inhibit phospho-Ser276 formation, which selectively inhibited IL-8, but not IκBα, gene expression.63 We interpret these findings to suggest that both glucocorticoids and antioxidants exert some of their anti-inflammatory actions by disrupting the NF-κB·P-TEFb interaction. The design of more potent or selective inhibitors of this association may be useful clinically to selectively disrupt subnetworks of NF-κB induced inflammation.
Similarly, disruption of STAT3 signaling may have specific applications in modulating vascular inflammation, a process where IL-6 may play a central role.5 Selective disruption of STAT3·P-TEFb may have significant effects on atherosclerosis and aortic remodeling in cardiovascular disease.
Summary
In this perspective, I have highlighted recent work illustrating the concept that P-TEFb plays a central role in mediating cytokine inducible gene expression. Although the bromodomain containing Brd4 protein appears to be important for P-TEFb activation, our studies also show that P-TEFb is targeted to specific inducible genes by nucleoplasmic association with activated NF-κB and STAT3 transcription factors. Currently we think P-TEFb forms complexes with NF-κB via distinct molecular contacts than those used for STAT3 interaction. A number of interesting questions are currently unresolved. What is the role of the nucleocytoplasmic shuttling form of CDK9? What are the full spectrum of substrates for the P-TEFb complex; and how does CDK9 phosphorylation affect their activity? Does P-TEFb play a role outside of simply inducing transcriptional elongation, for example, in 3’RNA processing? Is P-TEFb is a universal enhancer for STAT3 dependent transcription, or does it play a role in a subset of STAT3-dependent target genes? Further investigation into these questions and examination of the effect of disruption of this complex in inflammatory disease will be illuminating.
ACKNOWLEDGEMENTS
Work in my lab was supported by NHLBI 1 P50 HL083794, NHLBI RO1 HL70925. Core Laboratory support was from NIEHS grant P30 ES06676 (to J. Ward, UTMB) and BAA-HL-02-04 (A. Kurosky, UTMB).
REFERENCES
- 1.Kornberg RD. The molecular basis of eukaryotic transcription. Proceedings of the National Academy of Sciences. 2007 August 7;104(32):12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. [Review] [167 refs] Blood. 2000 February 1;95(3):745–755. [PubMed] [Google Scholar]
- 3.Brasier AR, Jamaluddin M, Casola A, Duan W, Shen Q, Garofalo R. A promoter recruitment mechanism for TNFα-induced IL-8 transcription in type II pulmonary epithelial cells: Dependence on nuclear abundance of Rel A, NF-kB1 and c-Rel transcription factors. J Biol Chem. 1998;273:3551–3561. doi: 10.1074/jbc.273.6.3551. [DOI] [PubMed] [Google Scholar]
- 4.Ramadori G, Christ B. Cytokines and the hepatic acute-phase response. Seminars in Liver Disease. 1999;19(2):141–155. doi: 10.1055/s-2007-1007106. [DOI] [PubMed] [Google Scholar]
- 5.Hou J, Tieu B, Ray S, Recinos AI, Cui R, Tilton R, Brasier AR. Roles of IL-6-gp130 Signaling in Vascular Inflammation. Current Cardiology. 2008;4:1–13. doi: 10.2174/157340308785160570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spencer CA, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–785. [PubMed] [Google Scholar]
- 7.Price DH. P-TEFb, a Cyclin-Dependent Kinase Controlling Elongation by RNA Polymerase II. Mol Cell Biol. 2000 April 15;20(8):2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grana X, Luca AD, Sang N, Fu Y, Claudio PP, Rosenblatt J, Morgan DO, Giordano a. PITALRE, a Nuclear CDC2-Related Protein Kinase that Phosphorylates the Retinoblastoma Protein in vitro. Proceedings of the National Academy of Sciences. 1994 April 26;91(9):3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napolitano G, Licciard P, Carbone R, Majello B, Lania L. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of CyclinT1 promotes its nuclear localization. J Cell Physiol. 2002;192:209–215. doi: 10.1002/jcp.10130. [DOI] [PubMed] [Google Scholar]
- 10.Hermann CH, Mancini MA. The CDK9 and cyclin t subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the Large Inactive P-TEFb Complex Indicates That It Contains One 7SK Molecule, a Dimer of HEXIM1 or HEXIM2, and Two P-TEFb Molecules Containing Cdk9 Phosphorylated at Threonine 186. J Biol Chem. 2005 August 5;280(31):28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 12.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Molecular Cell. 2005 August 19;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 13.Egloff S, Van Herreweghe E, Kiss T. Regulation of Polymerase II Transcription by 7SK snRNA: Two Distinct RNA Elements Direct P-TEFb and HEXIM1 Binding. Mol Cell Biol. 2006 January 15;26(2):630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong YW, Zhou Q. Relief of Two Built-In Autoinhibitory Mechanisms in P-TEFb Is Required for Assembly of a Multicomponent Transcription Elongation Complex at the Human Immunodeficiency Virus Type 1 Promoter. Mol Cell Biol. 2000 August 15;20(16):5897–5907. doi: 10.1128/mcb.20.16.5897-5907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, Yoon HG, Qin J, Wong J. Regulation of P-TEFb Elongation Complex Activity by CDK9 Acetylation. Mol Cell Biol. 2007 July 1;27(27):4641–4651. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabo A, Lusic M, Cereseto A, Giacca M. Acetylation of Conserved Lysines in the Catalytic Core of Cyclin-Dependent Kinase 9 Inhibits Kinase Activity and Regulates Transcription. Mol Cell Biol. 2008 April 1;28(7):2201–2212. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montanuy I, Torremocha R, Hernandez-Munain C, Sune C. Promoter Influences Transcription Elongation: TATA-BOX ELEMENT MEDIATES THE ASSEMBLY OF PROCESSIVE TRANSCRIPTION COMPLEXES RESPONSIVE TO CYCLIN-DEPENDENT KINASE 9. J Biol Chem. 2008 March 21;283(12):7368–7378. doi: 10.1074/jbc.M706243200. [DOI] [PubMed] [Google Scholar]
- 18.Hou T, Ray S, Brasier AR. The functional role of an IL-6 inducible CDK9-STAT3 complex in human ã-fibrinogen gene expression. J Biol Chem. 2007;282:37091–37102. doi: 10.1074/jbc.M706458200. [DOI] [PubMed] [Google Scholar]
- 19.Nowak D, Tian B, Jamaluddin M, Boldogh I, Vergara L, Choudhary JS, Brasier AR. Rel A Ser 276 Phosphorylation Is Required For Activation Of A Subset Of NF-kB Dependent Genes By Recruiting Cdk-9/Cyclin T1 Complexes. Molecular & Cellular Biology. 2008 doi: 10.1128/MCB.01152-07. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian B, Zhang Y, Luxon BA, Garofalo RP, Casola A, Sinha M, Brasier AR. Identification Of NF-κB Dependent Gene Networks In Respiratory Syncytial Virus-Infected Cells. J Virol. 2002;76:6800–6814. doi: 10.1128/JVI.76.13.6800-6814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian B, Brasier AR. Identification of an NF-kB Dependent Gene Network. Recent Progress in Hormone Research. 2003;58:95–130. doi: 10.1210/rp.58.1.95. [DOI] [PubMed] [Google Scholar]
- 22.Tian B, Nowak D, Brasier AR. A TNF Induced Gene Expression Program Under Oscillatory NF-κB Control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, Brasier AR. Microarrays and Transcription Factor Networks. Landes Bioscience; 2005. The Nuclear Factor-κB (NF-κB) Gene Regulatory Network. [Google Scholar]
- 24.Beg AA, Baldwin ASJ. The I kappa B proteins: multifunctional regulators of Rel/NF-kappa B transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 25.Brasier AR. The NF-κB Signaling Network: Insights from systems approaches. In: Brasier AR, Lemon SM, Garcia-Sastre A, editors. Cellular Signaling And Innate Immune Responses To RNA Virus Infections. American Society for Microbiology; 2008. pp. 119–135. [Google Scholar]
- 26.Brasier AR. The NF-κB Regulatory Network. Cardiovascular Toxicology. 2006;6(2):111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 27.Karin M. The beginning of the end: IkB kinase (IKK) and NF-kB activation. J Biol Chem. 1999;274(27339):27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 28.Choudhary S, Boldogh S, Garofalo RP, Jamaluddin M, Brasier AR. RSV influences NF-κB dependent gene expression through a novel pathway involving MAP3K14/NIK expression and nuclear complex formation with NF-κB2. J Virol. 2005;79:8948–8959. doi: 10.1128/JVI.79.14.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKalpha of a Second, Evolutionary Conserved, NF-kappa B Signaling Pathway. Science. 2001 August 24;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 30.Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A Fourth I[kappa]B Protein within the NF-[kappa]B Signaling Module. Cell. 2007 January 26;128(2):369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamaluddin M, Wang S, Boldogh I, Tian B, Brasier AR. TNF-α-Induced NF-κB/Rel A Ser 276 Phosphorylation And Enhanceosome Formation On The IL-8 Promoter Is Mediated By A Reactive Oxygen Species (ROS)-Dependent Pathway. Cellular Signalling. 2007;9:1419–1433. doi: 10.1016/j.cellsig.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 33.Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J Mol Biol. 2003 February 7;326(1):105–115. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 34.Poyet J-L, Srinivasula SM, Lin J-H, Fernandes-Alnemri T, Yamaoka S, Tsichlis PN, Alnemri ES. Activation of the IkB kinases by RIP via IKKg/NEMO-mediated oligomerization. J Biol Chem. 2000;275:37966–37977. doi: 10.1074/jbc.M006643200. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T. Catalysis by a multiprotein IkB kinase complex. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 36.Mercurio F, Zhu H, Murray BW, Shevchenko a, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkB kinases essential for NF-kB activation. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 37.Woronicz J, Gao X, Cao Z, Rothe M, Goeddel DV. IkB kinase-b: NF-kB activation and complex formation with IkB Kinase a and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 38.Karin M, Ben Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. [Review] [235 refs] Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto Y, Kim DW, Kwak YT, Prajapati S, Verma U, Gaynor RB. IKKgamma/NEMO facilitates the recruitment of the IkappaB proteins into the IkappaB kinase complex. J Biol Chem. 2001 September 28;276(39):36327–36336. doi: 10.1074/jbc.M104090200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang SQ, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity. 2000 March;12(3):301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
- 41.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-[kappa]B activation. Nat Cell Biol. 2006 April;8(4):398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 42.Ye J, Xie X, Tarassishin L, Horwitz MS. Regulation of the NF-kappaB activation pathway by isolated domains of FIP3/IKKgamma, a component of the IkappaB-alpha kinase complex. J Biol Chem. 2000 March 31;275(13):9882–9889. doi: 10.1074/jbc.275.13.9882. [DOI] [PubMed] [Google Scholar]
- 43.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex [see comments] Nature. 1998 September 17;395(6699):297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 44.Hai T, Yeung ML, Wood TG, Wei Y, Yamaoaka S, Gatalica Z, Jeang K-T, Brasier AR. An Alternative Splice Product of IκB Kinase (IKK)-γ, IKK γ−Δ, Differentially Mediates Cytokine And HTLV-I Tax Induced NF-κB Activation. J Virol. 2006;80(9):4227–4241. doi: 10.1128/JVI.80.9.4227-4241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkB kinase activity through IKKb subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 46.Han Y, Weinman S, Boldogh I, Walker RK, Brasier AR. Tumor necrosis factor-alpha-inducible IkappaBalpha proteolysis mediated by cytosolic m-calpain. A mechanism parallel to the ubiquitin-proteasome pathway for nuclear factor-kappab activation. J Biol Chem. 1999 January 8;274(2):787–794. doi: 10.1074/jbc.274.2.787. [DOI] [PubMed] [Google Scholar]
- 47.Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of direct genomic targets downstream of the NF-kappa B transcription factor mediating TNF signaling. J Biol Chem. 2005 February 18;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- 48.Vlahopoulos S, Boldogh I, Brasier AR. NF-κB dependent induction of interleukin-8 gene expression by tumor necrosis factor α: evidence for an antioxidant sensitive activating pathway distinct from nuclear translocation. Blood. 1999;94:1878–1889. [PubMed] [Google Scholar]
- 49.Zhong H, SuYahng H, Erdjument-Bromage H, Tempst P, Ghosh S. The Transcriptional Activity of NF-kB is Regulated by the IkB-associated PKAc Subunit through a Cyclic AMP-Indepdent Mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 50.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274(43):30353–30366. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 51.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Molecular Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 52.Chen Lf, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-{kappa}B RelA Phosphorylation Regulates RelA Acetylation. Mol Cell Biol. 2005 September 15;25(18):7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulanger MJ, Chow Dc, Brevnova EE, Garcia KC. Hexameric Structure and Assembly of the Interleukin-6/IL-6 {alpha}-Receptor/gp130 Complex. Science. 2003 June 27;300(5628):2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 54.Heinrich PC, Behrmann I, Haan S, Hermanns HM, M++Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003 August 15;374(1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurdi M, Booze GW. Can the Protective Actions of JAK-STAT in the Heart be Exploited Therapeutically? Parsing the Regulation of Interleukin-6-Type Cytokine Signaling. Journal of Cardiovascular Pharmacology. 2007;50(2):126–141. doi: 10.1097/FJC.0b013e318068dd49. [DOI] [PubMed] [Google Scholar]
- 56.Ray S, Boldogh S, Brasier AR. STAT3 NH2-Terminal Acetylation Is Activated By The Hepatic Acute-Phase Response And Required for IL-6 Induction of Angiotensinogen. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 57.Ray S, Sherman C, Lu M, Brasier AR. Angiotensinogen gene expression is dependent on STAT3-mediated p300/CBP coactivator recruitment and HAT activity. Mol Endocrinol. 2001;16:824–836. doi: 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- 58.Horvai A, Xu L, Korzus e. Nuclear integration of JAK/STAT and RAS/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 60.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Molecular Cell. 2001;3:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 61.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of Transcription, RNA Processing, and Surveillance by P-TEFb Kinase on Heat Shock Genes. Molecular Cell. 2004 January 16;13(1):55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 62.Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NF{kappa}B to effect promoter-specific transcriptional repression. Genes Dev. 2005 May 1;19(9):1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 Phosphorylation Is Required for Activation of a Subset of NF-{kappa}B-Dependent Genes by Recruiting Cyclin-Dependent Kinase 9/Cyclin T1 Complexes. Mol Cell Biol. 2008 June 1;28(11):3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]