Abstract
A novel apramycin resistance gene, apmA, was detected on the ca.-40-kb resistance plasmid pAFS11 from bovine methicillin-resistant Staphylococcus aureus (MRSA) of sequence type 398 (ST398). The apmA gene coded for a protein of 274 amino acids that was related only distantly to acetyltransferases involved in chloramphenicol or streptogramin A resistance. NsiI deletion of apmA resulted in a 16- to 32-fold decrease in the apramycin MICs. An apmA-specific PCR identified this gene in one additional bovine and four porcine MRSA ST398 isolates.
Methicillin-resistant Staphylococcus aureus (MRSA) of sequence type 398 (ST398) has been identified mainly as a colonizer of the skin and the mucosal surfaces of swine (6, 26, 28), although, more rarely, such isolates have also been found to be involved in infections of swine (9, 17, 18, 24). Moreover, MRSA ST398 has also been detected in other animals, such as cattle (8, 23), horses (25, 27), poultry (15), dogs (16), and rats (22), and in humans with exposure to MRSA ST398-colonized animals (7, 20, 28, 30). As a colonizer, MRSA ST398 is subject to selective pressure by antimicrobial agents that are not used primarily to control staphylococcal infections and, as a consequence, may acquire novel or uncommon resistance genes. One such example is provided by the observation that chloramphenicol-resistant MRSA ST398 isolates from swine (9) and cattle (8) did not carry any of the usually found staphylococcal cat genes for chloramphenicol resistance (19) but harbored the phenicol exporter gene fexA, which also confers resistance to florfenicol. Florfenicol is a fluorinated chloramphenicol derivative that is widely used for the control of respiratory tract infections in cattle and swine. Another example is apramycin resistance.
Apramycin is an aminocyclitol antibiotic that is used exclusively in veterinary medicine for the treatment of Escherichia coli infections in swine, cattle, sheep, poultry, or rabbits. Studies of apramycin-resistant Enterobacteriaceae identified the gene aac(3)-IV, which is located mostly on plasmids and confers resistance to apramycin and gentamicin (2, 3, 5, 21, 29). In contrast to the wealth of data available for apramycin resistance in Enterobacteriaceae (1, 31, 32), no information about apramycin resistance in staphylococci exists.
During two survey studies on MRSA ST398 from diseased swine and dairy cattle, 4/54 porcine and 2/16 bovine isolates revealed high apramycin MIC values of ≥32 μg/ml (8, 9). These isolates were tentatively classified as resistant, although no clinical breakpoints for apramycin approved by the Clinical Laboratory Standards Institute (CLSI) are currently available (4). One of these isolates, the bovine MRSA isolate 11, was chosen for further analysis of the genetic basis of apramycin resistance. The bovine MRSA isolate 11 carried a staphylococcal cassette chromosome mec element of type V (SCCmec V) and displayed the multilocus sequence type (MLST) ST398, the spa type t2576, and the dru type dt11a (8). Plasmid analysis identified the ca.-40-kb plasmid pAFS11, which, upon transformation into S. aureus RN4220, mediated a multiresistance phenotype (Table 1). The corresponding resistance genes were detected by specific PCR assays (8, 9, 14). In addition to kanamycin and neomycin resistance via aadD, macrolide-lincosamide-streptogramin B resistance via erm(B), tetracycline resistance via tet(L), and trimethoprim resistance via dfrK, plasmid pAFS11 conferred a high apramycin MIC of ≥128 μg/ml. The S. aureus RN4220 transformant carrying pAFS11, however, was classified as intermediate to gentamicin (MIC of 8 μg/ml) (Table 1).
TABLE 1.
Comparative analysis of the bovine MRSA ST398 isolate 11, S. aureus RN4220, and the S. aureus RN4220 transformant carrying the plasmid pAFS11
| Bacterial strain | Resistance genes | MIC (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| APR | GEN | ERY | CLI | TET | TMP | KAN | NEO | OXA | ||
| MRSA ST398 isolate 11 | apmA, erm(B), tet(L), tet(M), tet(K), dfrK, aadD, mecA, blaZ | ≥128 | 8 | ≥64 | ≥128 | 64 | ≥256 | ≥128 | 128 | 16 |
| S. aureus RN4220 | 1 | 0.25 | 0.12 | ≤0.12 | 0.12 | 0.5 | 4 | ≤1 | 0.12 | |
| S. aureus RN4220(pAFS11) | apmA, erm(B), tet(L), dfrK, aadD | ≥128 | 8 | ≥64 | ≥128 | 32 | ≥256 | 64 | 32 | 0.12 |
APR, apramycin; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin; TET, tetracycline; TMP, trimethoprim; KAN, kanamycin; NEO, neomycin; OXA, oxacillin.
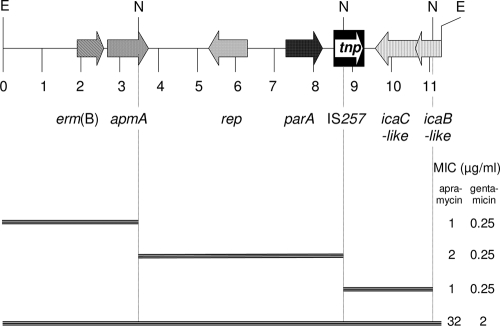
An 11,312-bp EcoRI fragment of pAFS11 was cloned into pBluescript II SK+ (Stratagene). Recombinant plasmids were transformed into E. coli strain JM101, and transformants were selected on apramycin-supplemented Luria-Bertani agar (15 μg/ml). Sequence analysis was conducted by primer walking starting with M13 universal and reverse primers. A schematic representation of the seven reading frames found on this EcoRI fragment is shown in Fig. 1. This segment comprised part of a Tn917 transposon with one terminal repeat and the entire erm(B) gene. A reading frame for a 315-amino-acid (aa) protein with 30.9 and 31.4% identity to distinctly larger chromosome replication initiation/membrane attachment proteins of Staphylococcus hominis (NCBI accession no. ZP_04059882) and Staphylococcus warneri (NCBI accession no. ZP_04678490), respectively, was detected. Further downstream was the reading frame for a 263-aa ParA protein that corresponded closely (96.2 and 95.1% identity, respectively) to the chromosome partitioning ATPases of Staphylococcus capitis (NCBI accession no. ZP_03614545) and S. aureus (NCBI accession no. ACY12632). A complete IS257 element was identified, but this did not exhibit 8-bp direct repeat sequences in the up- and downstream segments. The lack of these direct repeats suggested that recombination events via this insertion sequence have occurred. A complete reading frame for a 347-aa protein and the 3′ end of a reading frame (190 aa) showed 48.4% and 54.5% identity to IcaC (NCBI accession no. YP_189846) and IcaB (NCBI accession no. YP_189845), respectively, from a Staphylococcus epidermidis isolate.
FIG. 1.
Schematic presentation of the seven reading frames found on the 11,312-kb EcoRI fragment of pAFS11. The arrows indicate the extents and directions of transcription. A distance scale in kb is given below the map. The IS257 element is shown as a black box, with the white arrow indicating the transposase gene tnp. The MICs of apramycin and gentamicin conferred by the complete EcoRI fragment and the corresponding NsiI deletion clones are shown on the right-hand side. N, NsiI; E, EcoRI.
To confirm the role of the seventh reading frame, designated apmA, in apramycin resistance, the EcoRI fragment was digested with NsiI, which cuts once within the apmA reading frame, once within the IS257 sequence, and once within the icaB-like gene. Deletion clones in E. coli JM101 were tested for their apramycin MICs by broth microdilution according to the CLSI document M31-A3 (4). In comparison to clones carrying the original EcoRI fragment, all three deletion clones showed a 16- to 32-fold decrease in the apramycin MICs and also an 8-fold decrease in the gentamicin MICs. The apmA gene codes for a 274-aa protein that shows limited similarity to other proteins deposited in the databases. The best matches were 38.1% identity to a VatB-like xenobiotic acetyltransferase protein from Pasteurella multocida (NCBI accession no. NP_246134) and 33.3% identity to a putative chloramphenicol acetyltransferase from Escherichia fergusonii (NCBI accession no. YP_002383245). Based on the apmA sequence, a PCR assay using the primers apmA-fw (5′-CGTTTGCTTCGTGCATTAAA-3′) and apmA-rev (5′-TTGACACGAAGGAGGGTTTC-3′) (annealing temperature, 52°C; amplicon size, 656 bp) was developed and applied to MRSA ST398 isolates. While the remaining bovine and the four porcine apramycin-resistant isolates were positive for apmA, the isolates with MICs of ≤16 μg/ml were negative. All five additional isolates harbored SCCmec V and showed the spa type t011 and the dru type dt11a (8, 9). Transfer and hybridization experiments identified apmA in all five cases on plasmids of ca. 40 kb that were indistinguishable from or closely related to pAFS11 in their EcoRI, HindIII, BglII, and PvuI restriction patterns. All of these plasmids also harbored tet(L), dfrK, aadD, and erm(B) in addition to apmA.
Recent studies on antimicrobial resistance genes in MRSA ST398 led to the identification of a number of novel or unusual resistance genes, such as dfrK (10), vga(C) (11), erm(T) (12), and cfr (13). All of these genes were located on plasmids. Analysis of these plasmids suggested that recombination and cointegrate formation played a major role in the acquisition of novel resistance genes by MRSA ST398. In most of the described plasmids, insertion sequences, such as IS257 or ISSau10 (10, 12), seemed to be involved in recombination processes. This is, to the best of our knowledge, the first description of an apramycin resistance gene in Gram-positive cocci. The presence of apmA on the multiresistance plasmid pAFS11 enables its persistence and coselection under the selective pressure imposed by the use of kanamycin, neomycin, tetracyclines, macrolides, lincosamides, or trimethoprim.
Nucleotide sequence accession number.
The nucleotide sequence of the 11,312-bp EcoRI fragment of plasmid pAFS11 has been deposited in the EMBL database under accession number FN806789.
Acknowledgments
We thank Kerstin Meyer and Vera Nöding for excellent technical assistance.
This study was financially supported by internal funding from the Friedrich-Loeffler-Institut.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Boerlin, P., R. Travis, C. L. Gyles, R. Reid-Smith, N. Janecko, H. Lim, V. Nicholson, S. A. McEwen, R. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaslus-Dancla, E., J. L. Martel, C. Carlier, J. P. Lafont, and P. Courvalin. 1986. Emergence of aminoglycoside 3-N-acetyltransferase IV in Escherichia coli and Salmonella typhimurium isolated from animals in France. Antimicrob. Agents Chemother. 29:239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaslus-Dancla, E., P. Pohl, M. Meurisse, M. Marin, and J. P. Lafont. 1991. High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob. Agents Chemother. 35:590-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CLSI. 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals; approved standard—third edition. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Davies, J., and S. O'Connor. 1978. Enzymatic modification of aminoglycoside antibiotics: 3-N-acetyltransferase with broad specificity that determines resistance to the novel aminoglycoside apramycin. Antimicrob. Agents Chemother. 14:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Neeling, A. J., M. J. van den Broek, E. C. Spalburg, M. G. van Santen-Verheuvel, W. D. Dam-Deisz, H. C. Boshuizen, A. W. van de Giessen, E. van Duijkeren, and X. W. Huijsdens. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366-372. [DOI] [PubMed] [Google Scholar]
- 7.Denis, O., C. Suetens, M. Hallin, B. Catry, I. Ramboer, M. Dispas, G. Willems, B. Gordts, P. Butaye, and M. J. Struelens. 2009. Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg. Infect. Dis. 15:1098-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feßler, A., C. Scott, K. Kadlec, R. Ehricht, S. Monecke, and S. Schwarz. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619-625. [DOI] [PubMed] [Google Scholar]
- 9.Kadlec, K., R. Ehricht, S. Monecke, U. Steinacker, H. Kaspar, J. Mankertz, and S. Schwarz. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156-1164. [DOI] [PubMed] [Google Scholar]
- 10.Kadlec, K., and S. Schwarz. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadlec, K., and S. Schwarz. 2009. Identification of a novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53:3589-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadlec, K., and S. Schwarz. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehrenberg, C., C. Cuny, B. Strommenger, S. Schwarz, and W. Witte. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lüthje, P., and S. Schwarz. 2006. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 57:966-969. [DOI] [PubMed] [Google Scholar]
- 15.Nemati, M., K. Hermans, U. Lipinska, O. Denis, A. Deplano, M. Struelens, L. A. Devriese, F. Pasmans, and F. Haesebrouck. 2008. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 52:3817-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nienhoff, U., K. Kadlec, I. F. Chaberny, J. Verspohl, G.-F. Gerlach, S. Schwarz, D. Simon, and I. Nolte. 2009. Transmission of methicillin-resistant Staphylococcus aureus strains between humans and dogs: two case reports. J. Antimicrob. Chemother. 64:660-662. [DOI] [PubMed] [Google Scholar]
- 17.Pomba, C., F. Baptista, N. Couto, F. Loução, and H. Hasman. 2010. Methicillin-resistant Staphylococcus aureus CC398 isolates with indistinguishable ApaI restriction patterns in colonized and infected pigs and humans. J. Antimicrob. Chemother. 65:2479-2481. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz, S., K. Kadlec, and B. Strommenger. 2008. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius detected in the BfT-GermVet monitoring programme 2004-2006 in Germany. J. Antimicrob. Chemother. 61:282-285. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz, S., C. Kehrenberg, B. Doublet, and A. Cloeckaert. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28:519-542. [DOI] [PubMed] [Google Scholar]
- 20.Smith, T. C., M. J. Male, A. L. Harper, J. S. Kroeger, G. P. Tinkler, E. D. Moritz, A. W. Capuano, L. A. Herwaldt, and D. J. Diekema. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Threlfall, E. J., B. Rowe, J. L. Ferguson, and L. R. Ward. 1986. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J. Hyg. (Lond.) 97:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Giessen, A. W., M. G. van Santen-Verheuvel, P. D. Hengeveld, T. Bosch, E. M. Broens, and C. B. Reusken. 2009. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev. Vet. Med. 91:270-273. [DOI] [PubMed] [Google Scholar]
- 23.Vanderhaeghen, W., T. Cerpentier, C. Adriaensen, J. Vicca, K. Hermans, and P. Butaye. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144:166-171. [DOI] [PubMed] [Google Scholar]
- 24.van Duijkeren, E., M. D. Jansen, S. C. Flemming, H. de Neeling, J. A. Wagenaar, A. H. W. Schoormans, A. van Nes, and A. C. Fluit. 2007. Methicillin-resistant Staphylococcus aureus in pigs with exudative epidermitis. Emerg. Infect. Dis. 13:1408-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Duijkeren, E., M. Moleman, M. M. Sloet van Oldruitenborgh-Oosterbaan, J. Multem, A. Troelstra, A. C. Fluit, W. J. van Wamel, D. J. Houwers, A. J. de Neeling, and J. A. Wagenaar. 2010. Methicillin-resistant Staphylococcus aureus in horses and horse personnel: an investigation of several outbreaks. Vet. Microbiol. 141:96-102. [DOI] [PubMed] [Google Scholar]
- 26.Voss, A., F. Loeffen, J. Bakker, C. Klaassen, and M. Wulf. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther, B., S. Monecke, C. Ruscher, A. W. Friedrich, R. Ehricht, P. Slickers, A. Soba, C. G. Wleklinski, L. H. Wieler, and A. Lübke-Becker. 2009. Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 47:704-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witte, W., B. Strommenger, C. Stanek, and C. Cuny. 2007. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wray, C., R. W. Hedges, K. P. Shannon, and D. E. Bradley. 1986. Apramycin and gentamicin resistance in Escherichia coli and salmonellas isolated from farm animals. J. Hyg. (Lond.) 97:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wulf, M. W., E. Tiemersma, J. Kluytmans, D. Bogaers, A. C. Leenders, M. W. Jansen, J. Berkhout, E. Ruijters, D. Haverkate, M. Isken, and A. Voss. 2008. MRSA carriage in healthcare personnel in contact with farm animals. J. Hosp. Infect. 70:186-190. [DOI] [PubMed] [Google Scholar]
- 31.Yates, C. M., M. C. Pearce, M. E. Woolhouse, and S. G. Amyes. 2004. High frequency transfer and horizontal spread of apramycin resistance in calf faecal Escherichia coli. J. Antimicrob. Chemother. 54:534-537. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X. Y., L. J. Ding, and M. Z. Fan. 2009. Resistance patterns and detection of aac(3)-IV gene in apramycin-resistant Escherichia coli isolated from farm animals and farm workers in northeastern of China. Res. Vet. Sci. 87:449-454. [DOI] [PubMed] [Google Scholar]