Abstract
Eight rifampin-resistant streptococci of the mitis group were identified at the species level by using a concatenated 16S rRNA gene-sodA-rpoB-hlpA sequence. Characterization of their rpoB alleles showed single amino acid changes involved in rifampin resistance. Comparison of RpoB sequences from pneumococcal recombinant isolates, viridans isolates, and type strains revealed a species-specific amino acid signature, which allowed it to be ascertained that recombinant RpoBs were originated in genetic interchanges with Streptococcus mitis and Streptococcus oralis.
Viridans streptococci (VS) form part of the microbiota of the oropharynx and the gastrointestinal and female genital tracts (13, 37). However, they cause endocarditis in native valves and pneumonia in neutropenic cancer patients (7, 8, 43). By their 16S rRNA gene sequences, VS can be classified into five groups: mutans, salivarius, anginosus, sanguinis, and mitis (18). Species of the mitis group (SMG) include Streptococcus mitis, Streptococcus sanguinis, Streptococcus parasanguinis, Streptococcus gordonii, Streptococcus oralis, Streptococcus cristatus, Streptococcus infantis, Streptococcus peroris, Streptococcus pneumoniae, and Streptococcus pseudopneumoniae. Clinical features, together with their optochin susceptibility and bile solubility, distinguish S. pneumoniae bacteria from other SMG (27, 30, 39), although optochin-susceptible VS have been found (6, 32).
SMG isolated from blood cultures of cancer patients are commonly resistant to antibiotics (2, 16, 21, 22, 25, 42) and constitute a reservoir of resistance by acting as donors in the horizontal transfer of DNA to pneumococci, as observed for penicillin and fluoroquinolones (5, 17, 35, 38, 40). Rifampin is used in the treatment of tuberculosis and in meningitis caused by multiresistant pneumococcal strains, combined with either β-lactams or vancomycin (9, 31, 36). Rifampin binds to the DNA-dependent RNA polymerase (RpoB), inhibiting its function (10), which is essential for bacterial growth (15, 26). Resistance changes have been identified in four conserved regions (N, I, II, and III) of RpoB in several bacteria (3, 4, 14, 24, 34). This resistance in S. pneumoniae is due to spontaneous mutations, and it has been suggested to also be acquired by recombination with SMG (19). In this study, we have characterized rifampin-resistant SMG isolates, complementing the unique study of S. mitis (1), to ascertain the origin of the recombinant rpoB genes found in S. pneumoniae isolates.
Identification of viridans streptococci isolates to the species level.
Among 1,272 VS isolates collected from adult patients at Hospital de Bellvitge (Barcelona) during 10 years (1998 to 2007), 10 (0.79%) were rifampin resistant as determined by broth microdilution and agar dilution assays (11, 12). Eight of them with high resistance levels (MIC ≥ 32 μg/ml) were available for this study (Table 1). Although one VS isolate per patient was recovered, isolate 113 collected from patient 3 also yielded a rifampin-resistant S. anginosus isolate (113A) that was used for sequence comparisons. The global incidence of rifampin resistance observed in this study was similar to that found in Spain for S. pneumoniae (0.70%) (19), although a higher rate (3%) has been found in SMG isolated from hematologic cancer patients (1).
TABLE 1.
Summary of isolation data, resistance characteristics, and identification of isolates used in this study
| Isolate | Origina | Resistance patternb | Phenotypic characterization | Molecular characterizationc |
|---|---|---|---|---|
| 60 | DB | PEN, ERY, RIF | S. sanguinis | S. parasanguinis (98.6) |
| 79 | AF | ERY, CLI, SXT, RIF | S. sanguinis | S. oralis (97.6) |
| 113 | WS | PEN, RIF | S. oralis | S. oralis (98.1) |
| 395 | AF | RIF | S. sanguinis | S. gordonii (98.9) |
| 745 | AF | ERY, CLI, TET, SXT, RIF | S. sanguinis | S. oralis (98.4) |
| 779 | BL | RIF | S. mitis | S. mitis (98.7) |
| 889 | B | PEN, SXT, RIF | S. parasanguinis | S. parasanguinis (97.4) |
| 971 | E | ERY, TET, RIF | S. sanguinis | S. parasanguinis (95) |
DB, duodenal biopsy specimen; AF, ascitic fluid; WS, wound swab; BL, bronchoalveolar lavage fluid; B, blood; E, eye.
PEN, intermediate or highly resistant to penicillin (MIC ≥ 0.25 μg/ml); TET, resistant to tetracycline (MIC ≥ 8 μg/ml); ERY, resistant to erythromycin (MIC ≥ 1 μg/ml); CLI, resistant to clindamycin (MIC ≥ 1 μg/ml); SXT, resistant to trimethoprim-sulfamethoxazole (MICs ≥ 4 and 76 μg/ml); RIF, resistant to rifampin (MIC ≥ 4 μg/ml).
The species identification was based on clustering with type strains in a phylogenetic tree obtained with concatenated partial sequences of 16S rRNA genes, sodA, rpoB, and hlpA. Numbers in parentheses indicate the percentage of identity with the corresponding type strain.
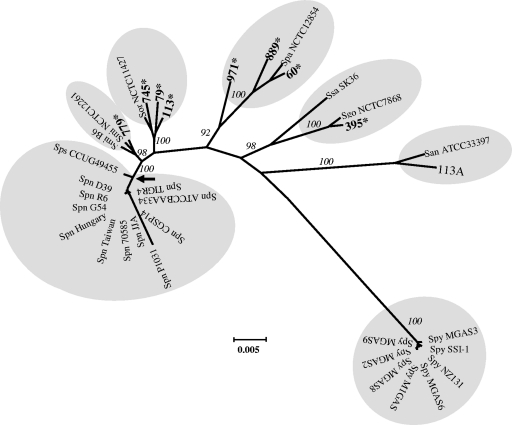
The 8 VS isolates were identified by phenotypic (39) and molecular methods. We used concatenated 16S rRNA gene-sodA-rpoB-hlpA sequences made with partial 16S rRNA genes, rpoB, and sodA (1,198, 344, and 324 bp, respectively) and the full-length hlpA (276 bp). To amplify the 16S rRNA genes and hlpA, we used the following primers: 16SDNAF1 (5′-GAGTTGCGAACGGGTGAGT-3′), 16SDNAR1 (5′-AGCGATTCCGACTTCAT-3′), huATG (5′-ATGGCAAACAAACAAGATT-3′), and huTAA (5′-TTATTTAACAGCGTCTTTAAGAGC-3′). The partial sodA and rpoB genes were amplified and sequenced as described previously (28, 19). These genes were selected for their polymorphism among streptococci and because they have been used as part of the ddl-gdh-rpoB-sodA sequence to differentiate SMG isolates (29). We assumed that hlpA (encoding the histonelike DNA binding protein HU) would improve our concatenated sequence discrimination capacity since HU, as an architectural cofactor, may require different DNA binding geometries (41) and, probably, sequence specificity. Clustering (bootstrap values, ≥92%) of the 2,142-bp 16S rRNA gene-sodA-rpoB-hlpA sequences of the eight isolates and type strains in a phylogenetic tree allowed species identification (Fig. 1 and Table 1). Accession numbers of the sequences used for comparisons can be found in Table S1 in the supplemental material. Among the six clusters observed (plus the out-group), all except S. pneumoniae/S. pseudopneumoniae and S. sanguinis/S. gordonii formed species-specific groups.
FIG. 1.
Phylogenetic tree of concatenated sequences of 16S rRNA genes, sodA, rpoB, and hlpA. Analysis was conducted with the MEGA program (version 4.0.2), using the Neighbor Joining algorithm. Bootstrap confidence intervals exceeding 90% are shown in italics. The scale bar calculated by the MEGA program indicates the genetic divergence. Eight S. pyogenes strains were used as the out-group. Shadowed in gray are clusters that identified S. pneumoniae (Spn) plus S. pseudopneumoniae (Sps), S. mitis (Smi), S. oralis (Sor), S. parasanguinis (Spa), S. sanguinis (Ssa) and S. gordonii (Sgo), S. anginosus (San), and S. pyogenes (Spy) strains. SMG isolates characterized in this work appear in boldface and followed by an asterisk. The arrow indicates the node that separates S. pneumoniae plus S. pseudopneumoniae from the rest of the clusters.
The within-group sequence diversity (mean ± standard deviation) for S. pneumoniae/S. pseudopneumoniae (0.4% ± 0.1%), S. mitis (1.2% ± 0.3%), and S. oralis (2.0% ± 0.5%) clusters reflected low sequence diversity. Our S. pneumoniae/S. pseudopneumoniae value was nearly half of that obtained using the ddl-gdh-rpoB-sodA concatenates (29), and for S. mitis, it was 4- to 5-fold lower than the value obtained by multilocus sequence typing (20, 23). Additionally, S. pneumoniae/S. pseudopneumoniae, S. mitis, and S. oralis clusters were clearly separated, as their genetic distances to the node formed with the branch of S. pneumoniae/S. pseudopneumoniae were 1.1% ± 0.7%, 1.9% ± 0.0%, and 3.2% ± 0.4%, which are statistically significant values (P < 0.0001).
Determination of mutations involved in rifampin resistance.
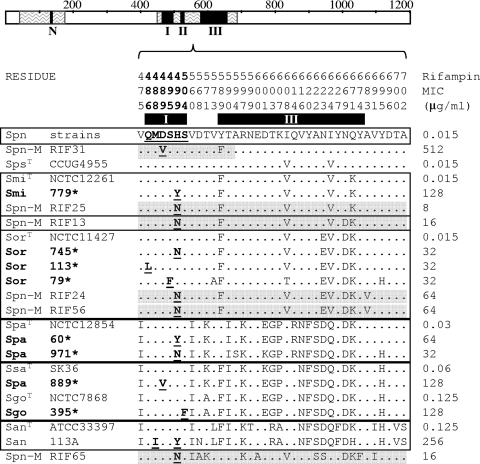
RpoB regions L42 to V175 and Q464 to T702 were sequenced as described previously (19) and compared. Changes were found in the Q464-to-T702 region (Fig. 2). Among them, only H499N had been described in rifampin resistance in SMG isolates (1), while the rest, with the exception of S504F (isolate 395), had been involved in resistance in S. pneumoniae (19). To test its role in resistance, transformation of S. pneumoniae R6 with the Q464-to-T700 fragment carrying S504F was performed as described previously (33). The transformant had the same rifampin MIC as isolate 395, showing that this change was indeed involved in resistance. Additional changes in cluster III, which were present in both susceptible and resistant strains (Fig. 2), are not believed to be involved in resistance.
FIG. 2.
Amino acid sequence variations in RpoB (V475 to A702) of rifampin-resistant recombinant isolates of S. pneumoniae (Spn-M) and SMG rifampin-resistant isolates characterized in this work (boldface and marked with an asterisk). RpoB is represented as a bar with clusters N, I, II, and III as black boxes and zigzagged areas showing sequenced areas. The amino acids present at each polymorphic site are shown in full for S. pneumoniae strains (R6, P1031, Hungary, Taiwan1, TIGR4, and JJA). For the other strains, only sites that differ from those are shown. Residue numbers are indicated vertically above the sequences, and black boxes below the numbers localize clusters I and III. Amino acid changes involved in rifampin resistance are shown in boldface and underlined. Species nomenclature is as defined in the Fig. 1 legend. A superscript T indicates a type strain. Recombinant sequences are shadowed in gray. Squares group sequences with the highest similarity according to scores obtained by ClustalW alignments.
RpoB sequence comparisons revealed that most changes not involved in rifampin resistance were conserved among the species (no more than two amino acid differences in regions I, II, and III) (Fig. 2). These changes could be considered a species-specific amino acid signature that give information about the phylogenetic origin of the isolates, as observed for ComC (29). On the basis of similarity scores with type strains (ClustalW), six groups could be deduced (Fig. 2), coinciding with the six clusters of the phylogenetic tree based on 16S rRNA gene-sodA-rpoB-hlpA sequences (Fig. 1). Two exceptions were observed: the S. pseudopneumoniae type strain that shared the same similarity with S. pneumoniae and S. mitis, and isolate 889 (S. parasanguinis by the concatenated sequence) that shared the same similarity with S. gordonii and S. sanguinis. Furthermore, this amino acid signature allowed us to ascertain the origin of recombinant RpoBs. Six rifampin-resistant S. pneumoniae recombinant isolates, which we had previously characterized (19), were compared with other VS. Four of them grouped with S. mitis and S. oralis (RIF13, -25, -24, and -56) (Fig. 2). The source for isolates RIF31 and RIF65 could not be deduced because of the partial recombinational nature of the first (19) and poor scores with any of the type strains for the second, due to either the donor not being included in this comparison or the occurrence of several recombination events. In conclusion, S. pneumoniae and SMG share the same mechanisms of rifampin resistance, and recombination events in S. pneumoniae take place mostly with S. mitis and S. oralis species.
Supplementary Material
Acknowledgments
This work was supported by Plan Nacional de I+D+I of Ministerio de Ciencia e Innovación (grant no. BIO2008-02154) and Comunidad de Madrid (grant no. COMBACT-S-BIO-0260/2006).
CIBER Enfermedades Respiratorias is an initiative from Instituto de Salud Carlos III.
Footnotes
Published ahead of print on 1 November 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Achour, W., O. Guenni, M. Fines, R. Leclercq, and A. Ben-Hassen. 2004. rpoB mutations in Streptococcus mitis clinical isolates resistant to rifampin. Antimicrob. Agents Chemother. 48:2757-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaide, F., M. A. Benítez, J. Carratala, F. Gudiol, J. Liñares, and R. Martin. 2001. In vitro activities of the new ketolide HMR 3647 (telithromycin) in comparison with those of eight other antibiotics against viridans group streptococci isolated from blood of neutropenic patients with cancer. Antimicrob. Agents Chemother. 45:624-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubry-Damon, H., M. Galimand, G. Gerbaud, and P. Courvalin. 2002. rpoB mutation conferring rifampin resistance in Streptococcus pyogenes. Antimicrob. Agents Chemother. 46:1571-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubry-Damon, H., C. J. Soussy, and P. Courvalin. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsalobre, L., M. J. Ferrándiz, J. Liñares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balsalobre, L., A. Hernández-Madrid, D. Llull, A. J. Martín-Galiano, E. García, A. Fenoll, and A. G. de la Campa. 2006. Molecular characterization of disease-associated streptococci of the mitis group that are optochin susceptible. J. Clin. Microbiol. 44:4163-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayer, A. S., and W. M. Scheld. 2000. Endocarditis and intravascular infections, p. 857-902. In G. Mandell et al. (ed.), Principles and practice of infectious diseases. Churchill-Livingstone, Philadelphia, PA.
- 8.Bochud, P. Y., P. H. Eggiman, T. H. Calandra, G. V. Melle, L. Saghafi, and P. Francioli. 1994. Bacteremia due to viridans streptococcus in neutropenic patients with cancer: clinical spectrum and risk factors. Clin. Infect. Dis. 18:25-31. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, J. S., and W. M. Scheld. 1997. The challenge of penicillin-resistant Streptococcus pneumoniae meningitis: current antibiotic therapy in the 1990s. Clin. Infect. Dis. 24:213-221. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility testing for bacteria that grow aerobically, 7th ed. Approved standard M7-A6. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Crowe, C. C., W. E. Sanders, and S. Longhey. 1975. Bacterial interference. II. The role of the normal throat flora in prevention of colonisation by group A streptococci. J. Infect. Dis. 128:527-532. [DOI] [PubMed] [Google Scholar]
- 14.Cruchaga, S., M. Pérez-Vázquez, F. Román, and J. Campos. 2003. Molecular basis of rifampicin resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 56:1011-1014. [DOI] [PubMed] [Google Scholar]
- 15.Das, A., J. De Vito, J. Sparkowski, and F. Warren. 1992. RNA synthesis in bacteria: mechanism and regulation of discrete biochemical events at initiation and termination. In J. Sutcliffe et al. (ed.), Emerging targets in antibacterial and antifungal chemotherapy. Chapman and Hall, New York, NY.
- 16.Douglas, C. W. I., J. Heath, K. K. Hampton, and F. E. Preston. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179-182. [DOI] [PubMed] [Google Scholar]
- 17.Enright, M., P. Zawadski, P. Pickerill, and C. G. Dowson. 1998. Molecular evolution of rifampicin resistance in Streptococcus pneumoniae. Microb. Drug Resist. 4:65-70. [DOI] [PubMed] [Google Scholar]
- 18.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrándiz, M. J., C. Ardanuy, J. Liñares, J. M. García-Arenzana, E. Cercenado, A. Fleites, A. G. de la Campa, and the Spanish Pneumococcal Infection Study Network G03/1036. 2005. New mutations and horizontal transfer of rpoB among rifampicin-resistant Streptococcus pneumoniae from four Spanish hospitals. Antimicrob. Agents Chemother. 49:2237-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, C., E. J. Alm, M. F. Polz, B. G. Spratt, and W. P. Hanage. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741-746. [DOI] [PubMed] [Google Scholar]
- 21.Gershon, A. S., J. C. de Azavedo, A. McGeer, K. I. Ostrowska, D. Church, D. J. Hoban, G. K. Harding, K. Weiss, L. Abbott, F. Smaill, M. Gourdeau, G. Murray, and D. E. Low. 2002. Activities of new fluoroquinolones, ketolides, and other antimicrobials against blood culture isolates of viridans group streptococci from across Canada, 2000. Antimicrob. Agents Chemother. 46:1553-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González, I., M. Georgiou, F. Alcaide, D. Balas, J. Liñares, and A. G. de le Campa. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. B 361:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera, L., S. Jiménez, A. Valverde, M. A. García-Aranda, and J. A. Sáez-Nieto. 2003. Molecular analysis of rifampicin-resistant Mycobacterium tuberculosis isolated in Spain (1996-2001). Description of new mutations in the rpoB gene and review of the literature. Int. J. Antimicrob. Agents 21:403-408. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, J. A., H. C. Schouten, E. E. Stobberingh, and P. B. Soeters. 1995. Viridans streptococci isolated from the blood stream. Relevance of species identification. Diagn. Microbiol. Infect. Dis. 22:267-273. [DOI] [PubMed] [Google Scholar]
- 26.Jin, D. J., and Y. N. Zhou. 1996. Mutational analysis of structure-function relationship of RNA polymerase in Escherichia coli. Methods Enzymol. 273:300-319. [DOI] [PubMed] [Google Scholar]
- 27.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 28.Kawamura, Y., R. A. Whiley, S. E. Shu, T. Ezaki, and J. M. Hardie. 1999. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 145:2605-2613. [DOI] [PubMed] [Google Scholar]
- 29.Kilian, M., K. Poulsen, T. Blomqvist, L. S. Håvarstein, M. Bek-Thomsen, H. Tettelin, and U. B. Sørensen. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lund, E., and J. Henrichsen. 1978. Laboratory diagnosis, serology and epidemiology of S. pneumoniae. Methods Microbiol. 12:241-262. [Google Scholar]
- 31.Martínez-Lacasa, J., C. Cabellos, A. Martos, A. Fernandez, F. Tubau, P. F. Viladrich, J. Liñares, and F. Gudiol. 2002. Experimental study of the efficacy of vancomycine, rifampicin and dexamethasone in the therapy of pneumococcal meningitis. J. Antimicrob. Chemother. 49:507-513. [DOI] [PubMed] [Google Scholar]
- 32.Martín-Galiano, A. J., L. Balsalobre, A. Fenoll, and A. G. de la Campa. 2003. Genetic characterization of optochin-susceptible viridans group streptococci. Antimicrob. Agents Chemother. 47:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martín-Galiano, A. J., and A. G. de la Campa. 2003. High-efficiency generation of antibiotic-resistant strains of Streptococcus pneumoniae by PCR and transformation. Antimicrob. Agents Chemother. 47:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, K., P. Hindersson, N. Hoiby, and J. M. Bangsborg. 2000. Sequencing of the rpoB gene in Legionella pneumophila and characterization of mutations associated with rifampin resistance in the Legionellaceae. Antimicrob. Agents Chemother. 44:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padayachee, T., and K. P. Klugman. 1999. Molecular basis of rifampin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:2361-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paris, M. M., S. M. Hickey, M. I. Uscher, S. Shelton, K. D. Olsen, and G. H. McCracken. 1994. Effect of dexamethasone on therapy of experimental penicillin- and cephalosporin-resistant pneumococcal meningitis. Antimicrob. Agents Chemother. 38:1320-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roos, K., S. E. Holm, E. Grahn, and L. Lind. 1993. Alpha streptococci as supplementary treatment of resistant streptococcal tonsillitis: a randomised placebo-controlled study. Scand. J. Infect. Dis. 25:31-36. [PubMed] [Google Scholar]
- 38.Schmitz, F. J., A. Fisher, M. Boos, S. Mayer, D. Milatovic, and A. C. Fluit. 2001. Quinolone-resistance mechanisms and in vitro susceptibility patterns among European isolates of Streptococcus mitis, Streptococcus sanguis, and Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 20:219-222. [DOI] [PubMed] [Google Scholar]
- 39.Spellerberg, B., and C. Brandt. 2007. Streptococcus, p. 412-429. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed, vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 40.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 41.Swinger, K. K., K. M. Lemberg, Y. Zhang, and P. A. Rice. 2003. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 22:3749-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng, L. J., P. R. Hsueh, Y. C. Chen, S. W. Ho, and K. T. Luh. 1998. Antimicrobial susceptibility of viridans group streptococci in Taiwan with an emphasis on the high rates of resistance to penicillin and macrolides in Streptococcus oralis. J. Antimicrob. Chemother. 41:621-627. [DOI] [PubMed] [Google Scholar]
- 43.Westling, K., P. Ljungman, A. Thalme, and I. Julander. 2002. Streptococcus viridans septicaemia: a comparison study in patients admitted to the departments of infectious diseases and haematology in a university hospital. Scand. J. Infect. Dis. 34:316-319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.