Abstract
We investigated an outbreak caused by carbapenem-resistant Acinetobacter baumannii carrying the blaOXA-23 gene. A novel insertion sequence (IS), named ISAba10, was found to be inserted into the ISAba1 element preceding the blaOXA-23 gene in a group of isolates showing higher carbapenem MICs. The presence of ISAba10 was associated with increased OXA-23 expression, likely by providing additional promoter sequences. ISAba10 was also inserted into the carO outer membrane protein gene in most of these isolates.
Multidrug-resistant Acinetobacter baumannii causes serious infections associated with high mortality rates, including septicemia, pneumonia, and urinary tract infections, especially in intensive care units (4, 12, 14). Carbapenem resistance in A. baumannii has mostly been ascribed to plasmid- and chromosome-encoded carbapenemases such as OXA carbapenemases and metallo-β-lactamases (MBLs) (10, 14). Loss of outer membrane proteins (OMPs), efflux pump overexpression, and alteration of penicillin-binding proteins have also been found to play roles in acquiring carbapenem resistance in A. baumannii (1, 5, 9).
Insertion sequences (ISs) may enhance β-lactamase gene expression by providing promoters. ISAba1 has frequently been found upstream of the ADC AmpC β-lactamase and OXA carbapenemase genes in A. baumannii (16). ISAba2, ISAba3, and ISAba4 elements have also been found to precede the blaOXA-58 and the blaOXA-23 genes in clinical isolates of A. baumannii (2, 15, 17).
In this work, we investigated an outbreak of carbapenem-resistant A. baumannii and identified a new IS that could be involved in carbapenem resistance by multiple mechanisms.
Consecutive nonreplicate carbapenem-resistant A. baumannii isolates were recovered from 23 patients hospitalized at a tertiary care hospital in Korea between May and July 2007. All isolates showed positive results with the modified Hodge test, suggesting carbapenemase production, but negative results with the EDTA-sodium mercaptoacetic acid double-disk synergy test for the screening of MBLs (6). All isolates were nonsusceptible to multiple drugs, including ampicillin, cefoxitin, ceftazidime, cefotaxime, cefepime, levofloxacin, amikacin, gentamicin, and tobramycin, by the CLSI disk diffusion method (3). SmaI macrorestriction anaylsis of 23 isolates exhibited genetic similarities of 70% to 100% by the unweighted pair group method with arithmetic average method (data not shown) (8, 18). Thirteen of the isolates (group I) showed identical SmaI macrorestriction patterns (Table 1).
TABLE 1.
Phenotypic and genotypic characteristics of carbapenem-nonsusceptible A. baumanniia
| Group (no. of isolates) | Isolate | IS preceding the blaOXA gene |
MIC (mg/liter) |
Expression level of blaOXA-23b | Status of CarO | |||
|---|---|---|---|---|---|---|---|---|
| ISAba1- blaOXA-23 | ΔISAba1-ISAba10- blaOXA-23 | ISAba1- blaOXA-66 | IMP | MER | ||||
| Group I (13) | SC0701 | − | + | − | 32 | 32 | 4.6 | Intact |
| SC0702 | − | + | − | 32 | 32 | 3.2 | Lost | |
| SC0703 | − | + | − | 32 | 32 | 2.2 | Lost | |
| SC0704 | − | + | − | 32 | 32 | 4.9 | Lost | |
| SC0705 | − | + | − | 32 | 32 | 3.4 | Lost | |
| SC0706 | − | + | − | 32 | 32 | 4.0 | Lost | |
| SC0707 | − | + | − | 32 | 32 | 2.3 | Lost | |
| SC0708 | − | + | − | 32 | 32 | 2.0 | Lost | |
| SC0709 | − | + | − | 32 | 32 | 4.6 | Lost | |
| SC0710 | − | + | − | 32 | 32 | 1.8 | Lost | |
| SC0711 | − | + | − | 32 | 32 | 1.5 | Lost | |
| SC0712 | − | + | − | 32 | 32 | 2.2 | Lost | |
| SC0713 | − | + | − | 32 | 64 | 2.7 | Lost | |
| Group II (7) | SC0721 | + | − | − | 16 | 16 | 1.0 | NT |
| SC0722 | + | − | − | 16 | 16 | 1.1 | NT | |
| SC0723 | + | − | − | 16 | 16 | 1.4 | NT | |
| SC0724 | + | − | − | 16 | 16 | 1.3 | NT | |
| SC0725 | + | − | − | 8 | 16 | 1.4 | NT | |
| SC0726 | + | − | − | 16 | 16 | 1.1 | NT | |
| SC0727 | + | − | − | 16 | 16 | 1.0 | NT | |
| Group III (3) | SC0731 | NT | NT | + | 8 | 16 | NT | NT |
| SC0732 | NT | NT | + | 8 | 16 | NT | NT | |
| SC0733 | NT | NT | + | 8 | 16 | NT | NT | |
| ATCC 19606T | NT | NT | − | 0.25 | 1 | NT | Intact | |
Abbreviations: ΔISAba1, disrupted ISAba1; IMP, imipenem; MER, meropenem; +, positive; −, negative; NT, not tested.
Expression levels of the blaOXA-23 gene were measured by real-time quantitative PCR and normalized against the 16S rRNA gene.
Genes encoding known carbapenemases were investigated as described previously (20). The naturally occurring blaOXA-66 gene, a member of the blaOXA-51 cluster, was detected in all 23 A. baumannii isolates. Furthermore, 20 of the 23 isolates showed positive results in PCR experiments for the detection of the blaOXA-23 gene. I-CeuI mapping experiments showed that a probe specific for blaOXA-23 hybridized with an approximately 500-kb I-CeuI chromosomal fragment that was also recognized by a probe specific for 16S rRNA genes, revealing a chromosomal location of the blaOXA-23 gene, as observed in recent studies (8, 11). Genes encoding OXA-24-like and OXA-58-like carbapenemases or MBLs such as IMP-1-like, VIM-2-like, and SIM-1 were not detected in any of the isolates.
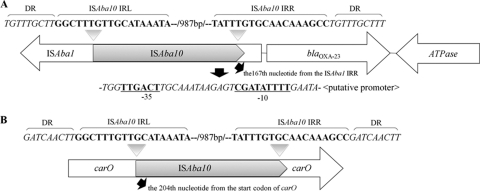
PCR experiments detected ISAba1 upstream of the blaOXA-23 gene in all the blaOXA-23-positive isolates (19, 20). However, the PCR product from 13 isolates (group I) was about 2.5 kb, which was larger than the expected size (1.4 kb), suggesting the insertion of additional DNA. Direct sequencing of PCR products showed the presence of a novel IS, named ISAba10, inserted at the 167th nucleotide from the right inverted repeat of the ISAba1 element. Although insertion of ISAba10 disrupted the ISAba1 element, promoter sequences for the blaOXA-23 gene within ISAba1 remained intact. The new ISAba10 element was 1,023 bp long, contained a 927-bp open reading frame (ORF) encoding a putative transposase, and was bounded by imperfect 18-bp inverted repeat sequences, which are common to members of the IS903 group (Fig. 1). A 9-bp duplication (5′-TGTTTGCTT-3′) flanked ISAba10 at the predicted insertion site. Amino acid sequence alignment using an online tool (http://www-is.biotoul.fr) showed that the ORF of the ISAba10 element shared some similarities to the transposases of ISs such as ISGNB1-1 (identity, 55%), ISJsp1 (52%), ISRusp5 (51%), ISAba7 (48%), and ISAba5 (45%) in the IS903 group.
FIG. 1.
Schematic representation of the genetic organization of ISAba10 disrupting ISAba1 adjacent to the blaOXA-23 gene in A. baumannii SC0701 (A) and ISAba10 disrupting the carO gene in A. baumannii SC0702 (B). Arrows designate transcription directions of genes. IRR, right inverted repeats; IRL, left inverted repeats; DR, direct repeat.
The remaining seven blaOXA-23-positive isolates (group II) carried an intact ISAba1 upstream of the blaOXA-23-like gene. In all group I and group II isolates, blaOXA-66 was not preceded by the ISAba1 element, while in the three blaOXA-23-negative isolates (group III) the blaOXA-66 gene was preceded by ISAba1.
Notably, group I isolates showed two to eight times higher MICs (32 to 64 mg/liter) for both imipenem and meropenem, compared to group II isolates, by agar dilution. Real-time quantitative PCR experiments with the primers and probes listed in Table 2 showed 2- to 5-fold higher blaOXA-23 gene expression in group I isolates than in group II isolates (Table 1) (8). Based on this, we speculated that the ISAba10 element may play a role in higher-level carbapenem resistance by conferring additional promoter sequences to the blaOXA-23 gene. Analyses using the online tool BPROM (Softberry, Inc., Mount Kisco, NY) suggested the presence of a putative promoter within the ISAba10 element (Fig. 1).
TABLE 2.
Primer and probe sequences used for real-time PCR in this study
| Primer or probe | Sequence (5′→3′) | Accession no. of reference gene |
|---|---|---|
| blaOXA-23 forward | TCTGGTTGTACGGTTCAG | FJ6281701 |
| blaOXA-23 reverse | TTTTTATCTGTTTGAATAACCAG | |
| blaOXA-23 probe | CCCGAGTCAGATTGTTCA | |
| 16S rRNA forward | AGCTAGAGTATGGGAGAGGATGG | EU030641 |
| 16S rRNA reverse | TTCGTACCTCAGCGTCAGTATTAG | |
| 16S rRNA probe | TGCCTTCGCCATCGGTATTCCTCCAGA |
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) experiments were performed as previously described (7). A. baumannii ATCC 19606 was used as a reference strain. All group I isolates, except one (isolate SC0701), lacked the 29-kDa CarO-like OMP, while the other OMPs, such as 33- to 36- and 43-kDa porins, were apparently similar to the wild-type strain (data not shown). In the 12 isolates lacking the 29-kDa CarO-like protein, PCR experiments for the carO gene yielded a PCR product about 1.7 kb in size, which was larger than the expected size (741 bp), suggesting the insertion of additional DNA (9). Direct amplicon sequencing confirmed the presence of the ISAba10 element within the carO gene in these isolates. ISAba10 inserted at the 204th base from the carO gene start codon (Fig. 1).
Disruption of the carO gene by the ISAba1, ISAba125, or ISAba825 element has previously been described (13, 16). Loss of the CarO OMP likely plays a role in carbapenem resistance in A. baumannii. Interestingly, however, the isolate SC0701, which carried the intact carO gene, exhibited similar MIC levels for imipenem and meropenem compared to the other 12 clonally related group I isolates. Our results suggest that, under similar conditions, loss of the CarO OMP had only a minor effect on carbapenem resistance in A. baumannii.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession numbers FJ998184 and GQ379223.
Acknowledgments
This work was supported by a grant (HMP-A090479) from the 2009 Good Health R&D Project, Ministry of Health & Welfare, South Korea.
We thank Young Hee Suh and Eun Hee Jeon for laboratory assistance.
Footnotes
Published ahead of print on 11 October 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bratu, S., D. Landman, D. A. Martin, C. Georgescu, and J. Quale. 2008. Correlation of antimicrobial resistance with beta-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob. Agents Chemother. 52:2999-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, T. L., R. C. Wu, M. F. Shaio, C. P. Fung, and W. L. Cho. 2008. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692-699. [DOI] [PubMed] [Google Scholar]
- 5.Hu, W. S., S. M. Yao, C. P. Fung, Y. P. Hsieh, C. P. Liu, and J. F. Lin. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, K., D. Yong, Y. S. Choi, J. H. Yum, J. M. Kim, N. Woodford, D. M. Livermore, and Y. Chong. 2007. Reduced imipenem susceptibility in Klebsiella pneumoniae clinical isolates with plasmid-mediated CMY-2 and DHA-1 beta-lactamases co-mediated by porin loss. Int. J. Antimicrob. Agents 29:201-206. [DOI] [PubMed] [Google Scholar]
- 8.Lee, Y., J. H. Yum, C. K. Kim, D. Yong, E. H. Jeon, S. H. Jeong, J. Y. Ahn, and K. Lee. 2010. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann. Clin. Lab. Sci. 40:43-48. [PubMed] [Google Scholar]
- 9.Lu, P. L., M. Doumith, D. M. Livermore, T. P. Chen, and N. Woodford. 2009. Diversity of carbapenem resistance mechanisms in Acinetobacter baumannii from a Taiwan hospital: spread of plasmid-borne OXA-72 carbapenemase. J. Antimicrob. Chemother. 63:641-647. [DOI] [PubMed] [Google Scholar]
- 10.Mendes, R. E., J. M. Bell, J. D. Turnidge, M. Castanheira, and R. N. Jones. 2009. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63:55-59. [DOI] [PubMed] [Google Scholar]
- 11.Mugnier, P. D., L. Poirel, T. Naas, and P. Nordmann. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Price, L. S., and R. A. Weinstein. 2008. Acinetobacter infection. N. Engl. J. Med. 358:1271-1281. [DOI] [PubMed] [Google Scholar]
- 13.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., S. Figueiredo, V. Cattoir, A. Carattoli, and P. Nordmann. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodford, N., M. J. Ellington, J. M. Coelho, J. F. Turton, M. E. Ward, S. Brown, S. G. Amyes, and D. M. Livermore. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351-353. [DOI] [PubMed] [Google Scholar]