Abstract
Gentamicin is a highly efficacious antibiotic against Gram-negative bacteria. However, its usefulness in treating infections is compromised by its poorly understood renal toxicity. Toxic effects are also seen in a variety of other organisms. While the yeast Saccharomyces cerevisiae is relatively insensitive to gentamicin, mutations in any one of ∼20 genes cause a dramatic decrease in resistance. Many of these genes encode proteins important for translation termination or specific protein-trafficking complexes. Subsequent inspection of the physical and genetic interactions of the remaining gentamicin-sensitive mutants revealed a network centered on chitin synthase and the Arf GTPases. Further analysis has demonstrated that some conditional arf1 and gea1 alleles make cells hypersensitive to gentamicin under permissive conditions. These results suggest that one consequence of gentamicin exposure is disruption of Arf-dependent protein trafficking.
Gentamicin is an important aminoglycoside antibiotic used for the treatment of serious infections with Gram negative bacteria. However, the use of gentamicin is limited by its associated toxicity, especially for the highly endocytic cells of the ear and kidney. Multiple studies in mammalian cells have begun to unravel the complex nature of aminoglycoside cellular toxicity (22, 46). Specifically, we have determined that aminoglycosides, once internalized, are distributed throughout proximal tubule cells to several organelles, including the endoplasmic reticulum (ER) and Golgi apparatus, via retrograde endosomal trafficking (37). Furthermore, this intracellular distribution and subsequent cytosolic release are necessary for cell toxicity (38, 43). However, the mechanistic basis of this toxicity remains enigmatic. Our interest in understanding the molecular mechanism of gentamicin toxicity is premised on the likelihood that an understanding of this process will lead to new therapeutic strategies that alleviate the toxic effects of gentamicin without reducing its effectiveness against Gram-negative bacteria.
Due to its well-characterized biochemistry and extensive genetic tools, we have chosen to use the model system Saccharomyces cerevisiae, or baker's yeast, to elucidate the basis of gentamicin toxicity. Wild-type yeast is insensitive to gentamicin; however, mutants that are highly sensitive have been found. An initial screen to identify genes necessary for gentamicin resistance was performed by Blackburn and Avery (3). These investigators examined the ∼4,800 strains within the yeast deletion collection for mutants sensitive to gentamicin and uncovered 17 mutants with increased gentamicin sensitivity. These mutants fall into several groups, the largest of which is associated with intracellular protein trafficking.
The lone chaperone gene uncovered was ZUO1, which encodes a DnaJ homolog (13). Along with Ssz1, it forms RAC, or ribosome-associated complex, which stimulates the ATPase activity of Ssb, an Hsp70 protein encoded by SSB1 and SSB2 (13, 17, 26). Mutants lacking any of these proteins are sensitive to gentamicin as well as to the aminoglycoside paromomycin and have increased termination readthrough (24, 33). These characteristics are also associated with cells mutant for the translation release factors Sup35 and Sup45 (48). Furthermore, Polevoda et al. recently demonstrated that Sup45 is methylated by Mtq2 and that cells lacking MTQ2 are also sensitive to the aminoglycosides paromomycin and geneticin (30).
Blackburn and Avery also noted that many of the gentamicin-sensitive mutants were associated with vacuolar and Golgi functions (3). Several of these mutants were defective in components of the C/HOPS (class C Vps/homotypic vacuole fusion and vacuole protein sorting) or GARP (Golgi-associated-retrograde protein) complexes. Upon closer inspection, we found that most mutants lacking components of the C/HOPS or GARP complexes have increased gentamicin sensitivity (47). The C/HOPS complex, consisting of Pep3, Pep5, Vps16, and Vps33, is necessary for efficient fusion of vesicles from the Golgi apparatus to endosomes (for a review, see reference 49). These proteins also form a larger complex with Vps39 and Vps41 at the vacuolar membrane (42). This complex acts as a guanine nucleotide exchange factor (GEF) for the Rab-like protein Ypt7 and is critical for proper tethering of endosomal vesicles to the vacuolar membrane. Mutants lacking one of the four members of the C/HOPS complex, as well as Vps39 or Vps41, have increased sensitivity to gentamicin. Cells lacking the C/HOPS-associated protein Vps8 also have increased sensitivity to gentamicin. Blackburn and Avery also found that cells lacking Luv1 (Vps54), a component of the GARP complex, have increased sensitivity to gentamicin (3). We subsequently demonstrated that cells lacking one of the other components of the GARP complex (Vps51, Vps52, or Vps53) are hypersensitive to gentamicin (47). GARP is a tethering complex that facilitates retrograde vesicle trafficking from the endosomes to the Golgi apparatus (35, 49). We also found that loss of two other proteins necessary for retrograde trafficking to the Golgi apparatus, the syntaxin-like t-snare Tlg2 and the Sec1/Munc homolog Vps45, leads to increased gentamicin sensitivity.
These results suggest that the still unidentified targets of gentamicin within mammalian cells are components of endocytic vesicle trafficking. However, the cytoplasmic concentration of gentamicin is an important consideration in toxicity. Thus, a related phenomenon is that defects in endocytic trafficking that increase the concentration of gentamicin in the cytoplasm may then cause an increase in the interactions of gentamicin with its true targets. However, these true targets could include endocytic proteins exposed to the cytoplasm. Defects in the GARP complex cause defects in retrograde trafficking back to the Golgi apparatus as well as vacuole fragmentation (35). Loss of the C/HOPS complex also causes vacuolar fragmentation (50). Loss of the Nhx1 Na+/H+ transporter leads to enlargement of the prevacuolar compartment and defective transport to the vacuole (4). One consequence of each of these defects could be an increase in the amount of gentamicin leaked from endocytic vesicles and released into the cytoplasm.
To determine whether functional relationships exist among any of the remaining gentamicin-sensitive mutants, we examined the literature for possible genetic or physical relationships between these genes. This reexamination allowed us to develop a network of biochemical, physical, and genetic interactions between the remaining sensitive mutants found by Blackburn and Avery or their encoded proteins and a key regulator of vesicular transport—the Arf1/2 family of GTPases.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. The yeast growth medium was prepared as previously described (20). Yeast strains were grown at 30°C on rich yeast extract-peptone-dextrose (YPD) medium (2% [wt/vol] peptone, 2% [wt/vol] glucose, and 1% [wt/vol] yeast extract) (unless otherwise specified). Gentamicin was added to the medium to the concentrations indicated in the figure legends. Solid medium was made with 2% (wt/vol) agar. Yeast strains containing temperature-sensitive plasmids of GCS1 (pPP805-3 and pPP805-28) were obtained from G. Johnston's lab (University of Halifax) (31, 32).
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| 121.13C | MATα ade2-101 his3-Δ200 leu2-3,112 lys2-801 ura3-52 arf1::HIS3 arf2::LEU2 (pJCB1-21*)Gal+ | 19 |
| APY022 | MATα ura3-52 leu2-3,112 his3-Δ200 lys2-801 ade2-101 gea1-6 gea2::HIS3 | 29 |
| BL2 | MATα segregant of FY24XFY86 | M. Goebl |
| BY1437 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 age2::KanMX4 | Open Biosystems |
| BY4073 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rub1::KanMX4 | Open Biosystems |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| BY5512 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 syt1::KanMX4 | Open Biosystems |
| BY5937 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 zuo1::KanMX4 | Open Biosystems |
| BY6121 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 glo3::KanMX4 | Open Biosystems |
| BY6829 | MATahis3Δ1 leu2Δ0met15Δ0ura3Δ0gea1::KanMX4 | Open Biosystems |
| BY23528 | MATa/MATα his3Δ1/his3Δ1leu2Δ0/leu2Δ0ura3Δ0/ura3Δ0met15Δ0/MET15lys2Δ0/LYS2sec7Δ::kanMX4/SEC7 | Open Biosystems |
| BY25401 | MATa/MATα his3Δ1/his3Δ1leu2Δ0/leu2Δ0ura3Δ0/ura3Δ0met15Δ0/MET15lys2Δ0/LYS2sec12Δ::kanMX4/SEC7 | Open Biosystems |
| BY7499684 | MATahis3Δ1leu2Δ0met15Δ0ura3Δ0ARF1::TAP-HIS3MX6 | Open Biosystems |
| CJY49-3-4 | MATα ura3-52leu2-3,112his3-Δ200lys2-801ade2-101 | 29 |
| CJY62-10-2 | MATα ura3-52leu2-3,112his3-Δ200lys2-801ade2-101gea1-4gea2::HIS3 | 29 |
| NY966 | MATα sec7-1ura3leu2his3 | P. Novick |
| NYY0-1 | MATaade2::ARF1::ADE2arf1::HIS3arf2::HIS3ura3lys2trp1his3leu2 | 53 |
| NYY11-2 | MATaade2::arf1-11::ADE2arf1::HIS3arf2::HIS3ura3lys2trp1his3leu2 | 53 |
| NYY16-1 | MATaade2::arf1-16::ADE2arf1::HIS3arf2::HIS3ura3lys2trp1his3leu2 | 53 |
| NYY18-1 | MATaade2::arf1-18::ADE2arf1::HIS3arf2::HIS3ura3lys2trp1his3leu2 | 53 |
| PPY164-5A | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1can1-100gcs1::URA3 pPP805-3 | 31 |
| PPY164-5B | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1can1-100age2::HIS3 pPP805-3 | 31 |
| PPY164-5C | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1can1-100 pPP805-3 | 31 |
| PPY164-5D | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1can1-100gcs1::URA3age2::HIS3 pPP805-3 | 31 |
| PPY147-28-2A | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1gcs1::URA3glo3::HIS3 pPP805-28 | 31 |
| PPY147-28-2C | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1gcs1::URA3 pPP805-28 | 31 |
| PPY147-28-2D | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1 pPP805-28 | 31 |
| PPY147-28-8A | MATα ura3-1leu2-3,112his3-11ade2-1trp1-1glo3::HIS3 pPP805-28 | 31 |
| RT166 | MATα his3-Δ200leu2-3,112lys2-801ura3-52arf1::HIS3arf2::LEU2 (pJCH2-8**)Gal+ | 19 |
*, bARF1 coding region behind the GAL1 promoter; **, hARF4 coding region behind the GAL1 promoter.
GTP-binding ARF1-TAP pulldown assay.
Plasmid pAB382 was kindly provided by P. Scott (University of Minnesota Medical School, Duluth) (56). Plasmid pAB382 consists of a truncated yeast GGA2 gene (encoding amino acids 1 to 326) fused in frame at the amino terminus to the glutathione S-transferase gene (GST). BL21(DE3) bacterial cells transformed with pAB382 were cultured at 37°C in 10 ml of Luria-Bertani (LB) medium containing 150 μg/ml ampicillin (LB-Amp) for 12 to 16 h to reach stationary phase. This preculture was then diluted to 500 ml with fresh LB-Amp and was grown until the culture reached an optical density at 600 nm (OD600) of 0.6 to 0.8. Isopropyl β-d-thiogalactoside (IPTG) was then added to a final concentration of 0.8 mM, and the cells were incubated at 37°C for 5 h. Cells were harvested by centrifugation for 10 min at 4,500 × g and 4°C. Cell pellets were washed once and were then resuspended in 7 ml of ice-cold lysis buffer containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.1% (wt/vol) Triton X-100, and 1× Complete protease inhibitor (Roche, IN). Cells were sonicated on ice 20 times for 15 s each time at 30 s intervals. The homogenate was then centrifuged for 60 min at 15,000 × g and 4°C, and the protein concentration of the supernatant was determined using the Bradford method (5). Ten MicroSpin GST purification modules (Amersham Pharmacia, NJ) with a 50-μl bed volume of glutathione Sepharose 4B were prewashed in 300 μl lysis buffer. One hundred seventy-four micrograms of the supernatant (adjusted to a final volume of 50 μl in lysis buffer) was gently mixed with each MicroSpin column and was incubated at 4°C with agitation for 50 min to ensure optimal binding of GST-Gga2p1-320 to the glutathione Sepharose 4B matrix. After incubation, the flowthroughs of each MicroSpin column and four subsequent washes with 150 μl lysis buffer were discarded. Two hundred micrograms (adjusted to a final volume of 250 μl) of a yeast ARF1-TAP strain whole-cell lysate prepared from cells grown to different cell densities in the absence or presence of gentamicin was added to the MicroSpin columns and was incubated at 4°C for 30 min with agitation. The flowthroughs of each MicroSpin column and four consecutive washes with 150 μl lysis buffer were discarded. The bound proteins were eluted by the addition of 40 μl of elution buffer (10 mM glutathione, 50 mM Tris-HCl [pH 8.0]) to the spin column, centrifugation, and collection of the eluant. The released protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and TAP immunoblotting.
Gentamicin-Sepharose 4B affinity column binding assay.
Two grams of CNBr-activated Sepharose 4B (GE Healthcare, NJ) was swollen in 10 ml of 1 mM HCl (to a final volume of 7 ml) and was washed in 400 ml of 1 mM HCl in several aliquots, followed by an additional wash in 20 ml of coupling buffer (0.1 M NaHCO3 [pH 8.3], 0.5 M NaCl). The resin was separated into a control resin group and a gentamicin-binding resin group. The control resin was mixed with 10 ml coupling buffer and was incubated at 4°C overnight with gentle agitation. The gentamicin-binding resin was mixed with 10 ml gentamicin coupling buffer (10 mM gentamicin, 0.1 M NaCO3 [pH 8.3], 0.5 M NaCl) and was incubated under the same conditions as the control resin. For the gentamicin-binding resin, the excess gentamicin was removed by a wash with 40 ml of coupling buffer. The remaining CNBr-activated groups on both resins were inactivated with blocking buffer (0.2 M glycine [pH 8.0], 1% bovine serum albumin) at 4°C overnight with gentle agitation. The resins were then washed with three cycles of alternating pH (0.1 M acetic acid-sodium acetate [pH 4.0] containing 0.5 M NaCl and 0.1 M Tris-HCl [pH 8.0] containing 0.5 M NaCl). Each resin was equilibrated with phosphate-buffered saline (PBS; 0.05 M NaH2PO4, 0.05 M Na2HPO4, 0.2 M NaCl [pH 7.4]) before use. Four hundred micrograms of yeast whole-cell lysate was made from the ARF1-TAP yeast strain grown to mid-log phase and was adjusted to a final volume of 2 ml using PBS. This extract was mixed with 1 ml of either control resin or gentamicin-binding resin and was incubated at 4°C for 2 h with gentle agitation. Each resin was washed four times with 10 ml PBS buffer before the final elution with 6 ml. The sample was concentrated to 40 μl in elution buffer (0.05 M NaH2PO4, 0.05 M Na2HPO4, 1 M NaCl [pH 7.4], 10 mM gentamicin) using a Vivaspin 500 column (molecular weight cutoff [MWCO], 10,000; GE Healthcare). The flowthroughs of the four washes and elution were collected for SDS-PAGE and Western blot analysis.
Isolation of rat kidney cortical lysate.
A rat kidney was removed and flushed with PBS (0.05 M NaH2PO4, 0.05 M Na2HPO4, 1 M NaCl [pH 7.4]). The outer cortex was dissected and homogenized using a Dounce homogenizer in homogenization buffer (HB, consisting of 0.25 M sucrose, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, and protease inhibitor cocktail [Calbiochem]). The homogenate was centrifuged for 15 min at 3,000 rpm and 4°C. The supernatant containing 7.5 mg/ml protein was collected and frozen in liquid nitrogen. The lysate was then used for the gentamicin binding assay as described above.
RESULTS
Arf1/2-dependent trafficking is a possible target of gentamicin.
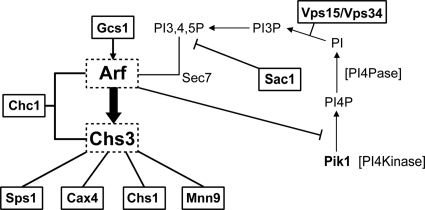
While the yeast Saccharomyces cerevisiae is relatively insensitive to gentamicin, mutations in any one of ∼20 genes cause a dramatic decrease in resistance. Many of these genes encode proteins important for translation termination or specific protein-trafficking complexes. However, roughly one-third of the mutants had no obvious relationship to translation or the two trafficking complexes C/HOPS and GARP. We therefore examined the literature for possible genetic or physical relationships between the genes of the remaining deletion mutants sensitive to gentamicin. This reexamination allowed us to develop a network of biochemical, physical, and genetic interactions between the remaining sensitive mutants found by Blackburn and Avery and yet another key component of protein trafficking—the Arf1/2 GTPases (Fig. 1).
FIG. 1.
Genetic/physical network based on SGD-defined relationships among genes whose functions are necessary for gentamicin resistance. Genes boxed with solid lines were found to exhibit gentamicin sensitivity by Blackburn and Avery (3). Pik1 would not have been identified in this screen, since its deletion causes inviability, while functional redundancy exists for members of the Arf and Chs protein families.
In S. cerevisiae, the Arf1/2 GTPases play a critical role in the production of the major cell wall component chitin by controlling the localization of the CHS3-encoded chitin synthase (45). Sps1 may also regulate the localization not only of Chs3 but also of the glucan synthase Gsc2 (18). CHS1 encodes another chitin synthase whose localization is regulated distinctly from that of Chs3 (57). CAX4 and MNN9 encode enzymes with a role in glycosylation (34). Furthermore, cells lacking either CAX4 or MNN9 are also sensitive to calcofluor white, an agent known to disrupt chitin deposition. GCS1 encodes an Arf-GAP (GTPase-activating protein), and cells doubly mutant for ARF1 as well as CHC1, GCS1, or PIK1 are inviable (6, 41, 55).
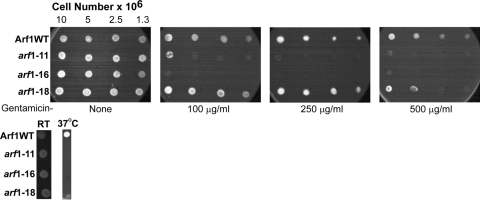
While Arf function is essential, Blackburn and Avery did not identify arf1Δ or arf2Δ mutants as hypersensitive to gentamicin, and upon close reexamination of these mutants, we also found no indication that these strains were hypersensitive to gentamicin (data not shown). We therefore examined whether the loss of Arf function causes yeast to become hypersensitive to gentamicin by the use of hypomorphic alleles of ARF1. Since the complete absence of Arf function is lethal, we utilized temperature-sensitive alleles of ARF1 (kindly provided by A. Nakano) to determine whether gentamicin affected cells with reduced Arf activity (53). These strains contain deletions of genomic ARF1 and ARF2 and have a specific arf1 mutant allele integrated at the ARF1 locus. As can be seen in Fig. 2, there is no detectable growth defect when these cells are grown at room temperature (23°C) in the absence of gentamicin. However, the strains containing mutant arf1 are hypersensitive to gentamicin. Interestingly, the gentamicin hypersensitivity generated by the three alleles used here, arf1-11, arf1-16, and arf1-18, is quite variable. The arf1-18 mutant has only a marginal growth defect when grown in the presence of 500 μg/ml gentamicin. However, the arf1-11 and arf1-16 mutants are very sensitive to gentamicin, even at a concentration of 100 μg/ml; the arf1-16 strain is unable to grow at all at 100 μg/ml gentamicin. Under our conditions in the absence of gentamicin, all three strains are temperature sensitive for growth at 37°C (Fig. 2).
FIG. 2.
Yeast cells mutant for ARF1 are hypersensitive to gentamicin. (Top) Wild-type (WT) cells (NYY0-1) or cells expressing only the indicated ARF1 mutation (NYY11-2, NYY16-1, or NYY18-1) were first grown overnight in YPD medium at 23°C, then transferred to solid YPD medium containing the indicated concentration of gentamicin, and finally incubated at 23°C overnight. (Bottom) The same strains were transferred to solid YPD medium and were grown overnight. Cells were then transferred to fresh YPD medium and were grown overnight at the indicated temperature, and images were captured. RT, room temperature.
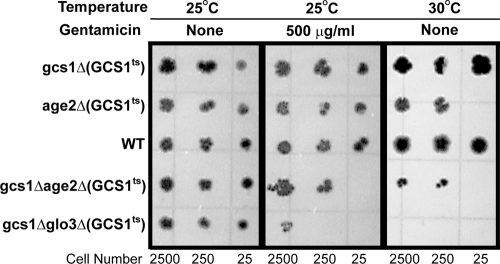
While Arf1 and Arf2 are relatively interchangeable, specificity for Arf function can be seen by examining the associated GAPs and guanine nucleotide exchange factors (GEFs) necessary for various steps in the secretory pathway. There are four GAPs associated with Arf function: Age1, Age2, Glo3, and Gcs1 (54). None of these proteins is essential, but several essential pairs of GAPs have been identified. Together, Age2 and Gcs1 are essential and affect movement from the trans-Golgi network (36), while Glo3 and Gcs1 also form an essential pair and are key regulators of retrograde trafficking from the Golgi apparatus to the ER (31). Furthermore, a gcs1Δ deletion strain was found to be weakly sensitive to gentamicin by Blackburn and Avery (3).
In order to assess the importance of GCS1 more definitively, we obtained a series of strains (kindly provided by G. Johnston) in which various combinations of Arf-GAP-encoding genes have been deleted and which are kept viable by the presence of a temperature-sensitive allele of GCS1 (Fig. 3; Table 1). Spot dilution assays of these cells were performed in the presence or absence of gentamicin, and duplicate plates were incubated at either 25°C or 30°C (Fig. 3) (see Materials and Methods). While each strain contains a temperature-sensitive allele of GCS1 (gcs1-3 or gcs1-28), PPY164-5D and PPY147-28-2A have gcs1::URA3 age2::HIS3 and gcs1::URA3 glo3::HIS3, respectively, and require the plasmid-borne copy of GCS1 for viability. As can be seen in Fig. 3, gcs1::URA3 age2::HIS3 and gcs1::URA3 glo3::HIS3 cells containing either gcs1-3 or gcs1-28 are sensitive to gentamicin at the permissive temperature of 25°C. Thus, defects in either the Arf GTPase or specific Arf-GAPs lead to increased gentamicin sensitivity.
FIG. 3.
Gentamicin sensitivities of yeast Arf GAP mutants. AGE1, AGE2, GLO3, and GCS1 encode the four GTPase-activating proteins (GAPs) associated with Arf function. None of these genes is essential, but the Age2/Gcs1 pair and the Glo3/Gcs1 pair are essential. Spot dilution assays were performed with the indicated numbers of cells, which were grown for 3 days on solid YPD medium. All strains contain a temperature-sensitive allele of GCS1, but only strains with defective GAP pairs are sensitive to gentamicin at the permissive temperature of 25°C.
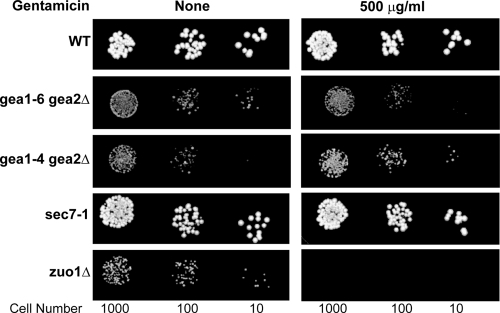
There are also several GEFs that function in different contexts with Arf1 and Arf2 (1). These include the nonessential Arf-GEF Syt1, which was not identified in the global screen for gentamicin sensitivity; the essential protein Sec7; and the essential pair of proteins Gea1 and Gea2. Since Sec7 and Gea1/Gea2 would not have been readily identified as having a role in gentamicin resistance, we examined the gentamicin sensitivities of a Sec7 temperature-sensitive strain (Table 1) and several strains that lacked wild-type GEA1 and GEA2 (Table 1) (29) but were kept alive by a copy of a temperature-sensitive allele of GEA1 (Table 1). As can be seen in Fig. 4, while the sec7ts mutant is unaffected by gentamicin, the gea1ts strain is sensitive to gentamicin. Thus, mutants dependent on defective alleles of ARF1 as well as GCS1 and GEA1 have increased gentamicin sensitivity.
FIG. 4.
Gentamicin sensitivities of yeast Arf-GEF mutants. GEA1, GEA2, SEC7, and SYT1 encode the four guanine nucleotide exchange factors (GEFs) associated with Arf function. SYT1 is not essential and was not identified in the global screen for gentamicin sensitivity. The GEA1/GEA2 pair and SEC7 are essential. Spot dilution assays were performed using the indicated cell numbers on solid YPD medium. Cells were grown for 2 days. These results show that while a sec7ts mutant is unaffected by gentamicin, the gea1ts (gea1Δ/gea2Δ) strain is sensitive to gentamicin.
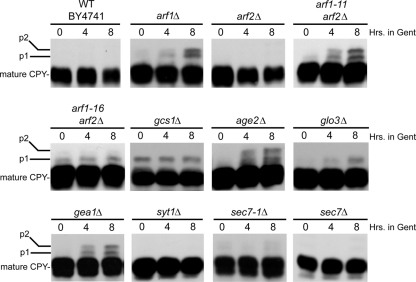
Arf activity is also important for the proper processing of carboxypeptidase Y (CPY) as it traffics through the secretory pathway. CPY is a soluble protein located in the lumen of the vacuole (44). Thus, we examined whether cells mutant for Arf or its regulators were defective in CPY processing in the presence of gentamicin. Interestingly, while arf1Δ and gea1Δ cells are both viable in the presence of gentamicin, both strains accumulate both p1 (ER) and p2 (Golgi complex) precursor forms of CPY (Fig. 5). As can also be seen in Fig. 5, when cells lacking both ARF1 and ARF2 that are kept alive by the presence of a temperature-sensitive allele of ARF1 (either arf1-11 or arf1-16) are treated with gentamicin, both the p1 (ER) and p2 (Golgi complex) precursor forms of CPY accumulate in arf1-11 cells, while arf1-16 and glo3Δ cells accumulate only modest levels of p1. Interestingly, age2Δ cells also accumulate CPY precursors in the presence of gentamicin. Age2 is an Arf-GAP that is a key regulator of trans-Golgi trafficking events. Thus, CPY trafficking and processing are also gentamicin sensitive in strains defective in ARF regulatory components.
FIG. 5.
Gentamicin disrupts CPY processing in cells mutant for Arf or Arf regulators. Strains were grown at 30°C to early-log phase (5 × 106 cells/ml) before gentamicin (to a final concentration of 500 μg/ml) was added to the medium. After 0, 4, or 8 h of additional incubation at 30°C, aliquots of cells were removed and prepared for Western blot analysis using anti-CPY primary antibodies. Gent, gentamicin.
Gentamicin influences GTP-bound Arf levels during a growth cycle.
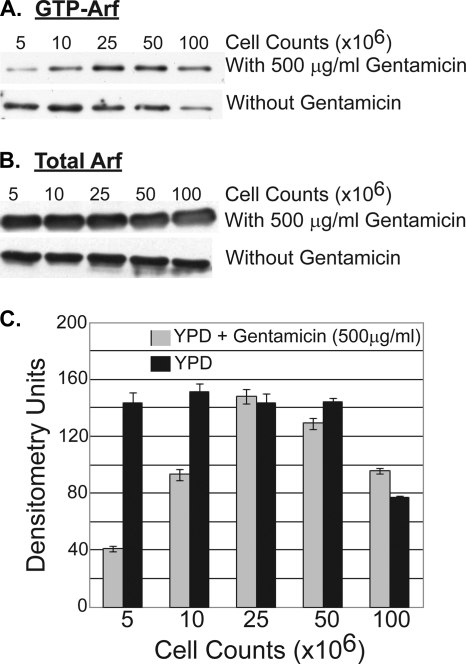
Given that not only cells defective in Arf1 but also cells defective in the GTPase regulators of Arf1 are sensitive to gentamicin, we examined whether gentamicin affected the levels of Arf1-GTP present in cells. The γ-ear adaptin proteins are a family of proteins that bind to both clathrin and Arf-GTP (16). The yeast homologs of this family are the proteins Gga1 and Gga2 (16). We therefore utilized the Arf-GTP binding specificity of Gga2 to measure Arf-GTP levels in cells growing in the presence of gentamicin. A GST-Gga2 protein was expressed in bacteria from plasmid pAB382 and was purified as described in Materials and Methods. The purified GST-Gga2 was bound to GST-Sepharose and was then incubated with Arf1-TAP yeast cell extracts grown in the presence or absence of gentamicin. The bound protein was eluted with glutathione and subjected to SDS-PAGE and TAP Western blot analysis. As can be seen in Fig. 6A and C, gentamicin alters Arf1-GTP levels in a manner dependent on the growth state of the cells. The relative abundance of Arf1-GTP is dramatically reduced in cells that are in early-logarithmic growth (cell densities, 5 × 106 and 107 cells/ml). However, as cells approach late-logarithmic growth (2.5 × 107 and 5 × 107 cells/ml), gentamicin has no detectable effect on Arf1-GTP levels. Finally, as cells enter stationary phase (108 cells/ml), gentamicin causes a modest increase in Arf1-GTP levels over those in cells grown in the absence of gentamicin. This effect is not caused by a change in the level of total Arf1, which remains constant in the presence or absence of gentamicin and across all growth phases of the cells (Fig. 6B). Thus, gentamicin can dramatically affect the level of Arf-GTP in a manner also influenced by the growth phase of the cells.
FIG. 6.
Arf1-GTP levels change while total Arf1 levels remain constant. (A and B) During different yeast cell growth phases, Arf1-GTP levels are lower at early- and mid-log phase in the presence (at 500 μg/ml) of gentamicin (A), while total Arf1 protein levels remain constant in the absence or presence (at 500 μg/ml) of gentamicin (B). (C) Scanning densitometry analysis of Arf1-GTP abundance in the presence (at 500 μg/ml) or absence of gentamicin.
Enrichment of Arf1 by gentamicin column chromatography.
One possible explanation for the effects of gentamicin on Arf activity is that gentamicin can bind directly to Arf or Arf-containing complexes. In order to test this hypothesis, yeast extracts from a TAP-ARF1 strain were passed over a gentamicin Sepharose column. The column was then washed four times, and bound protein was eluted with 10 mM gentamicin. The proteins present in this eluate were separated by SDS-PAGE and were then transferred to a polyvinylidene difluoride (PVDF) membrane. Western blot analysis was then performed using anti-TAP antibodies. As can be seen in Fig. 7, TAP-Arf1 was bound only by the gentamicin Sepharose column and not by the control beads. Further, this interaction could be enhanced by pretreating the extracts with GDP. However, bacterially expressed and purified Arf1 did not bind to gentamicin Sepharose (data not shown). This suggests that Arf1 binding either is indirect and/or requires a specific Arf1 conformation present only when it is interacting with one of its many regulatory proteins, i.e., Arf GAPs and ARF GEFs.
FIG. 7.
Enrichment of yeast Arf1 and rat Arf1 by gentamicin-Sepharose. (Top) Western blot analysis for yeast Arf1-TAP protein. From left to right, lanes show whole-cell lysate from an ARF1-TAP strain that was added to and incubated with gentamicin-Sepharose; four washes of the column after incubation with whole-cell lysate; and final elution from the column. (Bottom) Western blot analysis of rat Arf1 protein using preimmune serum or anti-rat Arf1 (FabGennix, Frisco, TX). Lanes are the same as explained above, except that the leftmost lane shows a loading control of rat kidney cortex lysates, which were added to and incubated with gentamicin-Sepharose.
While we did not detect the binding of unrelated TAP-tagged proteins to gentamicin-Sepharose, including TAP-Glo3 (data not shown), we wanted to determine whether the binding of Arf to gentamicin-Sepharose was a specific characteristic of yeast Arf or could be seen in other organisms. Therefore, we examined whether Arf1 from rat kidney cortex extracts also bound to gentamicin-Sepharose. Rat kidney cortex extracts were prepared as described in Materials and Methods and were passed over gentamicin-Sepharose. After several washes, the remaining proteins were eluted with gentamicin, separated by SDS-PAGE, and transferred to a PVDF membrane. In this case, Western blot analysis was performed using anti-rat Arf1 antibodies. As seen in Fig. 6, rat kidney Arf1 also bound to gentamicin-Sepharose. Thus, we can detect both yeast Arf and rat kidney Arf on gentamicin-Sepharose by either antibodies to an epitope tag (yeast) or antibodies directly recognizing Arf1 (rat). While other proteins were eluted from the gentamicin-Sepharose, mass spectrometry of these proteins showed they were not Arf regulators. These proteins are now the subject of further investigation.
Mammalian Arf activity is more sensitive to gentamicin inhibition than yeast Arf activity.
While wild-type yeast are resistant to gentamicin, mammalian cells can lose viability in the presence of therapeutic concentrations of gentamicin (10). Since endocytic functions are known to be affected by gentamicin in mammalian cells, we reasoned that one possibility is that Arf-dependent trafficking is more sensitive to disruption by the presence of gentamicin in mammalian cells than in yeast. To gain insight into whether mammalian Arf proteins might be at least partially responsible for this difference, we examined the gentamicin sensitivity of yeast cells in which the essential function provided by the ARF1/ARF2 gene pair is replaced by human ARF4 (hARF4) or bovine ARF1 (bARF1; the encoded product is identical to human Arf1). Yeast arf1Δ/arf2Δ cells expressing yeast ARF1 (NYY0-1 [53]), hARF4 (RT166 [19]), or bARF1 (121.13C [19]) (see Table 1) were grown in rich medium containing either 100 μg/ml or 500 μg/ml gentamicin. Cells expressing either hARF4 or bARF1 were sensitive to gentamicin at either concentration, although bARF1-utilizing cells were noticeably more sensitive to gentamicin than hARF4-utilizing cells (Fig. 8). It is noteworthy that the inhibitory concentration of 100 μg/ml is in the same range as that achieved in vivo when gentamicin is administered to patients (8, 9). Thus, it is possible that the increased sensitivity of mammalian cells to gentamicin may be at least partially due to the increased sensitivity of mammalian ARF to the presence of gentamicin.
FIG. 8.
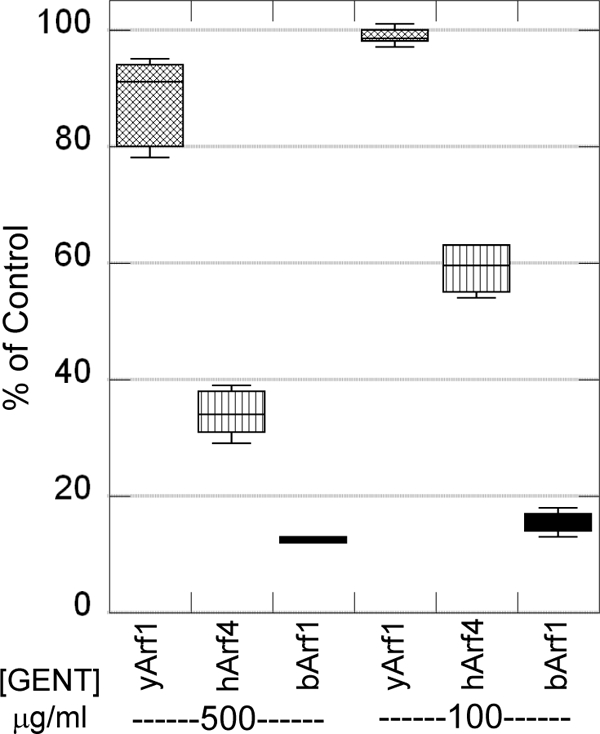
Expression of mammalian ARF genes changes the sensitivity of yeast cells to gentamicin. Yeast arf1Δ/arf2Δ cells expressing yeast ARF1 (yArf1), human ARF4 (hArf4), or bovine ARF1 (bArf1) were grown in YPD medium containing either 100 μg/ml or 500 μg/ml gentamicin and were incubated at 30°C overnight. Cells expressing either hARF4 or bARF1 were sensitive to gentamicin at either concentration.
DISCUSSION
The current literature raises critical questions regarding the understanding of aminoglycoside nephrotoxicity that center around the trafficking and compartmentation of aminoglycosides within the endosomal/lysosomal and cytosolic compartments. Further identification of the intracellular mechanisms and the proteins/pathways involved will play a pivotal role in developing needed therapeutic approaches. However, this is difficult in mammalian cells due to their inherent complexity. Therefore, given the tremendous genetic, genomic, and proteomic advantages of yeast and the demonstrated similarities between yeast and mammalian cells, we have begun to utilize yeast to understand the pathways important in inducing gentamicin toxicity.
In light of our findings, we reexamined the literature for potential relationships between gentamicin and the hypersensitive mutants uncovered by Blackburn and Avery that were not linked to the function of either the C/HOPS or GARP complexes or to chaperones critical for regulating translation. In this manner, we were able to formulate a network among these gentamicin-sensitive genes and the Arf1/2 GTPases based on both genetic and physical interactions (from the Saccharomyces Genome Database [SGD]). This network also includes the components of the phosphatidylinositol (PtdIns) pathway Sac1 and Pik1. Sac1 is a PtdIns(4)P phosphatase whose loss increases sensitivity to gentamicin (3), while Pik1 is a PtdIns(4)P kinase that generates a pool necessary for proper Golgi trafficking (11, 41, 51). Thus, it is likely that PtdIns(4)P is a key regulator of Arf-dependent Golgi function, possibly as a direct effector of Arf or its regulators, such as Gcs1, an Arf-GAP, or Gea1/2, which are Arf-GEFs. Interestingly, cationic amphiphilic compounds, such as gentamicin, are known to cause an accumulation of phospholipids in mammals (2). Based on these data and the role of Sac1 in yeast hypersensitivity to gentamicin, it will be informative to determine whether accumulation of phospholipids, especially that of PtdIns(4)P, plays a protective role in gentamicin exposure. Sps1 (18), Sac1 (40), and Chs1 are directly involved in the deposition of chitin, and Cax1 and Mnn9 are likely to be involved in the proper posttranslational modification of Chs3. Interestingly, although MNN9 was identified in the screen for mutants hypersensitive to gentamicin, it encodes a member of the α-1,6-mannosyltransferase complex; a deletion of any of the six genes (ANP1, HOC1, MNN9, MNN10, MNN11, VAN1) encoding components of this transferase confers sensitivity to calcofluor white, and many of these deletions are synthetically lethal with chitin synthase mutations (21).
As a direct test of a role for Arf in gentamicin toxicity, we found that cells expressing only a defective allele of ARF1 are hypersensitive to gentamicin (Fig. 2). Only some mutant alleles of ARF1 cause cells to become sensitive to gentamicin; however, arf1-16 strains are the most sensitive. Interestingly, the temperature-sensitive phenotype of a strain carrying arf1-16 is suppressed by overexpression of GLO3, which encodes an alternative Arf1 GTPase-activating protein to Gcs1, but that of the arf1-11 or arf1-18 strain is not (52). These data, coupled with the greater sensitivity of arf1-16 to gentamicin, suggest the particularly susceptible gentamicin target may be the interaction between Arf and Glo3 (and possibly Gcs1). In their original gentamicin sensitivity screen, Blackburn and Avery found gcs1Δ mutants to be hypersensitive to gentamicin (3). Originally, we found that the loss of GCS1 caused a subtle growth defect in response to gentamicin in our background. However, we have found that strains lacking either essential pair of ARF GTPase-encoding genes (gcs1Δ/age2Δ or gcs1Δ/glo3Δ) but kept viable by a partially defective allele of GCS1 are much more sensitive to gentamicin. Furthermore, cells lacking GEA2 and expressing only a temperature-sensitive allele of GEA1 are also sensitive to gentamicin, while sec7 mutants are not. These results suggest that gentamicin interferes with a subset of Arf functions. For example, the Arf-GEFs Gea1/Gea2 and the Arf-GAPs Gcs1/Glo3 regulate Arf function during the formation of COPI (coat protein complex I) vesicles for Golgi complex-to-ER retrograde trafficking (31). Robinson et al. (36) have shown that Arf and the other essential pair of GAPs, Age2 and Gcs1, are key regulators of endosome-to-Golgi complex trafficking. It has also been shown that the Gcs1-and-Age2 pair of Arf-GAPs is important in the trafficking of proteins from the trans-Golgi network to the plasma membrane (32). A conditional double mutant is defective in the secretion of invertase as well as the delivery of Ste3 to the plasma membrane and CPY to the vacuole. Thus, gentamicin appears to interfere with at least two distinct Arf functions—Golgi complex-to-ER retrograde trafficking and endosome-to-Golgi complex trafficking. A defect in Golgi complex-to-ER trafficking is also consistent with the sensitivity of CPY processing to gentamicin in arf1 mutant strains. Often defects in CPY processing are caused by improper recycling of the ER cargo receptors, such as Rer1 and Erv14, back to the ER (14, 27). Furthermore, GLO3 is a high-copy-number suppressor of arf1-16, the arf1 mutant found to be most sensitive to gentamicin (52). The genetic and physical interactions between the Arf pathway components and gentamicin are similar to those between the Arf pathway and brefeldin A. Brefeldin A interferes with Arf function by disrupting the Sec7-dependent exchange of GDP for GTP on Arf (23, 28, 39). Thus, it will be interesting to determine whether gentamicin disrupts Arf function via a similar mechanism involving the Gea proteins or one of the Arf-GAPs. Interestingly, the cluster of mutants that cause hypersensitivity to brefeldin A is very different from that involved in gentamicin hypersensitivity, possibly because of the specificity for either the Sec7-Arf function or the Gea1/2-Arf function (25).
A major effect of gentamicin in mammalian cells is the effect on the process of endocytosis (22). This is similar to the deposition of chitin, which is controlled by regulating the movement of Chs3 from internal membrane stores to the cell surface (7, 12). Both processes require Arf activity (45). Thus, the functions of Arf are likely to be very similar in yeast and mammalian cells. In fact, human Arf1 or human Arf4 can rescue an arf1 arf2 double yeast mutant (19). Thus, Arf function is highly conserved between yeast and humans. One can consider the activation of the cell wall stress response by gentamicin as simply a measure of Arf disruption. The gentamicin-inhibited trafficking events may be analogous to the defects seen during lysosomal vesicle fusion in the kidney proximal tubule cells of rats treated with gentamicin (15). While Arf will clearly be regulating a different set of endocytic events in mammals, if gentamicin directly interferes with Arf activity, it is likely that gentamicin will interfere with Arf function similarly in yeast and mammals. In fact, we demonstrate here that when yeast cells have the function of their essential pair of Arf1 and Arf2 replaced by mammalian ARF genes, these cells have the kind of gentamicin sensitivity more typically associated with mammals than with yeast.
Based on our recent findings (47) and those of Blackburn and Avery (3), we propose the following model for gentamicin toxicity (Fig. 9). Gentamicin enters a yeast cell; however, most of the gentamicin is delivered to the vacuole and is relatively nontoxic. In cells lacking a functional Nhx1 endocytic proton transporter, the C/HOPS complex or the GARP complex, vesicular movement of gentamicin to the vacuole is impaired, causing missorting of vesicles, ultimately resulting in an increase in the cytoplasmic gentamicin concentration, which is toxic. Translational termination has long been proposed to be a target of the aminoglycosides and of gentamicin in particular (see the introduction). Normally, the toxic effects of gentamicin are lessened through the activities of the chaperones associated with translational termination—Zuo1, Ssz1, Ssb1, and Ssb2. However, in their absence or in the presence of greater cytoplasmic gentamicin concentrations, gentamicin becomes toxic. Our data also suggest that the Arf1 protein itself or an associated protein or target of Arf1/2-dependent trafficking may be a direct target of gentamicin. This is suggested by the increased sensitivity seen in cells exposed to gentamicin that lack any one of a number of regulators of Arf activity. Finally, the C/HOPS and GARP complexes, as well as the regulators of translational termination and the Arf pathway, have mammalian homologs, suggesting that similar gentamicin interactions take place in mammalian cells. While we see no evidence for mitochondrial involvement in yeast resistance to gentamicin, it is well known that the mitochondrial translation machinery is typically sensitive to antibiotics affecting bacterial translation. Thus, a closer examination of the effects of gentamicin on mitochondrial translation is warranted.
FIG. 9.
Model for gentamicin targets in Saccharomyces.
Acknowledgments
This work was supported by National Institutes of Health grants P50-DK-61594 and P01-DK-53465, a Veterans Affairs Merit Review (to B. A. Molitoris), NSF grant MCB-0091317 (to M. G. Goebl), and an Indiana Genomics Initiative grant (INGEN) from the Lilly Endowment to the Indiana University School of Medicine.
We thank Barbara Sturonas-Brown for excellent assistance with the figures.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Anders, N., and G. Jürgens. 2008. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65:3433-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baronas, E. T., J.-W. Lee, C. Alden, and F. Y. Hsieh. 2007. Biomarkers to monitor drug-induced phospholipidosis. Toxicol. Appl. Pharmacol. 218:72-78. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, A. S., and S. V. Avery. 2003. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob. Agents Chemother. 47:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers, K., B. P. Levi, F. I. Patel, and T. H. Stevens. 2000. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 11:4277-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. Y., and T. R. Graham. 1998. An arf1Δ synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics 150:577-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang, J. S., and R. W. Schekman. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cojocel, C. 1997. Aminoglycoside nephrotoxicity, p. 495-523. In I. G. Sipes, C. A. McQueen, and A. J. Gandolfi (ed.), Comprehensive toxicology, vol. 7. Renal toxicology. Elsevier Science, Oxford, England. [Google Scholar]
- 9.DeBroe, M. E., and G. A. Porter. 2008. Clinical nephrotoxins: renal injury from drugs and chemicals, 3rd ed. Springer Science, New York, NY.
- 10.El Mouedden, M., G. Laurent, M.-P. Mingeot-Leclercq, and P. M. Tulkens. 2000. Gentamicin-induced apoptosis in renal cell lines and embryonic rat fibroblasts. Toxicol. Sci. 56:229-239. [DOI] [PubMed] [Google Scholar]
- 11.Faulhammer, F., S. Kanjilal-Kolar, A. Knödler, J. Lo, Y. Lee, G. Konrad, and P. Mayinger. 2007. Growth control of Golgi phosphoinositides by reciprocal localization of SacI lipid phosphatase and Pik1 4-kinase. Traffic 8:1554-1567. [DOI] [PubMed] [Google Scholar]
- 12.Flores Martinez, A., and J. Schwencke. 1988. Chitin synthetase activity is bound to chitosomes and to the plasma membrane in protoplasts of Saccharomyces cerevisiae. Biochim. Biophys. Acta 946:328-336. [DOI] [PubMed] [Google Scholar]
- 13.Gautschi, M., H. Lilie, U. Funfschilling, A. Mun, S. Ross, T. Lithgow, P. Rucknagel, and S. Rospert. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U. S. A. 98:3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillingham, A. K., A. H. Tong, C. Boone, and S. Munro. 2004. The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J. Cell Biol. 167:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giurgea-Marion, L., G. Toubeau, G. Laurent, J. A. Heuson-Stiennon, and P. M. Tulkens. 1986. Impairment of lysosome-pinocytic vesicle fusion in rat kidney proximal tubules after treatment with gentamicin at low doses. Toxicol. Appl. Pharmacol. 86:271-285. [DOI] [PubMed] [Google Scholar]
- 16.Hirst, J., W. W. Y. Lui, N. A. Bright, N. Totty, M. N. J. Seaman, and M. S. Robinson. 2000. A family of proteins with γ-adaptin and Vhs domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, P., M. Gautschi, W. Walter, S. Rospert, and E. A. Craig. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497-504. [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto, M. A., S. R. Fairclough, S. A. Rudge, and J. Engebrecht. 2005. Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis. Eukaryot. Cell 4:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahn, R. A., F. G. Kern, J. Clark, E. P. Gelmann, and C. Rulka. 1991. Human ADP-ribosylation factors. A functionally conserved family of GTP-binding proteins. J. Biol. Chem. 266:2606-2614. [PubMed] [Google Scholar]
- 20.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Lesage, G., J. Shapiro, C. A. Specht, A. M. Sdicu, P. Menard, S. Hussein, A. H. Tong, C. Boone, and H. Bussey. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mingeot-Leclercq, M. P., and P. M. Tulkens. 1999. Aminoglycosides: nephrotoxicity. Antimicrob. Agents Chemother. 43:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morinaga, N., S.-C. Tsai, J. Moss, and M. Vaughan. 1996. Isolation of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP ribosylation factor (ARF) 1 and ARF3 that contains a Sec7-like domain. Proc. Natl. Acad. Sci. U. S. A. 93:12856-12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muldoon-Jacobs, K. L., and J. D. Dinman. 2006. Specific effects of ribosome-tethered molecular chaperones on programmed-1 ribosomal frameshifting. Eukaryot. Cell 5:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murén, E., M. Öyen, G. Barmark, and H. Ronne. 2001. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast (Chichester, England) 18:163-172. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97-105. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa, S., and A. Nakano. 1993. Identification of a gene required for membrane protein retention in the early secretory pathway. Proc. Natl. Acad. Sci. U. S. A. 90:8179-8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyroche, A., B. Antonny, S. Robineau, J. Acker, J. Cherfils, and C. L. Jackson. 1999. Brefeldin A acts to stabilize an abortive ARF GDP Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3:275-285. [DOI] [PubMed] [Google Scholar]
- 29.Peyroche, A., R. Courbeyrette, A. Rambourg, and C. L. Jackson. 2001. The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114:2241-2253. [DOI] [PubMed] [Google Scholar]
- 30.Polevoda, B., L. Span, and F. Sherman. 2006. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 281:2562-2571. [DOI] [PubMed] [Google Scholar]
- 31.Poon, P. P., D. Cassel, A. Spang, M. Rotman, E. Pick, R. A. Singer, and G. C. Johnston. 1999. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon, P. P., S. F. Nothwehr, R. A. Singer, and G. C. Johnston. 2001. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155:1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakwalska, M., and S. Rospert. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:9186-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ram, A. F., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast (Chichester, England) 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 35.Reggiori, F., C. W. Wang, P. E. Stromhaug, T. Shintani, and D. J. Klionsky. 2003. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278:5009-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, M., P. P. Poon, C. Schindler, L. E. Murray, R. Kama, G. Gabriely, R. A. Singer, A. Spang, G. C. Johnston, and J. E. Gerst. 2006. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell 17:1845-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandoval, R. M., K. W. Dunn, and B. A. Molitoris. 2000. Gentamicin traffics rapidly and directly to the Golgi complex in LLC-PK(1) cells. Am. J. Physiol. Renal Physiol. 279:F884-F890. [DOI] [PubMed] [Google Scholar]
- 38.Sandoval, R. M., and B. A. Molitoris. 2004. Gentamicin traffics retrograde through the secretory pathway and is released in the cytosol via the endoplasmic reticulum. Am. J. Physiol. Renal Physiol. 286:F617-F624. [DOI] [PubMed] [Google Scholar]
- 39.Sata, M., J. G. Donaldson, J. Moss, and M. Vaughan. 1998. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc. Natl. Acad. Sci. U. S. A. 95:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schorr, M., A. Then, S. Tahirovic, N. Hug, and P. Mayinger. 2001. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11:1421-1426. [DOI] [PubMed] [Google Scholar]
- 41.Sciorra, V. A., A. Audhya, A. B. Parsons, N. Segev, C. Boone, and S. D. Emr. 2005. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol. Biol. Cell 16:776-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seals, D. F., G. Eitzen, N. Margolis, W. T. Wickner, and A. Price. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. U. S. A. 97:9402-9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Servais, H., P. Van Der Smissen, G. Thirion, G. Van der Essen, F. Van Bambeke, P. M. Tulkens, and M. P. Mingeot-Leclercq. 2005. Gentamicin-induced apoptosis in LLC-PK1 cells: involvement of lysosomes and mitochondria. Toxicol. Appl. Pharmacol. 206:321-333. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, T., B. Esmon, and R. Schekman. 1982. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 30:439-448. [DOI] [PubMed] [Google Scholar]
- 45.Trautwein, M., C. Schindler, R. Gauss, J. Dengjel, E. Hartmann, and A. Spang. 2006. Arf1p, Chs5p, and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 25:943-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verpooten, G., P. M. Tulkens, and B. A. Molitoris. 2003. Aminoglycosides and vancomycin, p. 151-170. In M. E. De Broe, G. A. Porter, W. M. Bennett, and G. A. Verpooten (ed.), Clinical nephrotoxins. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 47.Wagner, M. C., E. E. Molnar, B. A. Molitoris, and M. G. Goebl. 2006. Loss of the homotypic fusion and vacuole protein sorting or Golgi-associated retrograde protein vesicle tethering complexes results in gentamicin sensitivity in the yeast Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 50:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakem, L. P., and F. Sherman. 1990. Isolation and characterization of omnipotent suppressors in the yeast Saccharomyces cerevisiae. Genetics 124:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whyte, J. R., and S. Munro. 2002. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115:2627-2637. [DOI] [PubMed] [Google Scholar]
- 50.Wickner, W., and A. Haas. 2000. Yeast homotypic vacuole fusion: a window on organelle trafficking mechanisms. Annu. Rev. Biochem. 69:247-275. [DOI] [PubMed] [Google Scholar]
- 51.Wood, C. S., K. R. Schmitz, N. J. Bessman, T. G. Setty, K. M. Ferguson, and C. G. Burd. 2009. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187:967-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yahara, N., K. Sato, and A. Nakano. 2006. The Arf1p GTPase-activating protein Glo3p executes its regulatory function through a conserved repeat motif at its C-terminus. J. Cell Sci. 119:2604-2612. [DOI] [PubMed] [Google Scholar]
- 53.Yahara, N., T. Ueda, K. Sato, and A. Nakano. 2001. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol. Biol. Cell 12:221-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, C.-J., M. M. Cavenagh, and R. A. Kahn. 1998. A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:19792-19796. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, C. J., J. B. Bowzard, A. Anido, and R. A. Kahn. 2003. Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast (Chichester, England) 20:315-330. [DOI] [PubMed] [Google Scholar]
- 56.Zhdankina, O., N. L. Strand, J. M. Redmond, and A. L. Boman. 2001. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast (Chichester, England) 18:1-18. [DOI] [PubMed] [Google Scholar]
- 57.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]