Abstract
The proteomic response of Aspergillus fumigatus to caspofungin was evaluated by gel-free isobaric tagging for relative and absolute quantitation (iTRAQ) as a means to determine potential biomarkers of drug action. A cell fractionation approach yielding 4 subcellular compartment fractions was used to enhance the resolution of proteins for proteomic analysis. Using iTRAQ, a total of 471 unique proteins were identified in soluble and cell wall/plasma membrane fractions at 24 and 48 h of growth in rich media in a wild-type drug-susceptible strain. A total of 122 proteins showed at least a 2-fold change in relative abundance following exposure to caspofungin (CSF) at just below the minimum effective concentration (0.12 μg/ml). The largest changes were seen in the mitochondrial hypoxia response domain protein (AFUA_1G12250), the level of which decreased >16-fold in the secreted fraction, and ChiA1, the level of which decreased 12.1-fold in the cell wall/plasma membrane fraction. The level of the major allergen and cytotoxin AspF1 was also shown to decrease by 12.1-fold upon the addition of drug. A subsequent iTRAQ analysis of an echinocandin-resistant strain (fks1-S678P) was used to validate proteins specific to drug action. A total of 103 proteins in the 2 fractions tested by iTRAQ were differentially expressed in the wild-type susceptible strain but not significantly changed in the resistant strain. Of these potential biomarkers, 11 had levels that changed at least 12-fold. Microarray analysis of the susceptible strain was performed to evaluate the correlation between proteomics and genomics, with a total of 117 genes found to be changing at least 2-fold. Of these, a total of 22 proteins with significant changes identified by iTRAQ also showed significant gene expression level changes by microarray. Overall, these data have the potential to identify biomarkers that assess the relative efficacy of echinocandin drug therapy.
Aspergillus fumigatus is a leading cause of fungal morbidity and mortality in immunocompromised patients (12, 24). Early detection and treatment lead to improved clinical outcomes (12). Aspergillus fumigatus infections can be treated with triazole (posaconazole, itraconazole, and voriconazole), polyene (amphotericin B), or echinocandin (caspofungin, micafungin, and anidulafungin) drugs (5, 6, 17, 18). The echinocandins are the newest class of antifungal drugs and inhibit the biosynthesis of β-1,3,-d-glucan, which is a principal cell wall component (6, 20). They have a high level of efficacy against A. fumigatus (1, 13, 27, 31) and show limited clinical resistance (2, 30). Successful therapy is measured largely as a resolution of clinical symptoms. The use of biomarkers such as galactomannan or Aspergillus-specific nucleic acids is important for primary diagnosis, but their role in monitoring therapy is still evolving. Recently, novel biomarkers that can be used as drug targets or markers of effective treatment have been identified by microarray genomic analysis or other methods of RNA analysis (10, 37, 40). Proteomic analysis relying on two-dimensional (2D) gel electrophoresis has been used to identify novel allergens, evaluate changes in levels of expressed protein in response to amphotericin B, as well as build a reference map of different compartments of the cell (3, 15, 16, 22, 23, 26, 43). This conventional approach provides a basis for proteomic analysis but it is limited by the number of protein spots that can be resolved as well as the ability to excise, identify, and quantify the proteins (4). Furthermore, this approach for proteome studies suffers greatly from data compression when large genomes are evaluated, such as that of Aspergillus fumigatus, with >10,000 open reading frames (ORFs) (29).
To overcome these limitations, a cell fractionation approach can be used to better resolve total proteins (43). Furthermore, to obtain larger numbers of identified proteins and provide more in-depth analysis, a non-gel-based system of isobaric tagging for relative and absolute quantitation (iTRAQ) is now available. This system uses a series of isobaric tags that attach to all lysines and the N termini of each protein following proteolysis (34, 44). It has been used with other fungi, such as Saccharomyces cerevisiae and Fusarim graminearum, and has been used extensively for evaluating human and other mammalian diseases (32, 35, 38, 42). In the current study, the proteomic response of A. fumigatus to caspofungin was evaluated in four major subcellular fractions by iTRAQ in both a sensitive wild-type strain as well as an isogenic echinocandin-resistant strain; a comparative analysis of the genomic profiles was also conducted. The data highlight potential biomarker candidates to assess drug action.
MATERIALS AND METHODS
Strains, media, and antifungal susceptibility testing.
Isogenic A. fumigatus strain KU80 ΔakuB (9) and caspofungin-resistant strain EMFR-S678P (33) were used throughout this work. The strains were grown at 37°C on potato dextrose agar (PDA; Becton Dickenson, Sparks, MD) and in YPD broth (2% yeast extract, 4% Bacto peptone, 4% dextrose). Caspofungin (CSF; Merck & Co., Inc., Rahway, NJ) was obtained as a standard powder from the manufacturer. It was dissolved in sterile distilled water, and stock solutions were stored at −86°C. Broth microdilution susceptibility testing was performed in triplicate according to the directions of CLSI document M38-A2 (8). Aspergillus fumigatus strains KU80 ΔakuB and EMFR-S678P were subjected to growth inhibition testing of caspofungin using both RPMI and YPD media to yield a minimum effective concentration (MEC), the lowest concentration producing a significant change in hyphal morphology visible to the eye. The strains gave MEC values of 0.25 μg/ml and >16 μg/ml, respectively, in RPMI medium and 0.19 μg/ml and >16 μg/ml, respectively, in YPD broth.
Proteomic analysis. (i) Subcellular fraction recovery.
The isolates used in this work were grown with shaking (225 rpm) at 37°C in 50 ml of YPD broth containing 0.12 μg/ml of CSF. The inocula were adjusted to 1 × 105 conidia/ml using a hemocytometer. Mycelia were recovered by filtration through a Miracloth (CalBiochem, La Jolla, CA) after the allotted time. The mycelia were the washed using 250 ml cold sterile distilled water (dH2O), dried, and frozen at −80°C for future use. The filtrate was conserved and then concentrated by centrifugation using iCON concentrators with a molecular cutoff of 9 kDa (Thermo Scientific, Rockford, IL) to a volume of 4 ml and was considered the secreted fraction. The mycelium was defrosted on ice, and 2 volumes of 4°C lysis buffer (50 mM HEPES, 20% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM dithiothreitol [DTT]) was added. The obtained material was then disrupted in a French pressure cell at 20,000 lb/in2 5 times. The material was then spun at 5,000 × g for 10 min, and the pellet was washed 3 times with 40 ml of lysis buffer. This fraction was designated the cell wall/plasma membrane (CW/PM) fraction because of the tight association of plasma membranes. The supernatant was centrifuged at 100,000 × g for 1 h. The supernatant from this high-speed centrifugation was recovered and designated the cytoplasmic fraction. The pellet was washed and centrifuged again at 100,000 × g for 1 h and resuspended in 500 μl of lysis buffer and was designated the microsomal fraction, which contains all cell and organellar endomembranes. All protein fractions were quantified by using Coomassie Plus protein assay reagent (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions and stored at −80°C until analysis.
(ii) One-dimensional (1D) gel electrophoresis.
An aliquot of 1 μg of each fraction was resuspended in Novex Tris-glycine SDS sample buffer and run on 10% to 20% Tris-glycine gels (Invitrogen, Carlsbad, CA). The proteins were then visualized by using SilverSNAP Stain Kit II (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions.
(iii) 2D gel electrophoresis and protein quantification.
Acetone-precipitated proteins were resuspended in rehydration buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 100 mM DTT, 0.2% Biolytes (pH 5 to 8), 0.01% bromophenol blue}. For 2D gel electrophoresis, 50 μg of proteins in a total of 185 μl of the rehydration buffer was applied onto 11-cm Bio-Rad ReadyStrip IPG strips (pH 5 to 8) for overnight rehydration. First-dimension isoelectric focusing was performed with a Bio-Rad Protean IEF system for a total focusing time of 75,000 Vh. After the first dimension, the strips were equilibrated in buffer I (6 M urea, 0.375 M Tris-HCl [pH 8.8], 2% SDS, 20% glycerol, 2% [wt/vol] DTT) for 15 min. The strips were further equilibrated with buffer II (6 M urea, 0.375 M Tris-HCl [pH 8.8], 2% SDS, 20% glycerol, 2.5% [wt/vol] iodoacetamide) for 15 min and directly applied onto 12.5% SDS-PAGE gels for second-dimensional electrophoresis. After fixing, they were visualized by staining the gel with SYPRO ruby (Bio-Rad) on a 9400 Typhoon variable-mode imager (GE Healthcare) using a green laser (532 nm) and a 610BP30 emission filter.
(iv) iTRAQ labeling.
Cell wall and associated proteins were homogenized by using probe sonication at 50% duty for 3 cycles of 15 s with a 60-s incubation on ice between cycles. The supernatant was cleared by centrifugation at 19,000 × g for 30 min, and the pH was adjusted to 8.0 with 1 M triethylammonium biocarbonate (TEAB) buffer. The iTRAQ labeling procedures were performed according to the manufacturer's instructions (Applied Biosystems, ABI, Foster City, CA). After reduction and alkylation, tryptic digestion was initiated with the addition of 10 μg of trypsin (Promega Corporation, Madison, WI) to each of the eight samples, and the samples were incubated at 37°C overnight. Peptides derived from the soluble (S) and cell wall/plasma membrane fractions were labeled with different iTRAQ tags: tag 113 was the 24-h no-CSF-S fraction, tag 114 was the 24-h 0.12-μg/ml CSF-S fraction, tag 115 was the 48-h no-CSF-S fraction, tag 116 was the 48-h 0.12-μg/ml CSF-S fraction, tag 117 was the 24-h no-CSF-CW/PM fraction, tag 118 was the 24-h 0.12-μg/ml CSF-CW/PM fraction, tag 119 was the 48-h no-CSF-CW/PM fraction, and tag 121 was the 48-h 0.12-μg/ml CSF-CW/PM fraction for the sensitive strain; the resistant strain was the same with the exception that tags 113 to 116 were the CW/PM fraction in the resistant strain and tags 117 to 121 were the S fraction. The labeled samples were then mixed together and fractionated via two-dimensional chromatography according to procedures described previously (19). The high-performance liquid chromatography (HPLC) eluent was mixed with matrix solution (7 mg/ml alpha-cyano-4-hydroxycinnamic acid in 60% acetonitrile (ACN), 5 mM ammonium monobasic phosphate, and the internal mass calibrants consisting of 50 fmol/μl each GluFib [Glu1-fibrinopeptide B human] and ACTH [adrenocorticotropic hormone fragments 18 to 39]) through a 30-nl mixing tee and spotted directly onto matrix-assisted laser desorption ionization (MALDI) plates. The peptides were analyzed on a 4800 Proteomics Analyzer MALDI-tandem time of flight (TOF-TOF) mass spectrometer (ABI) in a data-dependent fashion using job-wide interpretation. Mass spectrometry (MS) spectra (m/z 800 to 3,600) were acquired in positive-ion reflection mode with internal mass calibration. A maximum of up to 15 of the most intense ions (signal-to-noise [S/N] ratio of >50) per spot were selected for subsequent tandem MS (MS/MS) analysis in the 2.0-keV mode. Each spectrum was averaged over 2,000 laser shots.
(v) Protein database search and bioinformatics.
TS2Mascot (Matrix Science, Inc., Boston, MA) was used to generate a peak list as a Mascot generic file (MGF) from tandem MS using the following parameters: a mass range from 20 to 60 Da below the precursor and a S/N ratio of 10. The MGF was submitted for an automated search using a local Mascot server (version 2.2) against the Aspergillus fumigatus protein database (9,630 entries) curated from the SwissProt protein database (ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase, downloaded on 21 August 2009). The following parameters were used: iTRAQ 8plex (K), iTRAQ 8plex (N terminal) and methylthio (C) as fixed modifications; iTRAQ 8plex (Y) and oxidation (M) as variable modifications; trypsin as an enzyme with a maximum of one missed cleavage allowed; monoisotopic peptide tolerance of 50 ppm; and MS/MS tolerance of 0.3 Da. Scaffold (version Scaffold_2_06_01; Proteome Software, Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater-than-95% probability as specified by the Peptide Prophet algorithm (21). Protein identifications were accepted if they could be established at a greater-than-95% probability and contained at least 2 identified peptides with a false discovery rate of ≤0.1%. Protein probabilities were assigned by the Protein Prophet algorithm (28). Proteins that contained similar peptides and that could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Peptides were quantitated by using the centroided reporter ion peak intensity. Intrasample channels were normalized based on the median ratio for each channel across all proteins. Multiple isobaric tag samples were normalized by comparing the median protein ratios for the reference channel. Protein quantitative values were derived from only uniquely assigned peptides. The minimum quantitative value for each spectrum was calculated as the 5% of the highest peak. Protein quantitative ratios were calculated as the median of all peptide ratios. Standard deviations were calculated as the interquartile range around the median. Quantitative ratios were log2 normalized for final quantitative testing. For each identified protein, associated gene ontology terms were automatically fetched from ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/UNIPROT/gene_association.goa_uniprot.gz by Scaffold software and plotted with respect to enrichment.
Microarray analysis. (i) Isolation of total RNA from Aspergillus fumigatus.
Total RNA was generated from frozen mycelia generated as described above with the RNeasy maxikit according to the manufacturer's instructions (Qiagen, Inc., Valencia, CA). RNA was DNase treated by using Turbo DNase (Ambion, Austin, TX), followed by heat inactivation of the enzyme. DNA-free RNA was then cleaned up by using the Qiagen RNeasy minikit and measured for quantity and purity by using RNA Nano Chips and the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
(ii) Labeling, prehybridization, and hybridization of DNA slides.
A. fumigatus total RNA (2 μg) was labeled by using protocols outlined by The Institute for Genomic Research (TIGR) standard operating procedure (SOP) M007 (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). All slides were whole-genome A. fumigatus DNA, version 3 (J. Craig Venter Institute, Rockville, MD). SuperScript III (Invitrogen, Carlsbad, CA) was used instead of PowerScript RT in the labeling reactions, as PowerScript RT has been discontinued. The hybridization of the labeled probes was performed as per SOP M008. The coverslips used were thick LifterSlip coverslips (Erie Scientific Company, Portsmouth, NH) with a hybridization chamber with an increased depth (Corning, Lowell, MA).
(iii) Image acquisition and data analysis.
All slides were scanned by using an Axon Instruments model 4000B scanner (Molecular Devices, Sunnydale, CA), with each channel being scanned individually. All scans used a 10-μm resolution and were converted into a resolution of 16 bits/pixel. All scanned images were then analyzed by using GenePix Pro 6.1 software. After global normalization with GenePix Pro, significance analysis of microarrays (SAM) was performed on all data by using TM4 software (36). The remaining data were then filtered by taking the mean of all data points for that spot (3 replicates with dye swap), and any gene with an expression value of ≥2 was considered significant.
RESULTS
Analysis of the Aspergillus fumigatus proteome by 2D gel electrophoresis.
A classical 2D gel electrophoresis system was used to evaluate the Aspergillus proteome from a whole-cell lysate. When this method was employed, the protein spots were too numerous and too clustered to get adequate resolution and identification of individual spots. To overcome this problem, a cell fractionation approach was taken to separate the secreted (S), cytoplasmic (C), microsomal (M), and cell wall and plasma membrane (CW/PM) fractions (Fig. 1). When each subcellular fraction was run separately, the protein spots were resolved at a greater level, and the identification of proteins of interest was more efficient and accurate. Each gel had between 400 and 700 easily resolvable protein spots with distinct protein profiles for each of 4 subcellular fractions (Fig. 2). The secreted fraction contains only proteins appearing in the medium; the microsomal fraction consists of plasma membranes, the endoplasmic reticulum, and subcellular organelle membranes, e.g., mitochondria, the Golgi apparatus, and the vacuole; the cytoplasmic fraction contains only soluble cytoplasmic proteins, while there was overlap between subcellular compartments in various fractions such as the cell wall, which contained both cell wall-specific and tightly associated membrane proteins. A total of 133 proteins were determined by mass spectroscopy following gel excision. The fractionation approach was highly reproducible. A number of the proteins were identified in multiple gels, and several tandem spots of the same molecular weight represented a single protein modified posttranslationally. Although the 2D gel system was highly reproducible and validated, the fractionation approach proved to be too cumbersome for a comprehensive analysis of potential biomarkers, which may be present in a low abundance. For this reason, the gel-free system of iTRAQ was utilized to analyze the Aspergillus proteome in the presence and absence of caspofungin.
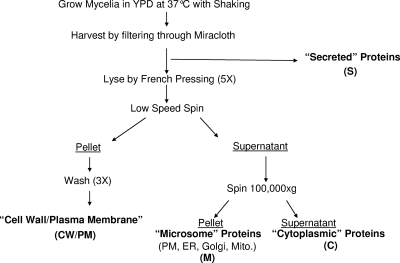
FIG. 1.
Subcellular fractionation strategy. Shown is the overall strategy used to disrupt cells and obtain different subcellular fractions for proteomic analysis. The filtrate was purified as the secreted (S) fraction. All cells were lysed by pressing through a French press at 20,000 lb/in2, and a low-speed spin separated microsomal (M) and cytoplasmic (C) fractions from the cell wall/plasma membrane (CW/PM) fraction. A high-speed spin then separated the final two fractions. PM, plasma membrane; ER, endoplasmic reticulum; Mito., mitochondria.
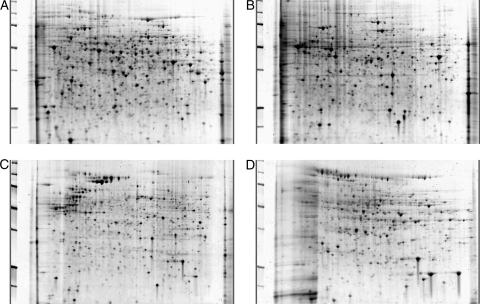
FIG. 2.
2D gels of cellular fractions. All subcellular fractions were subjected to 2-dimensional gel electrophoresis using strips of pI 5 to 8. (A) The cytoplasmic fraction tends to have the highest quantity of proteins of the four fractions. (B and C) The microsomal (B) and cell wall/plasma membrane (C) fractions may have some overlap due to incomplete separation. (D) Secreted proteins tend to be the fewest in number. All 4 fractions have individual patterns and proteins that are unique to that fraction alone as well as overlapping proteins that are consistent across fractions. All gels were stained with SYPRO ruby for visualization.
Proteomic analysis by iTRAQ.
The guiding principle of biomarker discovery used in this study is the identification of proteins that change in abundance in a susceptible strain but do not change in an isogenic resistant strain. To assess the specific behavior of caspofungin on the proteome of Aspergillus fumigatus, isogenic susceptible and echinocandin-resistant strains containing an S678P mutation in FKS1 (33) were grown in the presence of 0.12 μg/ml caspofungin, which is just below the MEC (0.19 μg/ml). Figure 3 illustrates that the colony size was reduced in the presence of drug and that cells displayed classical rosette-type structures. An fks1 mutant (fks1-S678P) (33) was phenotypically insensitive to drug and resembled the wild type in the absence of drug (Fig. 3). To expand the identification of proteins which may serve as potential biomarkers of drug action and to provide a more detailed evaluation of changes in protein levels in the presence of drug, an iTRAQ system was used to broadly identify proteins in the CW/PM and S fractions of isogenic susceptible and CSF-resistant strains of A. fumigatus at 24 and 48 h. These fractions were selected because they would be most likely to contribute to potential serum-based biomarkers.
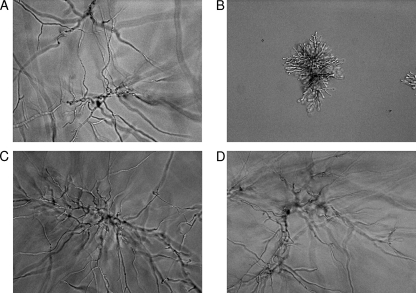
FIG. 3.
Cellular morphology. Distinct changes in morphology (40×) were observed as caspofungin was added to a final concentration of 0.12 μg/ml. (A and B) The susceptible strain KU80 ΔakuB showed fully formed hyphae with no drug (A) but was unable to develop fully when drug was present (B). (C and D) The resistant strain with a mutation in FKS1 did not show any changes in morphology in the absence (C) or presence (D) of drug.
In the susceptible strain a total of 471 proteins were identified from these fractions. However, using the strict criteria of 2 peptides with 95% confidence and a protein identification probability of 95%, 261 proteins were used for the final analysis (Table 1 and see Table S1 in the supplemental material). In the presence of drug (0.12 μg/ml caspofungin), abundance changes of 2-fold or greater were considered significant. A total of 56 proteins (26 up and 30 down) were significantly changed (>2-fold) in the CW/PM fraction at 24 h. A total of 81% (21/26) of the proteins that had increased levels in the CW component at 24 h were ribosomal proteins. The level of the chitinase ChiA1 was most decreased at 24 h in the cell wall of the susceptible stain, with a 12.1-fold change. The total number of proteins with significantly changing levels at 48 h was 9 (6 proteins reduced and 3 increased). The protein with the most significant change was a hypothetical protein (AFUA_8G07100), the level of which increased 5.28-fold at 24 h and >16-fold at 48 h. At 48 h the level of the stress response element Hsp98 was reduced 2.64-fold, and the level of Sba1 (an Hsp90 chaperone) was reduced 2.30-fold.
TABLE 1.
Potential biomarkers identified from iTRAQa
| Time (h) | Common name of target | Locus tag | Susceptible iTRAQ fold change | Fraction | Resistant iTRAQ fold change | Fold change by microarray |
|---|---|---|---|---|---|---|
| 24 | Mitochondrial hypoxia-responsive domain protein | AFUA_1G12250 | >−16 | Secreted | Too low | −4.5 |
| 24 | Class III chitinase ChiA1 | AFUA_5G03760 | −12.1 | CW/PM | NI | −11.7 |
| 24 | Mitochondrial hypoxia-responsive domain protein | AFUA_1G12250 | −6.1 | CW/PM | 1.15 | −4.5 |
| 24 | Plasma membrane H+-ATPase Pma1 | AFUA_3G07640 | −4.3 | CW/PM | −1.15 | −5.1 |
| 24 | 60S ribosomal protein L7 | AFUA_4G03880 | 2.0 | CW/PM | −1.07 | 3.8 |
| 24 | 60S ribosomal protein L23 | AFUA_5G05630 | 2.0 | CW/PM | NI | 4.5 |
| 24 | 60S ribosomal protein L22, putative | AFUA_3G12300 | 2.0 | CW/PM | 1.07 | 10.2 |
| 24 | 40S ribosomal protein S6 | AFUA_4G10800 | 2.0 | CW/PM | −1.07 | 3.7 |
| 24 | 40S ribosomal protein S9 | AFUA_3G06970 | 2.0 | CW/PM | −1.07 | 3.8 |
| 24 | 60S ribosomal protein P0 | AFUA_1G05080 | 2.1 | CW/PM | −1.23 | 4.8 |
| 24 | 60S ribosomal protein L11 | AFUA_4G07730 | 2.1 | CW/PM | NI | 8.3 |
| 24 | 60S ribosomal protein L44 | AFUA_2G08130 | 2.1 | CW/PM | 1.41 | 3.5 |
| 24 | 40S ribosomal protein S3Ae | AFUA_5G05450 | 2.3 | CW/PM | 1.07 | 4.9 |
| 24 | Ribosomal protein S5 | AFUA_7G01460 | 2.3 | CW/PM | No Change | 3.2 |
| 24 | Ribosomal L18ae protein family | AFUA_1G04530 | 2.3 | CW/PM | NI | 3.8 |
| 24 | 40S ribosomal protein S10b | AFUA_6G12660 | 2.3 | CW/PM | 1.07 | 3.8 |
| 24 | Ribosomal protein S13p/S18e | AFUA_6G13550 | 2.3 | CW/PM | −1.23 | 5.0 |
| 24 | 60S ribosomal protein L35Ae | AFUA_3G08460 | 2.3 | CW/PM | NI | 5.3 |
| 24 | Cytosolic large ribosomal subunit protein L7A | AFUA_6G12990 | 2.5 | CW/PM | 1.07 | 3.9 |
| 48 | BYS1 domain protein, putative | AFUA_5G01990 | −2.5 | CW/PM | 1.23 | 2.7 |
| 48 | Translation elongation factor eEF-3, putative | AFUA_7G05660 | 2.3 | CW/PM | 1.07 | −2.7 |
| 48 | GPI-anchored cell wall organization protein Ecm33 | AFUA_4G06820 | 2.3 | Secreted | 1.07 | −2.2 |
NI, not identified; too low, not enough signal to detect the reporter ion; GPI, glycosylphosphatidylinositol; CW/PM, cell wall/plasma membrane.
The secreted fraction of the susceptible strain showed a total of 14 proteins (8 proteins with decreased levels and 6 proteins with increased levels) that were altered at 24 h. The level of the major allergen and cytotoxin AspF1 was reduced >12-fold at 24 h (Fig. 4), along with the nuclear transport factor NTF-2, a PT repeat family protein, and cytochromes c and b5. Proteins with increased levels included two subunits of the ATP citrate lyase, Acl and subunit 1, both at more than 3-fold, as well as Hsp60, the allergen AspF4, and transaldolase. The secreted fraction at 48 h had a total of 43 proteins with a greater-than-2-fold change, including 32 proteins with decreased levels, while the remaining 11 proteins had increased levels. Seven of the 32 proteins with decreasing levels changed by more than 13-fold. These proteins include a putative 4-hydroxyphenylpyruvate dioxygenase and a hypothetical protein (AFUA_3G14940) at >16-fold as well as a nascent polypeptide-associated complex subunit and PT repeat family protein at 16-fold downregulated. The levels of two other proteins decreased 13.9-fold, which included the citrate synthase Cit1 and FKBP-type peptidyl-prolyl isomerase. The mitochondrial hypoxia response domain protein was the only protein with levels decreased by >16-fold at both 24 and 48 h in either fraction tested. Also of note is the fact that at 24 h, the level of AspF1 decreased more than 12-fold, while the level of AspF4 increased 2.3-fold.
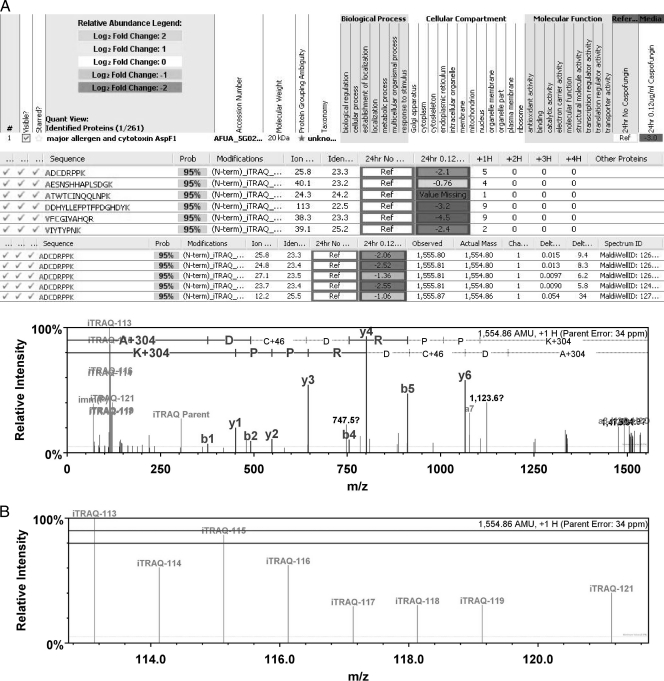
FIG. 4.
iTRAQ results for the major allergen and cytotoxin AspF1. (A) Typical Scaffold software result for iTRAQ showing the identification of proteins and accession numbers. (B) Peptides used to identify specific proteins and fold changes seen for each peptide. The number of times that a singly charged peptide was identified (+1H) is shown. Also, information from each peptide repeat and actual mass spectroscopy data are shown. Also shown is an enlargement of the reporter ion region depicting the level of each reporter ion used to generate the ratio of change for the protein. In this case, the changes seen are between ions 113 (no caspofungin) and 114 (0.12 μg/ml caspofungin) in the media at 24 h.
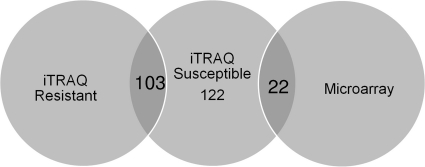
A parallel iTRAQ analysis was performed on an isogenic caspofungin-resistant strain (33) grown in the presence and absence of drug. Of the 261 proteins identified in the susceptible strain, 221 (84.7%) were also identified in the resistant strain (Fig. 5). Of the 122 proteins with changes of 2-fold or greater in the susceptible strain, 103 of these proteins were largely unchanged in abundance in the resistant strain, confirming them as potential biomarkers (Table 1). The most prominent potential biomarkers identified as changing 12-fold or more include the major allergen and cytotoxin AspF1, a PT repeat family protein, a subunit of the nascent polypeptide-associated complex, the citrate synthase Cit1, and FKBP-type peptidyl-prolyl isomerase along with the mitochondrial hypoxia response domain protein, 4-hydroxyphenylpyruvate dioxygenase, and one hypothetical protein. All of the proteins listed had decreased levels upon the addition of caspofungin. One hypothetical protein had an increase of greater than 16-fold upon the addition of drug but was not identified in the resistant strain and therefore does not strictly meet our definition of a potential biomarker. These proteins as well as several others warrant further study.
FIG. 5.
Venn diagram of the 122 proteins identified by iTRAQ as changing in the sensitive strain following drug exposure and 103 proteins identified in the resistant strain as being unchanged. A total of 22 genes encoding the proteins identified by iTRAQ were found to change significantly in the microarray analysis.
Microarray analysis.
To assess the relationship between the proteomic changes observed and global gene expression, a whole-genome array was evaluated. After 3 independent experiments with a dye flip for a total of 6 replicates, a total of 117 genes were considered to be significantly (>2-fold) differentially expressed in response to drug in the susceptible strain. Of these genes at 24 h, 18 were downregulated and 59 were upregulated (Table 1 and see Table S1 in the supplemental material). Of the 59 upregulated genes, 51 (86.4%) were ribosomal genes. This is consistent with the proteomics of the susceptible CW/PM fraction, which gave 81% ribosomal proteins. Of the remaining 8, 5 are hypothetical proteins. The 18 downregulated proteins include stress response genes such as Mn-superoxide dismutase and mitochondrial hypoxia response domain protein (consistent with iTRAQ) along with several transporter proteins (e.g., plasma membrane H+-ATPase and siderochrome-iron transporter). Three hypothetical proteins were also downregulated at 24 h. At 48 h, 33 genes were downregulated and 7 were upregulated at least 2-fold. None of the genes changing at 48 h were identified as being ribosomal. Of the 7 genes upregulated, 4 were hypothetical. The downregulated proteins include 7 hypothetical proteins along with 6 proteins that were also downregulated at 24 h (CFEM domain protein, carbonic anhydrase family protein, and 2 C2H2 transcription factors). The iTRAQ system identified a total of 471 proteins, 261 of which were used for further analysis in the secreted and cell wall and associated fractions. This compares to the 123 genes considered to be significant by microarray analysis.
A total of 22 proteins (22/122) with significant changes identified by iTRAQ in the susceptible strain also showed significant gene expression level changes by microarray analysis (Table 1 and see Table S1 in the supplemental material). At 24 h, all 19 differentially expressed gene products were consistent between the 2 methods. However, at 48 h, 3 proteins identified by iTRAQ had microarray changes of 2-fold in the opposite direction (either increasing or decreasing levels). This suggests that the gene/protein correlation breaks down significantly at 48 h compared with 24 h. The ChiA1 gene was downregulated 11.7-fold by microarray analysis, and the protein level decreased by 12.1-fold by iTRAQ analysis at 24 h. This pattern of decreased expression levels was also seen for the mitochondrial hypoxia response domain protein (>−16-fold by iTRAQ and −4.5-fold by microarray analysis) as well as the plasma membrane H+-ATPase (−4.3-fold by iTRAQ and −5.1-fold by microarray analysis). The 48-h time point showed consistent results between iTRAQ and microarray in that no ribosomal genes were shown to be significantly changing by microarray, and only 1 ribosomal protein was significantly upregulated by iTRAQ. These data suggest that ribosomal gene upregulation may occur as a burst at 24 h.
DISCUSSION
The Aspergillus proteome is large and complex, and classical 2D gel electrophoresis provides a convenient approach to assess global protein profiles and to identify prominently expressed proteins following gel excision and mass spectroscopy. However, the technique suffers from data compression when analyzing a large genome, like Aspergillus, with >10,000 open reading frames. This leads to difficulty in identifying potential biomarkers of drug action. By using a cell fractionation approach that emphasizes key cellular compartments and the comprehensive iTRAQ technique for protein analysis, these biomarkers can be more easily identified. In this report, we have used this approach to discover potential biomarkers that reflect the effect of the echinocandin drug caspofungin on A. fumigatus.
To date, most studies of drug action on Aspergillus have emphasized genomic approaches to infer changes in protein profiles (10, 14). Recently, the response of A. fumigatus to amphotericin B was looked at by proteomics via 2D gel electrophoresis and microarray analysis. That study found a total of 295 genes and 85 proteins that were differentially expressed. Of the 85 proteins, 48 were positively identified by mass spectrometry (15). In this study of caspofungin action, iTRAQ was used to positively identify 461 proteins from the S and CW/PM fractions. A number of the proteins were present in high abundance, such as AspF3 and the key glycolytic enzyme enolase, which were readily observed by 2D gel electrophoresis. These data indicate that the effect of caspofungin was not as a generalized metabolic poison to the cell, as would be expected from a fungicidal agent. The level of AspF3 was also found to increase 3.5-fold in the secreted fraction of the iTRAQ of the susceptible strain and to decrease 1.5-fold in the iTRAQ of the resistant strain at 48 h (Table 1). This allergen has been identified as a thioredoxin peroxidase and has been shown to have increased levels upon the addition of hydrogen peroxide (25). Enolase (AspF22) is also known to be an allergen and can stimulate a strong gamma interferon (IFN-γ) immune response in humans, and its homolog in Candida albicans was shown to provide partial protection as a vaccine candidate (7, 11).
Of the 261 proteins identified with a high level of confidence (see Table S1 in the supplemental material) in the cell wall/plasma membrane and secreted fractions of the susceptible iTRAQ, the proteins with the most value would be those with levels that are either highly increased or decreased following drug exposure and did not change in a resistant strain. Proteins from the CW/PM and S compartments were analyzed because they were most likely to interact with the host and could be identified as serum markers either directly or immunologically. iTRAQ was used to assess the total number of proteins differentially expressed in the presence of drug. These proteins were then assessed in the resistant strain to test if it was a general metabolic change or a change specific to caspofungin exposure. One of the most promising biomarkers is the mitochondrial hypoxia response domain protein (AFUA_1G12250), which was downregulated >16-fold at both 24 and 48 h in the secreted fraction. This is the only protein identified to have such a robust response. This protein was not seen when A. fumigatus was exposed to either voriconazole or amphotericin B, indicating its potential value as a biomarker specific to caspofungin (10, 15).
To examine the relationship between changes in the proteome and the genome under identical drug exposure conditions, a microarray analysis was conducted. Of the 117 genes identified as significantly changing, 59 (50.4%) correlated with the proteomics of the susceptible strain (Fig. 5). Most of these genes changing were ribosomal genes at 24 h. The microarray data showed an especially poor correlation (<35%) with corresponding proteins at 48 h. This may reflect a high level of transcript turnover as cells enter a type of stationary-phase growth. A total of 22 gene products were found to change in common in both the proteomic and genomic analyses (Table 1). These include the mitochondrial hypoxia response domain protein in both the cell wall and secreted fractions described above along with the chitinase ChiA1 and the plasma membrane H+-ATPase (Pma1), the levels of which were all decreasing. Previously, the Pma1 gene was shown to be upregulated 2-fold in response to amphotericin B, but our study showed a decrease of 4.3-fold by proteomic and 5.1-fold by microarray analyses in response to caspofungin (15). The changes in ChiA1 levels are consistent with studies of Candida, which responded to the inhibition of glucan synthase by cell wall remodeling involving changes in β-1-3-glucan and chitin as major cell wall components in response to caspofungin (39, 41).
One trend that emerged from the proteomics and genomics data is a strong increase in levels of ribosomal proteins at 24 h following drug exposure of the susceptible strain. A total of 58 ribosomal proteins were identified as changing significantly by either proteomics or genomics, suggesting that the cell is gearing up for reprogramming. This trend is clearly time sensitive, as only one ribosomal protein was significantly changing at 48 h. This trend was not as robust in the resistant strain, with 4/19 proteins (21%) identified as being ribosomal. Changes in levels of ribosomal proteins were seen previously in response to amphotericin B, but the levels of some were decreased, and all were seen only by microarray analysis in that study (15). No ribosomal proteins in this study were shown to be decreasing. The proteomics data showed several hypothetical proteins that have a significant change upon the addition of drug, but very little information can be garnered due to the lack of annotation in the database. These hypothetical proteins could be of great interest as biomarkers and clearly warrant further study.
Overall, the proteomic changes seen in response to caspofungin by both conventional 2D gel electrophoresis and iTRAQ provide insights into potential biomarkers of drug efficacy.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI069397 to D.S.P. and NS046593 to H.L.
We acknowledge Steven Park and Guillermo Garcia-Effron for their helpful discussions and suggestions and Yanan Zhao and Cristina Jimenez-Ortigosa for critical reading of the manuscript.
Footnotes
Published ahead of print on 25 October 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Antachopoulos, C., J. Meletiadis, T. Sein, E. Roilides, and T. J. Walsh. 2008. Comparative in vitro pharmacodynamics of caspofungin, micafungin, and anidulafungin against germinated and nongerminated Aspergillus conidia. Antimicrob. Agents Chemother. 52:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arendrup, M. C., G. Garcia-Effron, W. Buzina, K. L. Mortensen, N. Reiter, C. Lundin, H. E. Jensen, C. Lass-Florl, D. S. Perlin, and B. Bruun. 2009. Breakthrough Aspergillus fumigatus and Candida albicans double infection during caspofungin treatment: laboratory characteristics and implication for susceptibility testing. Antimicrob. Agents Chemother. 53:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asif, A. R., M. Oellerich, V. W. Amstrong, B. Riemenschneider, M. Monod, and U. Reichard. 2006. Proteome of conidial surface associated proteins of Aspergillus fumigatus reflecting potential vaccine candidates and allergens. J. Proteome Res. 5:954-962. [DOI] [PubMed] [Google Scholar]
- 4.Beranova-Giorgianni, S. 2003. Proteome analysis by two-dimensional gel electrophoresis and mass spectrometry: strengths and limitations. Trends Analyt. Chem. 22:273-281. [Google Scholar]
- 5.Brajtburg, J., and J. Bolard. 1996. Carrier effects on biological activity of amphotericin B. Clin. Microbiol. Rev. 9:512-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappelletty, D., and K. Eiselstein-McKitrick. 2007. The echinocandins. Pharmacotherapy 27:369-388. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhary, N., J. F. Staab, and K. A. Marr. 2010. Healthy human T-cell responses to Aspergillus fumigatus antigens. PLoS One 5:e9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, approved standard, 2nd ed., M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.da Silva Ferreira, M. E., M. R. Kress, M. Savoldi, M. H. Goldman, A. Hartl, T. Heinekamp, A. A. Brakhage, and G. H. Goldman. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva Ferreira, M. E., I. Malavazi, M. Savoldi, A. A. Brakhage, M. H. Goldman, H. S. Kim, W. C. Nierman, and G. H. Goldman. 2006. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50:32-44. [DOI] [PubMed] [Google Scholar]
- 11.Denikus, N., F. Orfaniotou, G. Wulf, P. F. Lehmann, M. Monod, and U. Reichard. 2005. Fungal antigens expressed during invasive aspergillosis. Infect. Immun. 73:4704-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 13.Diekema, D. J., S. A. Messer, R. J. Hollis, R. N. Jones, and M. A. Pfaller. 2003. Activities of caspofungin, itraconazole, posaconazole, ravuconazole, voriconazole, and amphotericin B against 448 recent clinical isolates of filamentous fungi. J. Clin. Microbiol. 41:3623-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner, R. E., P. Souteropoulos, S. Park, and D. S. Perlin. 2005. Characterization of Aspergillus fumigatus mutants with reduced susceptibility to caspofungin. Med. Mycol. 43(Suppl. 1):S299-S305. [DOI] [PubMed] [Google Scholar]
- 15.Gautam, P., J. Shankar, T. Madan, R. Sirdeshmukh, C. S. Sundaram, W. N. Gade, S. F. Basir, and P. U. Sarma. 2008. Proteomic and transcriptomic analysis of Aspergillus fumigatus on exposure to amphotericin B. Antimicrob. Agents Chemother. 52:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautam, P., C. S. Sundaram, T. Madan, W. N. Gade, A. Shah, R. Sirdeshmukh, and P. U. Sarma. 2007. Identification of novel allergens of Aspergillus fumigatus using immunoproteomics approach. Clin. Exp. Allergy 37:1239-1249. [DOI] [PubMed] [Google Scholar]
- 17.Herbrecht, R., D. W. Denning, T. F. Patterson, J. E. Bennett, R. E. Greene, J. W. Oestmann, W. V. Kern, K. A. Marr, P. Ribaud, O. Lortholary, R. Sylvester, R. H. Rubin, J. R. Wingard, P. Stark, C. Durand, D. Caillot, E. Thiel, P. H. Chandrasekar, M. R. Hodges, H. T. Schlamm, P. F. Troke, and B. de Pauw. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 347:408-415. [DOI] [PubMed] [Google Scholar]
- 18.Howard, S. J., D. Cerar, M. J. Anderson, A. Albarrag, M. C. Fisher, A. C. Pasqualotto, M. Laverdiere, M. C. Arendrup, D. S. Perlin, and D. W. Denning. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain, M. R., S. Bian, T. Liu, J. Hu, S. Elkabes, and H. Li. 2009. Altered proteolytic events in experimental autoimmune encephalomyelitis discovered by iTRAQ shotgun proteomics analysis of spinal cord. Proteome Sci. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahn, J. N., M. J. Hsu, F. Racine, R. Giacobbe, and M. Motyl. 2006. Caspofungin susceptibility in Aspergillus and non-Aspergillus molds: inhibition of glucan synthase and reduction of beta-D-1,3 glucan levels in culture. Antimicrob. Agents Chemother. 50:2214-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller, A., A. I. Nesvizhskii, E. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383-5392. [DOI] [PubMed] [Google Scholar]
- 22.Kniemeyer, O., F. Lessing, and A. A. Brakhage. 2009. Proteome analysis for pathogenicity and new diagnostic markers for Aspergillus fumigatus. Med. Mycol. 47(Suppl. 1):S248-S254. [DOI] [PubMed] [Google Scholar]
- 23.Kniemeyer, O., F. Lessing, O. Scheibner, C. Hertweck, and A. A. Brakhage. 2006. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus. Curr. Genet. 49:178-189. [DOI] [PubMed] [Google Scholar]
- 24.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lessing, F., O. Kniemeyer, I. Wozniok, J. Loeffler, O. Kurzai, A. Haertl, and A. A. Brakhage. 2007. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot. Cell 6:2290-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Medrano, R., M. C. Ovejero, J. A. Calera, P. Puente, and F. Leal. 1995. Aspergillus fumigatus antigens. Microbiology 141(Pt. 10):2699-2704. [DOI] [PubMed] [Google Scholar]
- 27.Maertens, J., I. Raad, G. Petrikkos, M. Boogaerts, D. Selleslag, F. B. Petersen, C. A. Sable, N. A. Kartsonis, A. Ngai, A. Taylor, T. F. Patterson, D. W. Denning, and T. J. Walsh. 2004. Efficacy and safety of caspofungin for treatment of invasive aspergillosis in patients refractory to or intolerant of conventional antifungal therapy. Clin. Infect. Dis. 39:1563-1571. [DOI] [PubMed] [Google Scholar]
- 28.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 29.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 30.Perlin, D. S. 2009. Antifungal drug resistance: do molecular methods provide a way forward? Curr. Opin. Infect. Dis. 22:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaller, M. A., L. Boyken, R. J. Hollis, J. Kroeger, S. A. Messer, S. Tendolkar, and D. J. Diekema. 2009. In vitro susceptibility of clinical isolates of Aspergillus spp. to anidulafungin, caspofungin, and micafungin: a head-to-head comparison using the CLSI M38-A2 broth microdilution method. J. Clin. Microbiol. 47:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham, T. K., and P. C. Wright. 2008. The proteomic response of Saccharomyces cerevisiae in very high glucose conditions with amino acid supplementation. J. Proteome Res. 7:4766-4774. [DOI] [PubMed] [Google Scholar]
- 33.Rocha, E. M., G. Garcia-Effron, S. Park, and D. S. Perlin. 2007. A Ser678Pro substitution in Fks1p confers resistance to echinocandin drugs in Aspergillus fumigatus. Antimicrob. Agents Chemother. 51:4174-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross, P. L., Y. N. Huang, J. N. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, S. Purkayastha, P. Juhasz, S. Martin, M. Bartlet-Jones, F. He, A. Jacobson, and D. J. Pappin. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3:1154-1169. [DOI] [PubMed] [Google Scholar]
- 35.Ruppen, I., L. Grau, E. Orenes-Pinero, K. Ashman, M. Gil, F. Algaba, J. Bellmunt, and M. Sanchez-Carbayo. 2010. Differential protein expression profiling by iTRAQ-two-dimensional LC-MS/MS of human bladder cancer EJ138 cells transfected with the metastasis suppressor KiSS-1 gene. Mol. Cell. Proteomics 9:2276-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 37.Schrettl, M., H. S. Kim, M. Eisendle, C. Kragl, W. C. Nierman, T. Heinekamp, E. R. Werner, I. Jacobsen, P. Illmer, H. Yi, A. A. Brakhage, and H. Haas. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70:27-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seenarain, V., H. M. Viola, G. Ravenscroft, T. M. Casey, R. Lipscombe, E. Ingley, N. G. Laing, S. D. Bringans, and L. C. Hool. 2010. Evidence for altered guinea pig ventricular cardiomyocyte protein expression and growth in response to 5 minute in vitro exposure to H2O2. J. Proteome Res. 9:1985-1994. [DOI] [PubMed] [Google Scholar]
- 39.Stevens, D. A., M. Ichinomiya, Y. Koshi, and H. Horiuchi. 2006. Escape of Candida from caspofungin inhibition at concentrations above the MIC (paradoxical effect) accomplished by increased cell wall chitin; evidence for beta-1,6-glucan synthesis inhibition by caspofungin. Antimicrob. Agents Chemother. 50:3160-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugui, J. A., H. S. Kim, K. A. Zarember, Y. C. Chang, J. I. Gallin, W. C. Nierman, and K. J. Kwon-Chung. 2008. Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS One 3:e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taib, M., J. W. Pinney, D. R. Westhead, K. J. McDowall, and D. J. Adams. 2005. Differential expression and extent of fungal/plant and fungal/bacterial chitinases of Aspergillus fumigatus. Arch. Microbiol. 184:78-81. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, R. D., A. Saparno, B. Blackwell, V. Anoop, S. Gleddie, N. A. Tinker, and L. J. Harris. 2008. Proteomic analyses of Fusarium graminearum grown under mycotoxin-inducing conditions. Proteomics 8:2256-2265. [DOI] [PubMed] [Google Scholar]
- 43.Vodisch, M., D. Albrecht, F. Lessing, A. D. Schmidt, R. Winkler, R. Guthke, A. A. Brakhage, and O. Kniemeyer. 2009. Two-dimensional proteome reference maps for the human pathogenic filamentous fungus Aspergillus fumigatus. Proteomics 9:1407-1415. [DOI] [PubMed] [Google Scholar]
- 44.Zieske, L. R. 2006. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57:1501-1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.