Abstract
Paromomycin, an aminoglycoside antibiotic having low mammalian cell toxicity, is one of the drugs currently used in the chemotherapy of cutaneous and visceral leishmaniasis. In order to understand the mode of action of this antibiotic at the molecular level, we have investigated the effects of paromomycin on protein synthesis in Leishmania and its mammalian hosts. We were able to demonstrate that in vivo protein synthesis in the promastigote stage of the parasite and its proliferation rate are markedly inhibited by paromomycin while being only slightly affected by other aminoglycoside antibiotics, such as streptomycin and neomycin B. Furthermore, both in vitro polypeptide synthesis induced by poly(U) as mRNA and accuracy of translation are significantly decreased by paromomycin in cell-free systems containing ribosomal particles of Leishmania promastigotes. Conversely, when ribosomes from mammalian cells are used instead of the protozoan particles, polyphenylalanine synthesis is only barely reduced by the antibiotic and the translation misreading remains almost unaltered. Surface plasmon resonance analysis of the interaction between paromomycin and protozoan or mammalian cell ribosomal RNAs shows a strong binding of antibiotic to the parasite ribosomal decoding site and practically no interaction with the mammalian cell counterpart. Our results indicating differential effects of paromomycin on the translation processes of the Leishmania parasite and its mammalian hosts can explain the therapeutic efficiency of this antibiotic as an antileishmaniasis agent.
Leishmaniasis, a parasitic disease caused by protozoa of the genus Leishmania, affects millions of people in different regions of the world, mainly in tropical and subtropical areas. The disease may reach an appreciable mortality rate and may adopt several forms in human patients, ranging from cutaneous and mucocutaneous lesions to a severe visceral form affecting various organs (7, 10, 20).
Leishmania has a digenetic life cycle involving an insect vector and a mammalian host. The parasite undergoes major morphological and physiological transformations during the different stages of its life cycle. Promastigotes are elongated and flagellated forms proliferating inside the sandfly, whereas amastigotes are rounded, flagellumless forms that multiply inside the mammalian host cells (20).
Over the years, a good number of drugs have been used in the clinical treatment of leishmaniasis. In general, these chemotherapies have usually been selected on empirical bases and not through an extensive knowledge of Leishmania metabolic selective features (20, 26). Up to the present time, several compounds have shown rather satisfactory results; however, most of these drugs have some limitations, such as high levels of toxicity in humans or the development of resistance mechanisms in parasites (27). Pentavalent antimonial compounds, pentamidine and amphotericin B, are very well known among the chemicals currently used in leishmaniasis therapy (20). Although their corresponding modes of action have not been completely elucidated yet, some possible mechanisms have been suggested. Pentavalent antimony undergoes an in vivo reduction into a trivalent form which is toxic to amastigotes and promastigotes. Pentamidine, which is able to substitute polyamines at their sites of binding to nucleic acids, might produce the inhibition of DNA replication and transcription (3), whereas amphotericin B is selectively toxic to parasites, probably due to its affinity to ergosterol, causing protozoan membrane damage (5).
Other chemicals have more recently been introduced in the therapy of leishmaniasis. Allopurinol and various purine analogs are effective because they inhibit the enzyme adenylosuccinate synthetase, which mediates the conversion of inosinic acid into AMP (19). Several natural products, such as quinones and terpene derivatives, have also shown antileishmanial activity (29, 30, 31, 35).
Recent studies on Leishmania biochemical pathways (26) have intended to detect metabolic processes occurring exclusively in the parasite and not in mammalian cells. In this way, it would be possible to find new strategies involving specific targets, such as trypanothione or glycosylphosphatidylinositol biosyntheses, which might be appropriate for the design of selective chemotherapies (38).
Several aminoglycoside antibiotics mainly used in bacterial infections have been tried as antiparasitic agents. Among them, paromomycin (aminosidine) has shown antileishmanial activity when used alone or in combination with other drugs (7, 10, 17, 18, 20). Topical treatments with paromomycin-containing ointments or intramuscular injections of the same antibiotic have been successfully used for a long time against cutaneous or visceral leishmaniasis, respectively (7, 23, 32, 34). However, very little is known about the differential effects of the drug on parasites and mammalian host cells. These selective activities might be related to the antibiotic actions on bacterial ribosomes causing the inhibition of their dissociation into subunits with the concomitant decrease of protein synthesis (17) and/or the induction of mistranslation errors by the specific interaction of paromomycin with the small rRNA decoding site (11, 36).
A multistep model has been proposed to explain the bactericidal effect of aminoglycosides, which involves antibiotic uptake, mistranslation by chain-elongating ribosomes, membrane damage, and subsequent ribosomal blockade preventing further protein synthesis (9). This model might also be relevant to the paromomycin mode of action on Leishmania, because the parasite ribosomes are good targets for the antibiotic (17).
In the present article, we show that paromomycin causes a strong inhibition of polyphenylalanine synthesis directed by poly(U) in a cell-free system prepared from Leishmania mexicana promastigotes and that this effect is considerably smaller when rat liver ribosomes replace the parasite particles. Moreover, the misreading frequency of leucine incorporation with poly(U) as a messenger is 10-fold higher when Leishmania mexicana ribosomes are used instead of those from mammalian systems. These results could explain the selective action of paromomycin on in vivo protein synthesis and protozoan proliferation. Surface plasmon resonance (SPR) measurements of the specific antibiotic binding to RNA oligonucleotides corresponding to a segment of the small ribosomal subunit decoding sites indicate that paromomycin interaction occurs only with the oligonucleotide from Leishmania ribosomes and not with that from mammalian particles. These results strongly suggest a selective antiparasitic effect of paromomycin.
MATERIALS AND METHODS
Chemicals.
The antibiotics paromomycin, neomycin, streptomycin, and ampicillin and reagents ATP, GTP, l-amino acids, bases, creatine phosphate, creatine phosphokinase, poly(U) as K salt, wheat germ tRNA, hemin, and HEPES buffer were obtained from Sigma (St. Louis, MO). Minimal essential medium (SMEM) and vitamins were from Gibco/BRL (Gaithersburg, MD). Fetal calf serum was purchased from Natocor (Carlos Paz, Córdoba, Argentina), and radioactive amino acids l-[U-14C]leucine (318 mCi/mmol), l-[U-14C]phenylalanine (496 mCi/mmol), and l-[35S]methionine (1,175 Ci/mmol) were obtained from Perkin-Elmer (Boston, MA).
Parasite cultures and cell extract preparation and fractionation.
Crithidia fasciculata (ATCC 11745) and Leishmania mexicana (MHOM/BZ/82/BEL21) were cultivated at 26 to 28°C in the semidefined medium SDM-79 (6) supplemented with hemin (20 mg/liter), 10% heat-inactivated fetal calf serum, and antibiotics (100 μg/ml streptomycin and 100 U/ml penicillin). Parasite growth was followed by cell counting, and all cultures reaching the stationary phase were diluted with fresh medium to (5 to 10) × 106 cells/ml. Parasites at the exponential phase of growth were collected by centrifugation at 1,000 × g for 10 min. Cells were resuspended in a solution containing 20 mM HEPES-KOH buffer, pH 7.5, 2 mM Mg acetate, 80 mM KCl, and 1 mM dithiothreitol (DTT) at a concentration of 2 × 109 cells/ml and then broken by freezing at −80°C and thawing for 4 consecutive times. Cell extracts were centrifuged at 4°C (20 min at 30,000 × g), and the supernatant fluids (S30) were dialyzed for 3 h against 10 mM HEPES buffer, pH 7.5, 1 mM Mg acetate, 20 mM KCl, and 1 mM DTT. S30 fractions were directly used to measure polypeptide synthesis induced by poly(U) as mRNA or for further isolation of S150 supernatant fractions (after a new centrifugation at 150,000 × g for 3 h) and subsequent purification of parasite cytoplasmic ribosomal particles by sucrose gradient centrifugation similar to that described for rat liver ribosomes.
Rat liver extracts and purification of the ribosomal fraction.
About 20 g of rat liver tissue was cut into small pieces and disrupted by 10 strokes in a tight-fitting Teflon-glass homogenizer using 80 ml of 0.25 M sucrose containing 50 mM Tris-HCl buffer, pH 7.5, 5 mM Mg acetate, 250 mM KCl, 2 mM DTT, and 0.5 mg heparin/ml as a nuclease inhibitor. After treatment at 4°C for 15 min with α-amylase (10 U/ml) in the presence of 0.1 mM CaCl2 for glycogen degradation, the liver extract was centrifuged at 12,000 × g for 15 min. The postmitochondrial supernatant fluid was treated with 1% Triton X-100 and 0.5% Na deoxycholate (final concentrations), and the mixture was placed on top of a two-step sucrose gradient made of 2 ml 1.5 M and 4 ml 2 M sucrose, both in a buffer solution containing 50 mM Tris-HCl (pH 7.5), 5 mM Mg acetate, 50 mM KCl, 2 mM DTT, and 0.1 mg of heparin/ml. Centrifugation was performed at 150,000 × g for 16 h in a Beckman Ti60 rotor. The ribosomal pellet was carefully rinsed with a solution containing 10 mM Tris-HCl (pH 7.5), 10 mM KCl, and 1.5 mM Mg acetate and then resuspended in the same solution to a concentration of 500 A260 U/ml and stored in a freezer at −80°C. The rat liver S150 supernatant fraction was obtained from dialyzed S30 extracts as described for parasite fractions.
In vivo protein synthesis in Leishmania mexicana promastigotes.
The rate of total protein synthesis was followed in aliquots of an exponentially growing parasite culture in the absence and presence of aminoglycoside antibiotics. The drug to be tested was added, and after a 15-min period to allow the antibiotic uptake, radioactive methionine was added (10 μCi/ml) and the incubation was continued at 37°C for 4 h. Samples were taken at the indicated times, and after treatment for 15 min at 37°C with an equal volume of 1 M NaOH containing 1 mM unlabeled methionine and albumin (20 μg/ml) as a carrier, proteins were precipitated with trichloroacetic acid (10% final concentration). After filtration through Whatman GF/C filters, the samples were washed with 10% trichloroacetic acid (TCA)-95% ethanol and then dried and counted in a scintillation spectrometer.
In vitro polypeptide synthesis induced by poly(U) in parasite and rat liver systems.
The reaction was carried out in a total volume of 0.1 ml containing 30 mM HEPES-KOH (pH 7.5), 50 mM K acetate, 5 to 10 mM Mg acetate, 0.1 mM EDTA, 1 mM ATP, 0.2 mM GTP, 8 mM creatine phosphate, 20 μg creatine phosphokinase, 2 mM DTT, 40 μg of stripped wheat germ tRNA, 50 μg of poly(U), radioactive phenylalanine (0.1 μCi, 20 μM [final concentration]), and S30 supernatant fraction (40 to 60 μg of protein), or 0.3 to 0.5 A260 units of purified ribosomes plus S150 supernatant fraction (20 to 40 μg of protein). The reaction was carried out for 40 min at 37°C and subsequently treated as described above. The addition of paromomycin at different concentrations allowed us to study the effect of this antibiotic on polypeptide synthesis.
Mistranslation frequency induced by antibiotics in parasite and mammalian systems.
The reaction mixture was similar to that described for poly(U)-dependent polypeptide synthesis, containing an S150 fraction from Leishmania mexicana and purified ribosomes obtained from parasites or rat liver. Different aliquots from the same preparations were used for the assays of radioactive phenylalanine or leucine incorporation in the presence of unlabeled leucine or phenylalanine, respectively. Where indicated, 20 μM paromomycin was added to the reaction mixtures. Mistranslation was measured as the percentage of leucine incorporation compared to that of phenylalanine.
Oligoribonucleotides corresponding to a segment of rRNA decoding region.
Polyribonucleotides with biotin followed by the TEG spacer at the 5′ end were prepared by Integrated DNA Technologies, Inc. (Coralville,IA). The polymers L and M have sequences corresponding to helix 44 within the decoding region of the cytoplasmic small ribosomal RNAs from Leishmania spp. and Homo sapiens (mammalian systems), respectively (14). The polymer N has a similar size but an unrelated sequence in the region relevant for antibiotic binding. The RNA oligonucleotide sequences are the following: L, 5′-/5BioTEG/rCrArCrCrGrCrCrCrGrUrCrGrUrUrGrUrUrUrCrCrArArGrUrCrGrGrCrGrArGrGrArArGrCrArArArArGrUrCrGrUrArArCrArArGrGrUrA-3′; M, 5′-/5BioTEG/rCrArCrCrGrCrCrCrGrUrCrGrCrUrArCrUrArCrCrArArGr. UrCrGrGrCrGrArGrGrArArGrUrArArArArGrUrCrGrUrArArCrArArGrGrUrA-3′; N, 5′-/5BioTEG/rCrArCrCrGrCrCrCrGrUrCrGrCrUrArCrUrArCrCrArArGrUrCrGrArCrUrUrGrGrUrArGrUrArGrCrGrArCrGrGrGrCrGrGrUrG-3′.
Immobilization of biotinylated RNAs on the sensor chips.
Streptavidin sensor chips were prepared from carboxymethyl dextran sensor chips (CM5) by amino coupling immobilization according to the manufacturer's directions. Briefly, streptavidin solution (1 mg/ml) was dialyzed against buffer acetate (10 mM; pH 4.5) and then injected over the four flow cells of the sensor chips prior to activation with N-hydroxysuccinimide (NHS) and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC). The free active sites after streptavidin binding were blocked with ethanolamine. An average of 5,000 resonance units (RU) of RNA oligomer was immobilized in each flow cell. Using a manual injection, three pulses of biotinylated RNA from Leishmania, a negative control RNA, or a human rRNA was injected over the flow cells. All the ligands were diluted at a final concentration of 5 pmol in 80 μl of buffer (10 mM HEPES, 0.1 mM EDTA, 100 mM NaCl, pH 6.8) as described by Hendrix et al. (13).
Surface plasmon resonance studies.
The interactions of immobilized RNA with antibiotics were measured by SPR using a Biacore T100 instrument (GE Healthcare, Uppsala, Sweden) that allows determination of real-time interactions between two molecules (16).
Micromolar concentrations of the aminoglycoside antibiotics paromomycin and neomycin were prepared by 2-fold dilution with HEPES-buffered saline (HBS) containing 150 mM NaCl and 0.005% surfactant P-20 (BIAcore) (pH 7.2). All binding experiments were performed at 25°C. Dissociation was carried out in HBS. Pulses of 300 mM Na2SO4 were used to regenerate the surface. SPR data were analyzed using Biacore T100 evaluation software. All the experiments were repeated at least three times, and residuals were less than 5%. Dissociation constants (KD) were determined under equilibrium binding conditions fitting a 1:1 binding nonlinear model after correction for nonspecific interaction, in which the aminoglycoside was passed over the unspecific RNA (N) used as a negative control.
RESULTS
Effects of paromomycin on Leishmania growth.
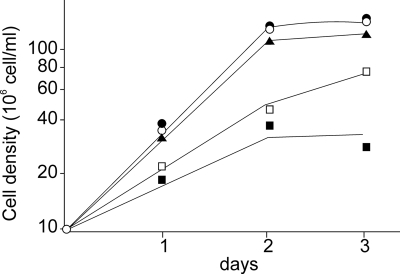
In order to investigate the selective effects of paromomycin on parasites of Leishmania spp., we followed the proliferation of Leishmania mexicana in SDM-79 medium. Cultures were performed in the absence and presence of different concentrations of the drug. We also used control cultures with the addition of other aminoglycoside antibiotics, such as streptomycin or neomycin B. Paromomycin inhibited Leishmania growth in a dose-dependent manner, as shown in Fig. 1, while streptomycin and neomycin B elicited a very small effect or no effect at all on parasite proliferation. The paromomycin concentration causing 50% of Leishmania growth inhibition (IC50) was about 200 μM, a rather high value in comparison with previously reported data (15). This discrepancy might be due to the fact that Leishmania promastigotes with lower paromomycin uptake and sensitivity than amastigote forms were used in our studies (15).
FIG. 1.
Inhibition of Leishmania mexicana growth by aminoglycoside antibiotics. Parasites were cultivated in SDM-79 medium in the absence of antibiotics (○) or with the addition of 0.4 mM streptomycin (•), 0.4 mM neomycin B (▴), or 0.2 mM (□) or 0.4 mM (▪) paromomycin. Parasite proliferation was followed for 72 h.
Effects of paromomycin on Leishmania protein synthesis in vivo.
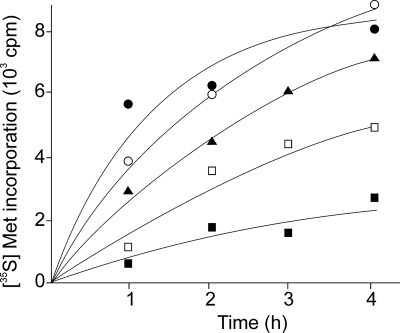
Total protein synthesis was measured in intact parasites labeled with radioactive methionine as described in Materials and Methods. Leishmania mexicana promastigotes were cultivated in the absence and presence of various paromomycin levels. In addition, different aliquots of the same parasite culture were incubated with streptomycin or neomycin B. The results shown in Fig. 2 agree with those observed for parasite proliferation. In vivo protein synthesis was markedly decreased by paromomycin but almost unaffected or slightly inhibited by streptomycin or neomycin B. The presence of streptomycin in the growth medium did not modify the inhibitory effects of paromomycin on Leishmania proliferation and protein synthesis.
FIG. 2.
Effect of aminoglycoside antibiotics on Leishmania mexicana protein synthesis in vivo. Parasites at the exponential phase of growth in the absence or presence of antibiotics were labeled with radioactive methionine (Met) and treated as indicated in Materials and Methods. Cultures without antibiotics (○) or in the presence of 0.4 mM streptomycin (•), 0.4 mM neomycin B (▴), or 0.2 mM (□) or 0.4 mM (▪) paromomycin were used.
It was ruled out that paromomycin could alter methionine uptake by Leishmania promastigotes because this amino acid transport gave very similar values in the absence and presence of the antibiotic (3.5 ± 0.6 and 3.9 ± 0.8 pmol/min/107 parasites, respectively). Therefore, the results shown in Fig. 2 are due to antibiotic inhibition of protein synthesis and not to effects on amino acid transport.
Paromomycin inhibition of polyphenylalanine synthesis in cell extracts or purified fractions from protozoa and rat liver.
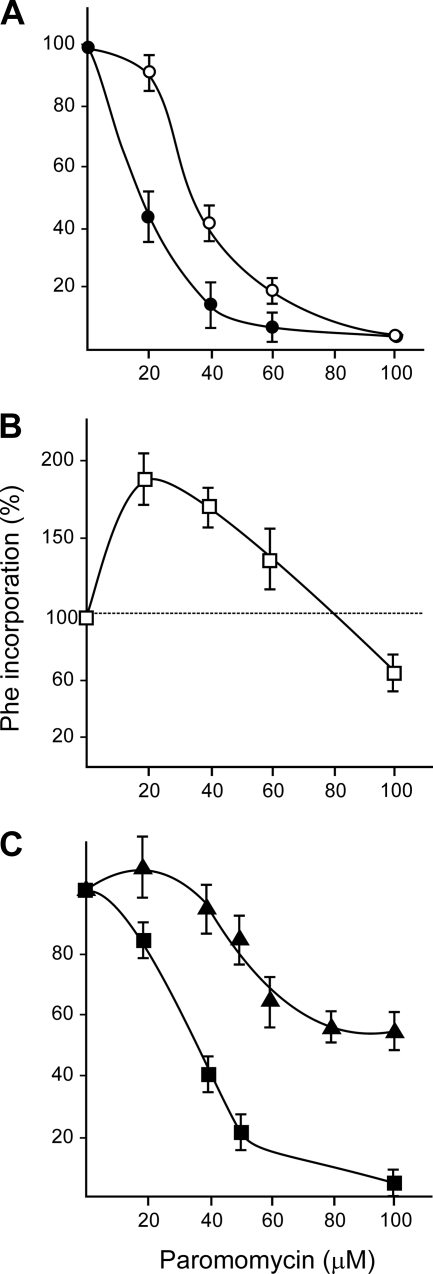
Cell extracts from different parasites and rat liver are able to efficiently translate synthetic polynucleotides. We studied the in vitro polypeptide synthesis directed by poly(U) as a messenger in systems prepared from Leishmania mexicana, Crithidia fasciculata, and rat liver. In the absence of mRNA, the incorporation of amino acids into polypeptides was negligible in all these systems. Upon the addition of the synthetic polynucleotide poly(U), the incorporation of phenylalanine (codons UUU and UUC) was stimulated between 50 and 100 times (1). Preliminary experiments have indicated that under our experimental conditions, the optimal Mg2+ concentration required for maximal phenylalanine incorporation into polypeptides was 7 mM for all the systems investigated (results not shown). The effects of paromomycin concentrations on polypeptide synthesis directed by poly(U) in cell-free systems (S30 fractions) from parasites and rat liver are shown in Fig. 3A and B, respectively. The antibiotic strongly inhibited polyphenylalanine formation in mixtures derived from Crithidia fasciculata and Leishmania mexicana, with a 50% inhibition at 16 and 34 μM, respectively (Fig. 3A). These lower IC50s are due to the fact that they correspond to an in vitro experiment where there is direct access of the drug to ribosomal particles; IC50s corresponding to in vivo assays (Fig. 1 and 2) are markedly higher. Unexpectedly, at low concentrations, the aminoglycoside presence enhances polypeptide synthesis in a reaction mixture containing an S30 supernatant fraction from rat liver, as can be seen in Fig. 3B. Since this remarkably different effect on rat liver systems might be due to specific features of the ribosomal particles or to properties of the enzymatic activities present in the supernatant fraction, we compared paromomycin effects on polypeptide synthesis in reaction mixtures differing only in the origin of ribosomes. The results shown in Fig. 3C indicate that while paromomycin elicited strong inhibition of polyphenylalanine synthesis performed by parasite ribosomes, as shown before, the antibiotic provoked only a slight decrease in the same reaction when mammalian particles were used. We obtained similar results using a different protocol to prepare parasite and liver fractions. This method did not include detergents to release ribosomes from membranes, so microsomes were discarded, and the resulting preparations correspond to purified free ribosomes.
FIG. 3.
Effect of paromomycin on polyphenylalanine synthesis in cell-free systems. (A and B) S30 cell fractions were obtained from the Crithidia fasciculata (•) or Leishmania mexicana (○) trypanosomatid parasites (A) or rat liver (B). (C) Polypeptide synthesis in reaction mixtures containing an S150 supernatant fraction from Leishmania mexicana and purified ribosomes prepared from rat liver (▴) or Leishmania (▪) extracts in the presence of different antibiotic concentrations. Samples were incubated and treated as described in Materials and Methods. All values correspond to the average ± SD of three determinations.
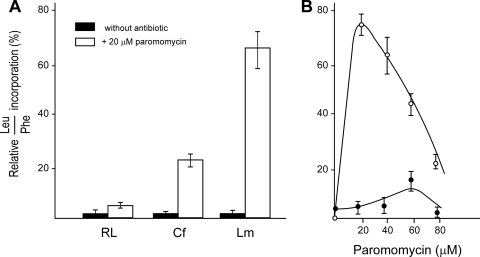
Effects of paromomycin on mistranslation in cell-free systems containing protozoan or mammalian ribosomes.
It is well known that aminoglycoside antibiotics can induce misreading of mRNA, thus decreasing the fidelity of the translation process (4, 28, 33). In the absence of antibiotics, the poly(U)-induced leucine misincorporation was very small because mRNA does not contain leucine codons (CUN and UUpurine). However, the addition of paromomycin elicited a very significant increase in errors when translation reactions were carried out in parasite extracts. We determined the frequency of mistranslation by calculating the ratio of leucine to phenylalanine incorporation into polypeptides induced by poly(U). We used cell-free systems containing purified ribosomes from Crithidia, Leishmania, or rat liver and the same S150 supernatant fraction prepared from Leishmania mexicana. The reactions were carried out in the absence or presence of various paromomycin concentrations. Different aliquots from each preparation were used to measure leucine and phenylalanine incorporation, as detailed in Materials and Methods. Figure 4A shows the misreading frequency in reaction mixtures with or without the addition of 20 μM paromomycin. The presence of the antibiotic markedly increased the misreading in mixtures containing ribosomal particles from protozoa compared to results in those assays with mammalian ribosomes. This effect was remarkably higher with ribosomes from Leishmania mexicana. The misreading frequencies with Leishmania and rat liver ribosomes were also measured at different paromomycin levels (Fig. 4B). Mistranslation was substantially higher when protozoan particles were used (15-fold at 20 μM paromomycin to 7-fold at 80 μM antibiotic). The marked misreading decrease at higher paromomycin concentrations agrees with the multistep model of aminoglycosides action on polypeptide synthesis (9). Thus, when ribosomes are strongly blocked by high levels of the drug, it could be expected that the residual protein synthesis might produce short polypeptides with lower misreading probability.
FIG. 4.
Effect of paromomycin on translation misreading. (A) Reaction mixtures containing an S150 supernatant fraction from Leishmania mexicana and purified ribosomal particles from trypanosomatid parasites (Crithidia fasciculata or Leishmania mexicana) or rat liver (mammalian cells system) were assayed in the absence and presence of 20 μM paromomycin. RL, Cf, and Lm, rat liver, Crithidia fasciculata, and Leishmania mexicana ribosomes, respectively. (B) The reaction mixtures contained an S150 fraction from Leishmania mexicana and purified ribosomes from Leishmania (○) or rat liver (•). Assays were carried out at different paromomycin concentrations. Misreading frequencies shown are the average values ± SD for three experiments.
Interaction between aminoglycoside antibiotics and protozoan or mammalian rRNA decoding site.
The described results related to the induction of translational misreading seem to indicate a strong selective interaction between aminoglycoside antibiotics and Leishmania cytoplasmic ribosomes. For a long time, it has been known that these drugs bind specifically to a region of the small subunit rRNA decoding A site (11, 12, 24). This rRNA segment is involved in the codon-anticodon recognition occurring during the translation mechanism. The structure of RNA-paromomycin complexes has been determined by chemical footprinting techniques and more recently by nuclear magnetic resonance spectroscopy and X-ray crystallography (11, 21, 36). Based on previous results with Escherichia coli ribosomes, we chose model oligomers for our studies on paromomycin affinities to rRNA from Leishmania mexicana (oligomer L) and mammalian cells (oligomer M). The oligoribonucleotides used correspond to the eukaryotic counterparts of the antibiotic binding site in these organisms (14). We included in our experiments a similar oligomer (N) with an unrelated sequence around the antibiotic binding site as a negative control. The oligoribonucleotides containing biotin at their 5′-terminal end were immobilized on streptavidin-coated sensor chips as described in Materials and Methods. An average coating of 1,500 RU of oligomers was achieved, and the surface plasmon resonance analysis was performed with serial dilutions of various antibiotics used as ligands. The sensorgrams for interaction between the antibiotics and rRNA are depicted in Fig. 5. Association and dissociation were achieved rapidly, and the regeneration of the surface was conducted using Na2SO4. A strong binding of paromomycin and neomycin B to Leishmania rRNA can be seen (Fig. 5A and B). The corresponding steady-state affinity analysis is shown in Fig. 5C (paromomycin) and D (neomycin B). The apparent KD values of these interactions determined by nonlinear regression were (1.7 ± 0.3) × 10−3 and (6.2 ± 0.7) × 10−4 M, respectively. Attempts to determine the association and dissociation rates for paromomycin binding to Leishmania rRNA were unsuccessful due to the extremely fast processes. However, neomycin B showed lower rates for both, which could be determined as an association rate (kon) of 267.9 M−1 s−1 and a dissociation rate (koff) of (1.66 ± 0.35) × 10−1 s−1. Conversely to the case with aminoglycosides, the β-lactam antibiotic ampicillin showed no binding to Leishmania mexicana RNA (Fig. 5F) or mammalian RNA (not shown). We also analyzed binding of paromomycin to immobilized human rRNA and found that none of the paromomycin concentrations showed specific binding (Fig. 5E). These results can explain the selective effects of paromomycin on Leishmania ribosomes.
FIG. 5.
Surface plasmon resonance analysis (SPR) of the interaction of aminoglycoside antibiotics with L. mexicana rRNA. (A) SPR sensorgram of the interactions between paromomycin (8 to 0.06 mM) and immobilized L. mexicana rRNA (1,450 RU) after correction of nonspecific binding. (B) SPR sensorgram of the interaction of neomycin B (2 to 0.03 mM) and immobilized L. mexicana rRNA (1,560 RU) after correction of nonspecific binding. (C and D) Nonlinear analysis for the determination of the apparent KD of the interaction between immobilized L. mexicana rRNA and aminoglycoside antibiotics, paromomycin, and neomycin B, respectively. (E) SPR sensorgram of the interactions between paromomycin (8 to 0.06 mM) and immobilized human rRNA (1,400 RU). (F) SPR sensorgram of the interactions between ampicillin (8 to 0.06 mM) and immobilized L. mexicana rRNA (1,510 RU). The sensorgrams are representative of at least three different experiments.
DISCUSSION
Increasing levels of Leishmania resistance to the antimonial compounds used for many years as antiparasitic agents have led to the search for new and less toxic drugs potentially useful in leishmaniasis chemotherapy. Paromomycin, an aminoglycoside antibiotic originally used as an antibacterial agent, has been shown to be effective against Leishmania infections when used either alone or in combined formulations with other drugs (7, 20).
In order to understand the selective activity shown by paromomycin against parasites at the molecular level, with almost no effect on host cell metabolism and survival, we have investigated the process of protein synthesis in both cell systems and how they are affected by aminoglycoside antibiotics. In vivo translation experiments with Leishmania represent total protein synthesis carried out by cytoplasmic and mitochondrial ribosomes. In contrast, in vitro assays were performed with postmitochondrial extracts, and therefore, their results correspond only to cytoplasmic ribosome polypeptide synthesis.
It is worth mentioning that even using Leishmania promastigotes, which are less sensitive to paromomycin than amastigotes (15), we detected a strong antibiotic inhibition on polypeptide synthesis with systems containing parasite ribosomes, while only a small effect on mammalian cell ribosomal particles was obtained under the same conditions (Fig. 3).
The marked differential inhibition by paromomycin of polyphenylalanine synthesis induced by synthetic mRNA in protozoan extracts seems to be related to the stability of ribosomal monomers. It has been shown that particle dissociation into ribosomal subunits at low Mg2+ concentration in Leishmania was almost completely abolished in the presence of paromomycin (17). Under these conditions, ribosomes released after each round of mRNA translation might occur mainly as monomers which are unable to again bind the poly(U) molecules and therefore cannot reinitiate protein synthesis. Conversely, ribosomes from mammalian cells are more stable than their parasite counterparts, and the corresponding polypeptide synthesis seems to be less affected by the antibiotic.
Misreading is a normal event in protein synthesis leading to the incorporation of an erroneous amino acid in the growing polypeptide chain due to a mismatch in the codon-anticodon recognition step on the ribosome. Basal misreading in cell-free systems occurs at a frequency of 1 to 5 × 10−3, but it can be altered by ribosomal mutations, changes in salt concentrations in the intracellular pool, or the presence of some antibiotics (28). The normal misreading frequency may reach values as high as 1 to 2% when working with eukaryotic systems in the presence of a synthetic mRNA, such as poly(U). The most likely misreading errors involve a single base-pair mismatch between codon and anticodon, and this relationship is maintained in the presence of error-inducing antibiotics (8). A mismatch of more than one base pair in the same codon has very low probability. The standard assay to calculate the frequency of mistranslation in a cell-free system involves the measurement of the relative incorporation of leucine compared to that of phenylalanine when poly(U) is used as mRNA. Codons for phenylalanine are UUU and UUC, and those for leucine are CUU, CUC, UUA, UUG, CUA, and CUG. Misreadings induced by aminoglycoside antibiotics are usually errors at the first or third position of the codon (28). These errors should cause the incorporation of leucine instead of phenylalanine in the reaction mixtures used in the present study. Figure 4A shows that the misreading frequency induced by paromomycin in the mammalian system was slightly higher than the basal error level. Conversely, the erroneous incorporation of leucine in the reaction mixture containing Leishmania ribosomes was remarkably higher at different concentrations of the antibiotic (Fig. 4A and B), with misreading frequencies reaching values between 25 and 75%.
We have shown that paromomycin drastically alters the accuracy of protein synthesis in Leishmania, markedly increasing the misreading levels. Some defective proteins formed under these conditions might cause deleterious effects on parasite survival. Our results indicate that the antibiotic binds to the parasite ribosomal decoding site, modifying the codon-anticodon recognition process. In order to investigate the specificity of the interaction between paromomycin and the A-site of Leishmania rRNA, we carried out a surface plasmon resonance analysis using different oligoribonucleotides as putative targets (Fig. 5). The sensorgrams obtained show that while paromomycin displays a rather high affinity for the polymer corresponding to the decoding site of Leishmania rRNA (oligomer L), the interactions with rRNA from mammalian cells (oligomer M) or a negative control (N) were negligible (Fig. 5). After sequence alignment of paromomycin binding sites on Leishmania and prokaryotic ribosomes, Hobbie et al. (14) have concluded that the substitution of a single base G for A in a bulged region of bacterial small subunit rRNA helix 44 is able to elicit a remarkable increase in paromomycin affinity for bacterial particles.
It is interesting to point out that although neomycin B, a very closely related paromomycin analog, showed a higher affinity for the Leishmania rRNA decoding site (Fig. 5B), it could not inhibit either in vivo protein synthesis or proliferation in cultures of Leishmania mexicana promastigotes (Fig. 1 and 2). This neomycin deficiency might be due to poor uptake of the antibiotic by parasites (15) or to its chemical inactivation occurring intracellularly (17). On the other hand, streptomycin, another aminoglycoside antibiotic used in this work, also did not affect either protein synthesis or parasite proliferation. These results agree with those of previous reports concluding that streptomycin binds rRNA in a region not involving the A site (37). In addition, recent studies on crystallized ribosome-antibiotic complexes have allowed new insights into the specific recognition details at the antibiotic-binding pockets of ribosomal particles (2, 22, 25).
All the results described in the present work strongly suggest that paromomycin can discriminate between ribosomes of Leishmania and mammalian cells and that this marked differential effect may account for the effectiveness shown by the antibiotic against leishmaniasis.
Acknowledgments
We are indebted to Sara H. Goldemberg and Maximiliano Juri Ayub for helpful discussions.
This work was supported by the University of Buenos Aires, the National Research Council of Argentina (CONICET), and Agencia Nacional de Promoción Científica y Técnica, grants PICT-06450 (to M.M.F), and PICT-05 38293 and PICT-06 608 (to E.L.M.). E.L.M. is also supported by the Fogarty International Center (TW007972) and International Center for Genetic Engineering and Biotechnology (CRP/ARG09-02).
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Algranati, I. D. 1980. Inhibition of polypeptide synthesis initiation by a change of Mg2+ concentration in wheat germ cell-free systems. Biochem. Biophys. Res. Commun. 96:54-60. [DOI] [PubMed] [Google Scholar]
- 2.Bashan, A., and A. Yonath. 2008. Correlating ribosome function with high-resolution structures. Trends. Microbiol. 16:326-335. [DOI] [PubMed] [Google Scholar]
- 3.Basselin, M., M. A. Badet-Denisot, F. Lawrence, and M. Robert-Gero. 1997. Effects of pentamidine on polyamine level and biosynthesis in wild-type, pentamidine-treated, and pentamidine-resistant Leishmania. Exp. Parasitol. 85:274-282. [DOI] [PubMed] [Google Scholar]
- 4.Beakier-Gingras, L., and P. Phoenix. 1984. The control of accuracy during protein synthesis in Escherichia coli and perturbations of this control by streptomycin, neomycin or ribosomal mutations. Can. J. Biochem. Cell Biol. 62:231-244. [DOI] [PubMed] [Google Scholar]
- 5.Brajtburg, J. S., S. Elberg, D. R. Schwarts, A. Vertut-Croquin, D. Schlessinger, G. S. Kobayashi, and G. Medoff. 1985. Involvement of oxidative damage in erythrocyte lysis by amphotericin B. Antimicrob. Agents Chemother. 27:172-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun, R., and M. Schonenberger. 1979. Cultivation and in-vivo cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 7.Bryceson, A. D. M., A. Murphy, and A. H. Moody. 1994. Treatment of “Old World” cutaneous leishmaniasis with aminosidine ointment: results of an open study in London. Trans. R. Soc. Trop. Med. Hyg. 88:226-228. [DOI] [PubMed] [Google Scholar]
- 8.Davies, J., L. Gorini, and B. D. Davis. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93-106. [PubMed] [Google Scholar]
- 9.Davis, B. D., L. Chen, and P. C. Tai. 1986. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 83:6164-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-on, J., G. P. Jacobs, and L. Weinrauch. 1988. Topical chemotherapy of cutaneous leishmaniasis. Parasitol. Today 4:76-81. [DOI] [PubMed] [Google Scholar]
- 11.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A-site of Escherichia coli 16 S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367-1370. [DOI] [PubMed] [Google Scholar]
- 12.Fourmy, D., S. Voshizawa, and J. D. Puglisi. 1998. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 277:333-345. [DOI] [PubMed] [Google Scholar]
- 13.Hendrix, M., E. S. Priestley, G. F. Joyce, and C.-H. Wong. 1997. Direct observation of aminoglycoside-RNA interactions by surface plasmon resonance. J. Am. Chem. Soc. 119:3641-3648. [DOI] [PubMed] [Google Scholar]
- 14.Hobbie, S. N., S. K. Kalapala, S. Akshay, C. Bruell, S. Schmidt, S. Dabow, A. Vasella, P. Sander, and E. C. Böttger. 2007. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 35:6086-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhingran, A., B. Chawla, S. Saxena, M. P. Barret, and R. Madhubala. 2009. Paromomycin: uptake and resistance in Leishmania donovani. Mol. Biochem. Parasitol. 164:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson, R., A. Michaelsson, and L. Mattsson. 1991. Kinetic analysis of monoclonal antigen-antibody-antigen interactions with a new biosensor based analytical system. J. Immunol. Methods 145:2415-2419. [DOI] [PubMed] [Google Scholar]
- 17.Maarouf, M., F. Lawrence, S. L. Crofr, and M. Robert-Gero. 1995. Ribosomes of Leishmania are a target for the aminoglycosides. Parasitol. Res. 81:421-425. [DOI] [PubMed] [Google Scholar]
- 18.Maarouf, M., F. Lawrence, S. Brown, and M. Robert-Gero. 1997. Biochemical alterations in paromomycin-treated Leishmania donovani. Parasitol. Res. 83:198-202. [DOI] [PubMed] [Google Scholar]
- 19.Marr, J. J., and R. L. Berens. 1997. Antileishmanial effect of allopurinol II. Relationship of adenine metabolism in Leishmania species to the action of allopurinol. J. Infect. Dis. 136:724-732. [DOI] [PubMed] [Google Scholar]
- 20.Mishra, J., A. Saxena, and S. Singh. 2007. Chemotherapy of leishmaniasis: past, present and future. Curr. Med. Chem. 14:1153-1169. [DOI] [PubMed] [Google Scholar]
- 21.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16 S ribosomal RNA. Nature 327:387-394. [DOI] [PubMed] [Google Scholar]
- 22.Moore, P. B., and T. A. Steitz. 2005. The ribosome revealed. Trends Biochem. Sci. 30:201-203. [DOI] [PubMed] [Google Scholar]
- 23.Neal, R. A., A. G. Murphy, P. Olliaro, and S. L. Croft. 1994. Aminosidine ointments for the treatment of experimental cutaneous leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 88:223-225. [DOI] [PubMed] [Google Scholar]
- 24.Ogle, J. M., A. P. Carter, and V. Ramakrishnan. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259-266. [DOI] [PubMed] [Google Scholar]
- 25.Ogle, J. M., and V. Ramakrishnan. 2005. Structural insights into translational fidelity. Annu. Rev. Biochem. 74:129-177. [DOI] [PubMed] [Google Scholar]
- 26.Opperdoes, F. R., and G. H. Coombs. 2007. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 23:149-158. [DOI] [PubMed] [Google Scholar]
- 27.Ouellette, M. 2001. Biochemical and molecular mechanisms of drug resistance in parasites. Trop. Med. Int. Health 6:874-882. [DOI] [PubMed] [Google Scholar]
- 28.Parker, J. 1989. Errors and alterations in reading the universal genetic code. Microbiol. Rev. 53:273-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray, S., B. Hazra, B. Mittra, A. Das, and H. K. Majumder. 1998. Diospyrin, a bisnaphtoquinone: a novel inhibitor of type I DNA topoisomerase of Leishmania donovani. Mol. Pharmacol. 54:994-999. [DOI] [PubMed] [Google Scholar]
- 30.Sülsen, V. P., S. I. Cazorla, F. M. Frank, F. C. Redko, C. A. Anesini, J. D. Coussio, E. L. Malchiodi, V. S. Martino, and L. V. Muschietti. 2007. Trypanocidal and leishmanicidal activities of flavonoids from Argentine medicinal plants. Am. J. Med. Trop. Med. Hyg. 77:654-659. [PubMed] [Google Scholar]
- 31.Sülsen, V. P., F. M. Frank, S. I. Cazorla, C. A. Anesini, E. L. Malchiodi, B. Freixa, R. Vila, L. V. Muschietti, and V. S. Martino. 2008. Trypanocidal and leishmanicidal activities of sesquiterpene lactones from Ambrosia tenuifolia Sprengel (Asteraceae). Antimicrob. Agents Chemother. 52:2415-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundar, S., T. K. Jha, C. P. Thakur, P. K. Sinha, and S. K. Bhattacharya. 2007. Injectable paromomycin for visceral leishmaniasis in India. N. Engl. J. Med. 356:2571-2581. [DOI] [PubMed] [Google Scholar]
- 33.Tenson, T., and A. Mankin. 2006. Antibiotics and the ribosome. Mol. Microbiol. 59:1664-1679. [DOI] [PubMed] [Google Scholar]
- 34.Thakur, C. P., P. Olliaro, S. Gothoskar, S. Bhowmick, B. K. Choudhury, S. Prasad. M. Kumar, and B. B. Verma. 1992. Treatment of visceral leishmaniasis (kala-azar) with aminosidine (paromomycin)-antimonial combinations, a pilot study in Bihar, India. Trans. R. Soc. Trop. Med. Hyg. 86:615-616. [DOI] [PubMed] [Google Scholar]
- 35.Tiuman, T. S., T. Ueda-Nakamura, D. A. García Cortez, B. P. Dias Filho, J. A. Morgado-Díaz, W. de Souza, and C. V. Nakamura. 2005. Antileishmanial activity of parthenolide, a sesquiterpene lactone isolated from Thanacetum parthenium. Antimicrob. Agents Chemother. 49:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 37.Wong, C.-H., M. Hendrix, E. S. Priestley, and W. A. Greenberg. 1998. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem. Biol. 5:397-406. [DOI] [PubMed] [Google Scholar]
- 38.Wyllie, S., M. L. Cunningham, and A. H. Fairlamb. 2004. Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J. Biol. Chem. 279:39925-39932. [DOI] [PubMed] [Google Scholar]