Abstract
The production of metallo-β-lactamase (MBL) is an important mechanism of resistance to β-lactam antibiotics, including carbapenems. Despite the discovery and emergence of many acquired metallo-β-lactamases, IMP-type determinants (now counting at least 27 variants) remain the most prevalent in some geographical areas. In Asian countries, and notably Japan, IMP-1 and its closely related variants are most widespread. Some other variants have been detected in other countries and show either an endemic (e.g., IMP-13 in Italy) or sporadic (e.g., IMP-12 in Italy or IMP-18 in the United States) occurrence. The IMP-18-producing Pseudomonas aeruginosa strain PS 297 from the southwestern United States carried at least two class 1 integrons. One was identical to In51, while the other, named In133 and carrying the blaIMP-18 gene cassette in the third position, showed an original array of five gene cassettes, including aacA7, qacF, aadA1, and an unknown open reading frame (ORF). Interestingly. In133 differed significantly from In96, the blaIMP-18-carrying integron identified in a P. aeruginosa isolate from Mexico. The meropenem and ertapenem MIC values were much lower for Escherichia coli strains producing IMP-18 (0.06 and 0.12 μg/ml, respectively) than for strains producing IMP-1 (2 μg/ml for each). Kinetic data obtained with the purified enzyme revealed lower turnover rates of IMP-18 than of other IMP-type enzymes with most substrates.
IMP-type enzymes were the first acquired metallo-β-lactamases (MBLs) to be detected in clinically relevant Gram-negative pathogens such as Enterobacteriaceae and Pseudomonas aeruginosa (15, 19). IMP-1 was first described in imipenem-resistant isolates from Japan in the 1990s (19). Other IMP-type variants were later identified in Europe (5). Although other types of acquired subclass B1 MBLs subsequently appeared (e.g., VIM-type MBLs, SPM-1, GIM-1, SIM-1, KHM-1, TMB-1, NDM-1, and DIM-1) (23, 25, 27, 29, 32), IMP-type enzymes remain among the most prevalent and widely spread MBL determinants, especially in some geographical settings (25). The clinical relevance of these determinants is also shown by the relatively high prevalence of MBL producers among carbapenem-resistant organisms. More specifically, IMP-type enzymes represent the major acquired MBL in Japan (almost 80% of MBL-producing P. aeruginosa strains in some regions [13]) and are most common in the Asian-Pacific region, while they remain only sporadically encountered in North America and in some European countries (e.g., Romania and Italy) (7, 9, 10, 17, 20, 24, 30). Recent reports also documented the import of blaIMP-carrying strains from Asia (a region of higher endemicity of carbapenem-resistant strains) to northern Europe, where antimicrobial resistance is limited (26).
The genes encoding IMP-type MBLs, as well as some other subclass B1 enzymes (e.g., VIM-type enzymes), are found in gene cassettes inserted in plasmid- or chromosome-borne integron/transposon structures. These latter structures play an important role in the spread of these resistance determinants among bacterial isolates, often resulting in a multidrug resistance profile, as integrons frequently carry various kinds of resistance determinants (15, 22).
The IMP sublineage has at least 27 unique variants (http://www.lahey.org/studies) differing by up to 22% amino acid sequence divergence (between IMP-9 and IMP-19) that exhibit important structural and functional differences from each other or from enzymes of other sublineages. IMP-type MBL determinants are mostly found in P. aeruginosa, Acinetobacter baumannii, and Enterobacteriaceae isolates, although they have also been sporadically identified in other organisms (e.g., Pseudomonas putida) (25). They confer resistance to most β-lactam antibiotics, including β-lactamase inhibitor-β-lactam combinations, oxyiminocephalospsorins, and carbapenems. However, these enzymes differ from other acquired MBLs in that they are apparently unable to hydrolyze temocillin, a derivative of ticarcillin presenting an α-methoxy substituent (7, 14).
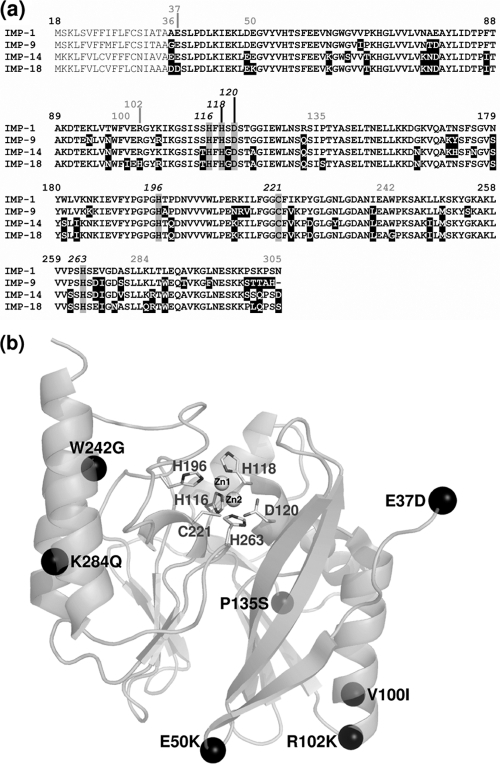
The IMP-18 determinant was first identified in the Unites States and then in Mexico and, more recently, in Puerto Rico (10, 11, 31). In P. aeruginosa isolate 4660 recovered from Mexico, the blaIMP-18 determinant was found in the blaIMP-18-aadA1b array (In96). To our knowledge, no detailed biochemical characterization of IMP-18 is currently available. However, a rationale for a detailed biochemical characterization of IMP-18 could be found in the sequence comparison of the various IMP-type enzymes (Fig. 1a). Indeed, such an alignment highlights that IMP-18 was divergent from the other IMP-type MBLs by at least 8.9% (i.e., 22 amino acids) and that some substitutions occurs in unique positions (Fig. 1b). Specifically, one of these substitutions affects position 242, where a tryptophan (replaced by a glycine in IMP-18) was invariantly found not only in all other IMP-type variants but also in all subclass B1 enzymes reported thus far. In this work, we investigated the genetic context of the blaIMP-18 determinant identified in P. aeruginosa isolate PS 297 recovered in the United States and determined the biochemical properties of its gene product.
FIG. 1.
(a) Amino acid sequence alignment of selected IMP variants. Substitutions with IMP-1 are shown with a black background. The sequence corresponding to the mature enzyme is boldfaced (the first 18 amino acids being the signal peptide). The numbering is according to the standard scheme proposed for class B β-lactamases (8), and amino acid substitutions that are unique in IMP-18 are indicated by gray numbers. (b) Cartoon representation of the IMP-1 X-ray crystal structure (Protein Data Bank code 1DDK), showing the locations of the unique amino acid substitutions found in IMP-18. Substitutions A36D and N305S, located at the amino- and carboxy-terminal extremities of the protein, are not shown because the corresponding residues are not included in the IMP-1 structure.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. aeruginosa PS 297, used as the source of the MBL gene, was isolated from a tracheal aspirate in the southwestern United States; the properties of this strain have been described previously (11). Escherichia coli DH5α (Gibco Life Technologies, Gaithersburg, MD) was used as a host for recombinant plasmids, and E. coli BL21(DE3) (Stratagene, La Jolla, CA) was used for overproduction of the IMP-18 enzyme by using a T7 promoter-based expression system. Bacteria were always grown aerobically at 37°C. E. coli DH5α derivative strains were cultured in Mueller-Hinton broth (Oxoid Ltd., Basingstoke, United Kingdom), and E. coli BL21(DE3) was grown in ZYP-5052 medium for production of the recombinant protein (28). Medium, when necessary, was supplemented with the appropriate antibiotics (kanamycin at 50 μg/ml or chloramphenicol at 85 μg/ml).
Determination of cassette array sequences.
PCR amplification of class 1 integron variable regions (cassette arrays) was performed using primers designed to target the 5′-CS and 3′-CS regions as previously described (24). Five hundred nanograms of purified total DNA of P. aeruginosa PS 297 was used as the template. The resulting PCR products were purified and completely sequenced on both strands by using a primer walking approach.
Cloning of the MBL gene.
The complete blaIMP-18 gene from P. aeruginosa PS 297 was amplified by PCR using oligonucleotides IMP-18-EXP-Fw (5′-AGGGTACATATGAAAAAATTATTTGTTTTATGTG) and IMP-18-EXP-Rev (5′-CCGGATCCTTAGCTACTTGGCTGTAACG) (the underlines indicate the NdeI and BamHI restriction sites, respectively) as previously reported (6, 7). The amplified DNA was digested with NdeI and BamHI restriction enzymes and cloned into vector pLB-II (a pBC-SK derivative [Stratagene, La Jolla, CA] modified in our laboratory [1]) and into the T7 promoter-based expression vector pET-9a (Novagen, Madison, WI) previously digested with the same enzymes. The resulting constructs were named pLBII-IMP-18 and pET-IMP-18, respectively. A strain producing the IMP-1 MBL [E. coli DH5α(pLB-II-IMP-1)] was also prepared by adopting the same cloning strategy and using PCR primers IMP-1-EXP-Fw (5′-CGGAATTCATATGAGCAAGTTATCTGTATTCTTTATA, which includes restriction sites for EcoRI [boldface] and NdeI [underlined]) and IMP-1-EXP-Rev (5′-CCGGATCCTTAGTTGCTTGGTTTTGATGG, which includes a BamHI restriction site [underlined]).
Antimicrobial susceptibility testing.
The in vitro antimicrobial susceptibility profiles of E. coli DH5α derivatives were determined with the microdilution broth method as recommended by the Clinical and Laboratory Standards Institute (3), using Mueller-Hinton broth with a bacterial inoculum of 5 × 104 CFU/well. MICs were recorded after 18 h at 37°C.
Production and purification of IMP-18.
IMP-18 was produced from a culture of E. coli BL21(DE3)(pET-IMP-18) grown for 24 h at 37°C. Bacterial cells were harvested by centrifugation (10,000 × g, 30 min, 4°C) and lysed using a cell disruption system (Constant Systems Ltd., Daventry, United Kingdom). The sample was first clarified by centrifugation in order to eliminate the cellular debris and then desalted using a HiPrep 26/10 desalting column (GE Healthcare, Uppsala, Sweden) with 20 mM HEPES containing 50 μM ZnSO4 (pH 7.0) buffer. The resulting sample was loaded at a flow rate of 2 ml/min onto an SP Sepharose high-performance column (bed volume, 5 ml) (GE Healthcare), previously equilibrated with the same buffer, and bound proteins were eluted using a linear NaCl gradient (0 to 1 M in 100 ml). β-Lactamase-containing fractions were pooled, concentrated to 1.2 mg/ml, and stored at −20°C.
Protein analysis techniques.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed according to the method of Laemmli (13a), using final acrylamide concentrations of 12% and 5% (wt/vol) for the separating and the stacking gels, respectively. After electrophoresis, the protein bands were stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA).
Determination of kinetic parameters and inactivation by chelating agents.
The hydrolysis of β-lactam substrates was monitored by measuring the absorbance variation under the experimental conditions reported previously (6, 14). All measurements were performed on a Cary 100 UV-Vis spectrophotometer (Varian, Walnut Creek, CA) at 30°C in 10 mM HEPES buffer (pH 7.5) containing 50 μM ZnSO4, using a reaction volume of 500 μl. The purified IMP-18 was diluted in the same buffer supplemented with 20 μg/ml bovine serum albumin (BSA) to prevent enzyme denaturation. The steady-state kinetic parameters (kcat and Km) were calculated after direct fit of the initial rates to the Henri-Michaelis-Menten equation or using the Hanes-Woolf linearization. Km values lower than 10 μM were measured as inhibition constants by using a competitive inhibition model and 100 μM nitrocefin as the reporter substrate. The inactivation of IMP-18 by chelating agents was investigated as previously described (12).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL/GenBank/DDBL sequence databases and assigned the accession numbers FN556189 and FN556190.
RESULTS AND DISCUSSION
Genetic context of the blaIMP-18 determinant in P. aeruginosa strain PS 297.
PCR experiments designed to amplify the variable region of class 1 integrons yielded two amplification products that significantly differed in size (3.9 and 1.5 kb).
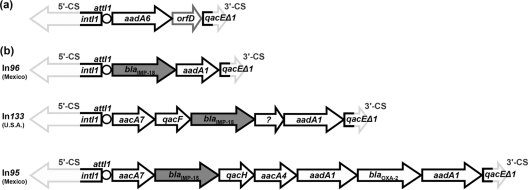
The smaller cassette array was identical to the aadA6-orfD array (In51) previously found in a VEB-1-producing P. aeruginosa isolate from France (18). This array seems to be widespread and has been found in, e.g., Brazil, Mexico, Costa Rica, India, Iran, and China (http://www.ncbi.nlm.nih.gov), in some cases with minor differences. An aadA6-orfD cassette array (In97, 4 nucleotide differences with the In51 cassette array) was also found in P. aeruginosa strain 4677 from Mexico, which also carries the IMP-15-encoding array designated In95 (Fig. 2) (9).
FIG. 2.
Structure of the cassette arrays of the two class 1 integrons found in P. aeruginosa PS 297. Gene cassettes are represented by arrows (the sequences of the gray-outlined portions of intI1 and qacEΔ1 were not determined). (a) Structure of the shorter cassette array (accession no. FN556189), identical to In51. (b) Structure of the blaIMP-18-carrying cassette array (In133, accession no. FN556190). The putative gene cassette of unknown function (fourth position) is indicated by a question mark. The structure of In133 is compared to that of other blaIMP-containing cassette arrays (In95 and In96) identified in clinical isolates from Mexico, highlighting the differences between In133 and In96 (both carrying a blaIMP-18 gene cassette) and the presence of similar gene cassettes in In133 and In95 (which carries a blaIMP-15 determinant) (10, 11).
The larger cassette array (named In133), aacA7-qacF-blaIMP-18-unknown gene cassette-aadA1b, has not been reported before (Fig. 2). It was associated with the strong version of the Pc promoter located in the integrase gene (4). In agreement with the resistance to gentamicin and amikacin exhibited by isolate PS 297, two cassettes (in first and last positions) were found to encode aminoglycoside resistance factors (the AACA7 acetyltransferase and the AADA1 adenylyltransferase), the first being compatible with the resistance to gentamicin and amikacin exhibited by strain PS 297. The qacF-like gene encodes a quaternary ammonium compound resistance protein variant identical to a QacF variant reported only once in the database (accession no. DQ149925) encoded by a plasmid-borne cluster of three integrons in several isolates of Vibrio cholerae (2) and showing two amino acid substitutions (L7M and S10A) compared with the QacF determinant encoded by In40 from Enterobacter aerogenes BM2688 (21). It encodes an efflux protein belonging to the small multidrug resistance family and shows sequence similarity with other Qac/Erm-type drug:proton antiporters that have been reported in other integrons or hosts (sequence identity ranging from 79.3 to 35.1% with QacG and QacC, respectively) (15, 16).
The IMP-18-encoding cassette is found in the third position, which suggests that other cassette integration events occurred after the insertion of the blaIMP determinant in this integron. This observation is rather unusual, as MBL-encoding gene cassettes are more frequently found in either first or second position in the integron structure (25, 30).
The attC site (59-base element) of the blaIMP-18 gene cassette is 134 bp long and thus closer to the attC sites found associated with most blaIMP alleles (e.g., blaIMP-1-like, blaIMP-4, blaIMP-7, and blaIMP-16) compared to those of the blaIMP-2 and blaIMP-8 gene cassettes, the latter exhibiting a much shorter attC site (7). Compared to In96, the blaIMP-18 gene cassette attC site differs by a single deletion (T1291 of sequence accession no. EF184215). The blaIMP-18 attC site also exhibits the largest divergence with other blaIMP-associated attC sites of similar lengths (nucleotide identities range from 69 to 81%, while other attC sites share identities in the range of 75 to 98%).
The nature of the putative gene cassette found in the fourth position remains unknown. Indeed, the corresponding 411-bp DNA fragment did not show any similarity with sequences present in the NCBI/EMBL/DDJB databases, nor does it contain any open reading frame, but the sequence does present the typical features of a gene cassette with its attC site that includes both the core and inverse core site for recombination.
Molecular cloning of blaIMP-18 and contribution to β-lactam resistance.
In order to compare the contributions of IMP-18 and IMP-1 to β-lactam resistance, laboratory strains where the MBL gene was cloned under the transcriptional control of the Plac promoter were obtained. The production of IMP-18 in E. coli resulted in higher MIC values for most tested antibiotics but did not affect aztreonam (which is not a substrate of MBLs), piperacillin, and temocillin, the latter apparently behaving as poor substrates of IMP-18 (Table 1). A similar behavior was also observed, to some extent, with the IMP-1-producing recombinant strain, indicating that rather low MICs are commonly observed with these agents when IMP-type determinants are transferred in E. coli laboratory strains. The MIC values for oxyiminocephalosporins were increased 16- to 128-fold, the major changes being observed with cefotaxime, ceftazidime, and cefotetan.
TABLE 1.
In vitro susceptibility profiles of E. coli DH5α derivatives carrying the cloned blaIMP-18 genes and E. coli DH5α carrying the empty plasmida
| β-Lactam | MIC (μg/ml) |
||
|---|---|---|---|
| E. coli DH5α (pLBII-IMP-18) | E. coli DH5α (pLBII-IMP-1) | E. coli DH5α (pBC-SK) | |
| Ampicillin | 64 | 256 | 4 |
| Piperacillin | 1 | 1 | 1 |
| Ticarcillin | 256 | 512 | 8 |
| Temocillin | 16 | 32 | 16 |
| Cephalothin | 32 | 256 | 4 |
| Cefoxitin | 256 | >256 | 4 |
| Cefuroxime | 64 | 128 | 4 |
| Cefotaxime | 32 | 8 | 0.25 |
| Ceftazidime | 32 | 32 | 0.25 |
| Ceftriaxone | 4 | 8 | 0.25 |
| Cefepime | 2 | 4 | 0.25 |
| Cefotetan | 32 | >32 | 0.25 |
| Imipenem | 1 | 2 | 0.06 |
| Meropenem | 0.06 | 2 | ≤0.015 |
| Ertapenem | 0.12 | 2 | ≤0.015 |
| Aztreonam | 0.12 | 0.12 | 0.12 |
Strains producing the IMP-1 enzyme or carrying the empty vector are shown for comparison.
As already reported in previous studies, the production of either IMP-1 or IMP-18 did not confer full resistance to carbapenems (per CLSI breakpoints) but resulted in increases in MIC values of up to seven 2-fold dilutions. Interestingly, the contributions of IMP-1 and IMP-18 to carbapenem MICs were very different (Table 1). While IMP-1 yields similar MIC values with all tested carbapenems, the production of IMP-18 was associated with much lower MICs for meropenem and ertapenem. This most likely reflects a difference in the hydrolytic activity toward these compounds ( Table 2).
TABLE 2.
Kinetic parameters of selected IMP-type MBLs for the hydrolysis of various β-lactam substratesd
| Substrate |
kcat (s−1) |
Km (μM) |
kcat/Km (M−1·s−1) |
|||
|---|---|---|---|---|---|---|
| IMP-1 | IMP-18 | IMP-1 | IMP-18 | IMP-1 | IMP-18 | |
| Ampicillin | 950 | 65 | 200 | 103 | 4.8 × 106 | 6.3 × 105 |
| Benzylpenicillin | 320 | 54 | 520 | 90 | 6.2 × 105 | 6.2 × 105 |
| Piperacillina | >10 | >200 | 7.2 × 105 | 5.1 × 104 | ||
| Ticarcillin | 1.1 | 30 | 740 | 180 | 1.5 × 103 | 1.5 × 105 |
| Temocillin | >0.75 | >2,000 | >1,000 | <102 | 7.5 × 102b | |
| Cephalothin | 48 | 0.4 | 21 | 0.1 | 2.4 × 106 | 4.0 × 106 |
| Cefoxitin | 16 | 2 | 8 | 11 | 2.0 × 106 | 1.8 × 105 |
| Cefuroxime | 8 | 0.9 | 37 | 7 | 2.2 × 105 | 1.3 × 105 |
| Cefotaxime | 1.3 | 0.7 | 4 | 3 | 3.5 × 105 | 2.3 × 105 |
| Ceftazidime | 8 | 1 | 44 | 1.3 | 1.8 × 105 | 7.7 × 105 |
| Cefepime | 7 | 0.35 | 11 | 0.8 | 6.6 × 105 | 4.4 × 105 |
| Nitrocefin | 63 | 55 | 27 | 9 | 2.3 × 106 | 6.1 × 106 |
| Imipenem | 46 | 17 | 39 | 7 | 1.2 × 106 | 2.4 × 106 |
| Meropenem | 5 | 0.05 | 10 | 8.4 | 5.0 × 105 | 6.0 × 103 |
| Ertapenem | 16 | 0.03 | 21 | 2.6 | 7.6 × 105 | 1.2 × 104 |
| Aztreonam | <0.01 | <0.01c | >1,000 | >1,000 | ||
First-order kinetics were observed with this substrate.
With temocillin, the standard deviation on kinetic parameters was 20%.
No hydrolysis was detected by using an enzyme concentration of 1.8 μM in the reaction mixture.
Data for IMP-1 (except ertapenem [this study]) are from reference 14. Individual kinetic parameters are the means of three measurements, and standard deviations were always <10%, unless otherwise specified.
Purification and biochemical characterization of IMP-18.
Thanks to the high yield of MBL production (approximately 100 mg/liter of culture), a purified IMP-18 preparation (purity, >98%) could be obtained after a single cation exchange chromatography step. In kinetic assays, IMP-18 was able to efficiently hydrolyze most tested β-lactam compounds except aztreonam (which is not recognized by MBLs) and temocillin, which typically is a poor substrate for IMP-type enzymes (7, 14). Most observed catalytic efficiencies were greater than 105 M−1·s−1, except for piperacillin, meropenem, and ertapenem (Table 2). Surprisingly, the lowest turnover rates and catalytic efficiencies measured were those for the latter carbapenems. These data represent a striking feature of IMP-18 compared with other IMP variants. These data are also in agreement with the low MIC values exhibited by the recombinant E. coli clone producing IMP-18 for these compounds. Km values were overall lower for cephalosporins and carbapenems (Km, 0.1 to 11 μM) than for penicillin substrates (Km, ≥90 μM) (Table 2). Although the highest turnover rates were measured using benzylpenicillin, ampicillin, ticarcillin, and imipenem, IMP-18 shows strikingly low kcat values for the other substrates, including expanded-spectrum cephalosporins (e.g., cefotaxime and cefepime) and 1-β-methyl carbapenems (meropenem and ertapenem). The turnover rate of IMP-18 with these substrates was 2 orders of magnitude lower than that of IMP-1 or IMP-12. A similarly low kcat value was measured for cephalothin, although a concomitant decrease of the Km value of this substrate yielded a catalytic efficiency close to that of IMP-1. However, the comparison of the MIC values of the strains producing IMP-1 and IMP-18 would indicate that, at least with this compound, the kcat value (rather than kcat/Km) would influence the resulting susceptibility of the strain. Cefoxitin was also hydrolyzed by IMP-18, although less efficiently than by IMP-1. It remains unclear why this substrate is overall a good substrate for IMP-type enzymes, while no interaction with temocillin was observed even though this compound shows the same typical α-methoxy substituent.
IMP-18 was efficiently inactivated by metal chelators, with dipicolinic acid showing higher pseudo-first-order inactivation rates than those of EDTA (k2/K, >40 and 0.15 M−1·s−1 for dipicolinic acid and EDTA, respectively). This behavior is consistent with that observed with other IMP-type enzymes and, generally, with all other subclass B1 MBLs.
Concluding remarks.
The present study further highlights the heterogeneity and complexity shown by the cassette arrays of MBL-encoding class 1 integrons. Although the epidemiological relationship between the recently reported IMP-18-producing isolates has not been specifically investigated, it is noteworthy that the same MBL determinant has completely different genetic backgrounds, suggesting different sources of acquisition of the MBL gene cassette. Moreover, the structure of In133 is quite unusual as the MBL-encoding cassette was not found at the first or second position as in most MBL-encoding integrons, reflecting the possibility that current selective pressure might have induced the acquisition of other resistance traits. This finding indicates that this integron substantially evolved its structure after the acquisition of the MBL gene, as gene cassettes appear to integrate at the front of the array (recombination events could also alter cassette order). The complexity of this cassette array might also have played a crucial role in the coselection of resistance genes, including blaIMP-18, which conferred carbapenem resistance. Therefore, the selective pressure driving the acquisition of blaIMP-18 may not be a consequence of direct selection by β-lactam agents but possibly a consequence of selection by other antimicrobial agents or disinfectants. These data raise an important question on how to practically avoid such resistance phenotypes in the clinical setting, if such clinical strains that are resistant to last-resort drugs could be maintained by rather ordinary and commonly used antimicrobial agents and/or disinfectants.
From a functional standpoint, IMP-18 is characterized by overall lower turnover rates than those of other IMP-type variants, especially toward the carbapenem meropenem. The unprecedented replacement of the highly conserved Trp242 residue with a glycine might impact on the overall structure and stability of IMP-18, thus contributing to its lower reactivity. Further investigation of its structural and biochemical properties should be carried out to elucidate this point.
Acknowledgments
This work was funded in part by a grant from the Italian Ministero dell'Istruzione, Università e Ricerca (MIUR; contract number 2005061894_004).
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Borgianni, L., J. Vandenameele, A. Matagne, L. Bini, R. A. Bonomo, J. M. Frere, G. M. Rossolini, and J. D. Docquier. 24 May 2010. Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed]
- 2.Ceccarelli, D., A. M. Salvia, J. Sami, P. Cappuccinelli, and M. M. Colombo. 2006. New cluster of plasmid-located class 1 integrons in Vibrio cholerae O1 and a dfrA15 cassette-containing integron in Vibrio parahaemolyticus isolated in Angola. Antimicrob. Agents Chemother. 50:2493-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 7th ed. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemtoher. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornaglia, G., M. L. Riccio, A. Mazzariol, L. Lauretti, R. Fontana, and G. M. Rossolini. 1999. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet 353:899-900. [DOI] [PubMed] [Google Scholar]
- 6.Docquier, J. D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J. M. Frere, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Docquier, J. D., M. L. Riccio, C. Mugnaioli, F. Luzzaro, A. Endimiani, A. Toniolo, G. Amicosante, and G. M. Rossolini. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J. M. Frere. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garza-Ramos, U., R. Morfin-Otero, H. S. Sader, R. N. Jones, E. Hernandez, E. Rodriguez-Noriega, A. Sanchez, B. Carrillo, S. Esparza-Ahumada, and J. Silva-Sanchez. 2008. Metallo-β-lactamase gene blaIMP-15 in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob. Agents Chemother. 52:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garza-Ramos, U., P. Tinoco, J. Silva-Sanchez, R. Morfin-Otero, E. Rodriguez-Noriega, G. Leon-Garnica, H. S. Sader, and R. N. Jones. 2008. Metallo-β-lactamase IMP-18 is located in a class 1 integron (In96) in a clinical isolate of Pseudomonas aeruginosa from Mexico. Int. J. Antimicrob. Agents 31:78-80. [DOI] [PubMed] [Google Scholar]
- 11.Hanson, N. D., A. Hossain, L. Buck, E. S. Moland, and K. S. Thomson. 2006. First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-β-lactamase, IMP-18. Antimicrob. Agents Chemother. 50:2272-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez Valladares, M., M. Kiefer, U. Heinz, R. P. Soto, W. Meyer-Klaucke, H. F. Nolting, M. Zeppezauer, M. Galleni, J. M. Frere, G. M. Rossolini, G. Amicosante, and H. W. Adolph. 2000. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett. 467:221-225. [DOI] [PubMed] [Google Scholar]
- 13.Kouda, S., M. Ohara, M. Onodera, Y. Fujiue, M. Sasaki, T. Kohara, S. Kashiyama, S. Hayashida, T. Harino, T. Tsuji, H. Itaha, N. Gotoh, A. Matsubara, T. Usui, and M. Sugai. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J. Antimicrob. Chemother. 64:46-51. [DOI] [PubMed] [Google Scholar]
- 13a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frere, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Littlejohn, T. G., D. DiBerardino, L. J. Messerotti, S. J. Spiers, and R. A. Skurray. 1991. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene 101:59-66. [DOI] [PubMed] [Google Scholar]
- 17.Mereuta, A. I., D. T. Buiuc, G. M. Rossolini, and J. D. Docquier. 2007. Simultaneous detection of VIM-2 and IMP-13-producing carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Romania, abstr. C2-1498, p. 127. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 18.Naas, T., L. Poirel, and P. Nordmann. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445-451. [DOI] [PubMed] [Google Scholar]
- 19.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagani, L., C. Colinon, R. Migliavacca, M. Labonia, J. D. Docquier, E. Nucleo, M. Spalla, B. M. Li, and G. M. Rossolini. 2005. Nosocomial outbreak caused by multidrug-resistant Pseudomonas aeruginosa producing IMP-13 metallo-β-lactamase. J. Clin. Microbiol. 43:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ploy, M. C., P. Courvalin, and T. Lambert. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob. Agents Chemother. 42:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., J. D. Pitout, and P. Nordmann. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2:501-512. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., J. M. Rodriguez-Martinez, N. Al Naiemi, Y. J. Debets-Ossenkopp, and P. Nordmann. 2010. Characterization of DIM-1, an integron-encoded metallo-β-lactamase from a Pseudomonas stutzeri clinical isolate in The Netherlands. Antimicrob. Agents Chemother. 54:2420-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossolini, G. M., and J. D. Docquier. 2007. Class B β-lactamases, p. 115-144. In R. A. Bonomo and M. E. Tolmasky (ed.), Enzyme-mediated resistance to antibiotics: mechanisms, dissemination and prospects for inhibition. ASM Press, Washington, DC.
- 26.Samuelsen, O., M. A. Toleman, A. Sundsfjord, J. Rydberg, T. M. Leegaard, M. Walder, A. Lia, T. E. Ranheim, Y. Rajendra, N. O. Hermansen, T. R. Walsh, and C. G. Giske. 2010. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob. Agents Chemother. 54:346-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi, J. I., K. Morita, T. Kitao, N. Watanabe, M. Okazaki, T. Miyoshi-Akiyama, M. Kanamori, and T. Kirikae. 2008. KHM-1, a novel plasmid-mediated metallo-β-lactamase from a Citrobacter freundii clinical isolate. Antimicrob. Agents Chemother. 52:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier, F. W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 29.Walsh, T. R. 2008. Clinically significant carbapenemases: an update. Curr. Opin. Infect. Dis. 21:367-371. [DOI] [PubMed] [Google Scholar]
- 30.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolter, D. J., N. Khalaf, I. E. Robledo, G. J. Vazquez, M. I. Sante, E. E. Aquino, R. V. Goering, and N. D. Hanson. 2009. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: dissemination of KPC and IMP-18 β-lactamases. Antimicrob. Agents Chemother. 53:1660-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yong, D., M. A. Toleman, C. G. Giske, H. S. Cho, K. Sundman, K. Lee, and T. R. Walsh. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]