Abstract
The sequence of pR3521, a self-transmissible plasmid from Escherichia coli, was determined. pR3521 (110,416 bp) comprised a contiguous IncB sequence (84,034 bp) sharing extensive similarities with IncI replicons and an acquired region (26,382 bp) carrying sequences of diverse origin, containing blaACC-4, blaSCO-1, blaTEM-1b (two copies), strA, strB, sul2, and aacC2.
Multidrug resistance among enterobacteria is commonly due to acquisition of plasmids carrying various resistance determinants (4). We have previously identified pR3521, a self-transferable and multiresistant plasmid from an Escherichia coli isolate from a hospitalized patient in Greece (18). Work had been largely focused on the characterization of loci containing blaACC-4, an ampC gene encoding ACC-4, an extended-spectrum variant of the ACC-1 cephalosporinase (19), and blaSCO-1, a novel gene of unknown origin coding for an RTG-type carbenicillinase (18, 21). blaACC-4 was included in a sequence derived from the chromosome of Hafnia alvei and linked with ISEcp1. blaSCO-1 was included in a segment of likely chromosomal origin that was associated with IS26 (18, 19). Plasmid-mediated production of SCO-1 and ACC-1 has also been described for Klebsiella pneumoniae, Proteus mirabilis, and Salmonella enterica serovar Livingstone in Europe and North Africa, but detailed characteristics of the respective plasmids were not reported (6, 13).
In this study, the complete nucleotide sequence of pR3521 is presented. pR3521 belonged to incompatibility (Inc) group B, which is closely related to the IncI family of replicons (5, 17). A hypothesis as to the mechanisms of accumulation of diverse resistance genes in pR3521 is also discussed.
General features of pR3521.
An E. coli K-12 transconjugant clone was used as a source of pR3521 (18). Plasmid pR3521, purified by CsCl gradient ultracentrifugation, was partially digested with Sau3A, and the fragments were ligated into the chloramphenicol-resistant vector pBCSK(+) (Stratagene, La Jolla, CA). The recombinant plasmids were used to transform E. coli DH5α. Transformants were selected with chloramphenicol (20 μg/ml). Recombinant plasmids were purified with a Qiagen plasmid midi kit (Qiagen, Hilden, Germany), and the nucleotide sequences of the inserts were determined using an ABI 377 sequencer (Applied Biosystems, Foster City, CA). Sequence gaps were filled by primer walking and sequencing of PCR products using primers hybridizing to known regions (18, 19). Contigs were assembled using the Laser Gene software program (DNASTAR, Madison, WI). For sequence analysis and annotation, the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST/), an insertion sequence (IS) finder (www-is.biotoul.fr/), an open reading frame (ORF) finder (www.bioinformatics.org/sms/), and the Artemis software program (www.sanger.ac.uk/) were utilized.
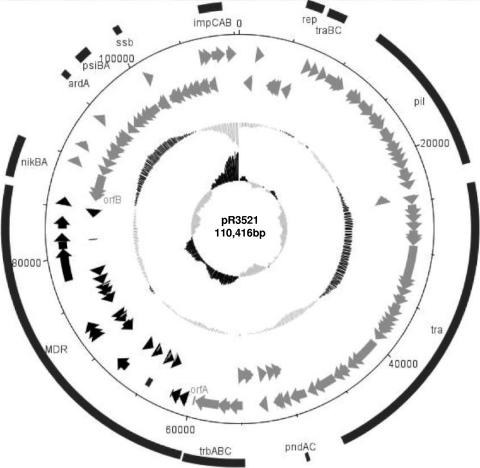
The sequence of pR3521 comprised 110,416 bp (G+C content, 52.6%) and included 124 coding sequences (119 complete and 5 truncated). The ORFs and their characteristics are presented in Table 1. A circular map of pR3521 is shown in Fig. 1. The plasmid was composed of two distinct parts: a contiguous plasmidic segment of 84,034 kb (G+C content, 53%), sharing similarities with replicons of complex I, and an acquired sequence of 26,382 bp (G+C content, 51.3%), containing eight antibiotic resistance genes (blaACC-4, blaSCO-1, blaTEM-1b [two copies], strA, ΔstrB, aacC2, and sul2), intact (n = 7) and defective (n = 1) mobile elements (including four IS26 elements, IS26-1 to IS26-4), single copies of ISKpn11 and ISKpn12, a ΔISEcpI element, and one Tn2 transposon as well as sequences of diverse chromosomal origins.
TABLE 1.
Names, coordinates, and putative functions of ORFs identified in the IncB plasmid pR3521a
| Gene name | Coordinatesb | Putative function(s) |
|---|---|---|
| yagA | Compl. 533-1627 | Hypothetical protein |
| yafB | 1981-2583 | Hypothetical protein |
| orf1 | Compl. 3279-3560 | Hypothetical protein |
| orf2 | Compl. 3666-3941 | Plasmid stabilization protein (RelE/ParE family) |
| orf3 | Compl. 3941-4219 | CopG family protein |
| repA | Compl. 5133-6209 | Replication initiation protein |
| traB | 6866-7507 | F pilus assembly |
| traC | 7648-8310 | F pilus assembly |
| yqiJ | 8577-9197 | Putative adhesin |
| yqiK | 9224-10918 | Putative adhesin |
| pilI | 10999-11241 | Type IV prepilin cluster |
| pilL | 11942-13012 | Type IV prepilin cluster; lipoprotein |
| pilM | 13016-13453 | Type IV prepilin cluster |
| pilN | 13485-15104 | Type IV prepilin cluster; secretin protein |
| pilO | 15125-16420 | Type IV prepilin cluster |
| pilP | 16410-16868 | Type IV prepilin cluster |
| pilQ | 16971-18479 | Type IV prepilin cluster; ATP-binding protein |
| pilR | 18481-19575 | Type IV prepilin cluster; membrane protein |
| pilS | 19703-20173 | Type IV prepilin cluster; prepilin |
| pilT | 20220-20705 | Type IV prepilin cluster |
| pilU | 20721-21347 | Type IV prepilin cluster; prepilin peptidase |
| pilV | 21364-22725 | Type IV prepilin cluster |
| orf4 | 23183-23473 | Hypothetical protein |
| orf5 | Compl. 23565-23885 | Hypothetical protein |
| traE | 24147-24968 | F pilus assembly |
| traF | 25070-26272 | F pilus assembly |
| traH | 26376-26834 | F pilus assembly |
| traI | 26831-27667 | DNA helicase |
| traJ | 27651-28799 | ATP-binding protein |
| traK | 28796-29086 | F pilus assembly |
| sogL | 29150-33211 | DNA primase |
| sogS | 29339-33211 | Regulator of SogL |
| traL | 33228-33578 | F pilus assembly |
| traM | 33590-34285 | Mating signal |
| traN | 34296-35255 | Aggregate stability |
| traO | 35259-36590 | Hypothetical protein |
| traP | 36587-37300 | Conjugal transfer protein |
| traQ | 37297-37827 | Conjugal transfer protein |
| traR | 37874-38272 | Hypothetical protein |
| traS | 38329-38580 | Surface exclusion |
| traT | 38687-39313 | Surface exclusion |
| traU | 39608-42652 | F pilus assembly |
| traV | 42652-43272 | F pilus assembly |
| traW | 43395-44435 | F pilus assembly |
| traX | 44432-45001 | F pilin acetylation |
| traY | 45076-47247 | Integral membrane protein |
| excA | 47545-47982 | Surface exclusion |
| orf6 | 48256-49842 | Hypothetical protein |
| pndA | Compl. 49980-50132 | Postsegregation killing |
| pndC | Compl. 49987-50280 | Counter protein for PndA |
| orf7 | 50384-50704 | Hypothetical protein |
| orf8 | Compl. 50749-51066 | Hypothetical protein |
| orf9 | 51035-51361 | Hypothetical protein |
| orf10 | 51365-51580 | Hypothetical protein |
| orf11 | Compl. 51778-52332 | Hypothetical protein |
| yeaH | 52712-53029 | Putative nuclease |
| orf12 | Compl. 53419-53715 | Hypothetical protein |
| orf13 | Compl. 54250-54357 | Hypothetical protein |
| trbA | 54734-56050 | Conjugal transfer protein |
| trbB | 56047-57171 | Conjugal transfer protein |
| trbC | 57152-59455 | Conjugal transfer protein |
| orfA*1 | 59569-59735 | Hypothetical protein |
| sul2 | 60319-61134 | Dihydropteroate synthase |
| strA | 61195-61998 | Streptomycin phosphotransferase |
| strB*1 | 61998-62106 | Streptomycin phosphotransferase (nonfunctional) |
| blaTEM-1 | Compl. 62254-63114 | TEM-1 β-lactamase precursor |
| tnpA | Compl. 63253-63957 | TnpA transposase of IS26-1 |
| orfA*2 | Compl. 64047-64182 | Hypothetical protein |
| tnpB*1 (rcr2) | 64299-64771 | Putative transposase (nonfunctional) |
| tnpA | Compl. 64824-65528 | TnpA transposase of IS26-2 |
| ΔtnpR | 65642-66003 | Resolvase (nonfunctional) |
| blaSCO-1 | Compl. 66139-67005 | SCO-1 carbenicillinase precursor |
| Glycosidase-like gene | 67167-68366 | Cellulase-like protein |
| umuC | Compl. 69483-70760 | UV protection |
| umuD | Compl. 70766-71194 | UV protection |
| dbp | Compl. 71290-71562 | Putative DNA-binding protein |
| tnpA | 71687-72037 | Putative transposase of ISKpn12 |
| tnpA | 72058-72447 | Putative transposase of ISKpn12 |
| σ′70 gene | 72515-73066 | σ′70 factor-like protein |
| tnpA | Compl. 73591-74421 | Putative transposase of ISKpn11 |
| tnpA | Compl. 74418-74750 | Putative transposase of ISKpn11 |
| aacC2 | Compl. 75088-75948 | Gentamicin-(3)-N-acetyl-transferase |
| blaTEM-1 | Compl. 76090-76950 | TEM-1 β-lactamase precursor |
| tnpR | Compl. 77133-77726 | TnpR resolvase of Tn2 |
| tnpA | 77695-80859 | TnpA transposase of Tn2 |
| strB*2 | 80893-81625 | Streptomycin phosphotransferase (nonfunctional) |
| tnpB*2 | Compl. 81597-81668 | Putative transposase (nonfunctional) |
| tnpA | 81732-82436 | TnpA transposase of IS26-3 |
| ΔtnpA | 82489-82669 | TnpA transposase of ISEcp1 (nonfunctional) |
| blaACC-4 | 82963-84123 | ACC-4 cephalosporinase precursor |
| gdhA | Compl. 84186-85100 | Glutamate dehydrogenase |
| ΔtnpA | Compl. 85123-85297 | TnpA transposase of Tn2 (nonfunctional) |
| tnpA | 85361-86065 | TnpA transposase of IS26-4 |
| orfB | Compl. 86177-86626 | Hypothetical protein |
| nikB | Compl. 86694-89405 | Relaxase |
| nikA | Compl. 89417-89836 | Relaxosome component protein |
| yggA | 89976-90317 | Hypothetical protein |
| ydiA | Compl. 90403-91254 | Hypothetical protein |
| ydhA | Compl. 91376-91747 | Hypothetical protein |
| ygeA | 91831-92082 | Hypothetical protein |
| orf14 | Compl. 92113-92295 | Hypothetical protein |
| ydgA | Compl. 92358-93272 | Hypothetical protein |
| ccgAII | Compl. 93269-93658 | Prevention of RecA overproduction |
| ygdA | Compl. 93835-94197 | Hypothetical protein |
| ygcA | Compl. 94194-94628 | Hypothetical protein |
| orf15 | 94747-95244 | Hypothetical protein |
| ardA | Compl. 95360-95860 | Anti-restriction protein |
| ygaA | Compl. 96330-96926 | Hypothetical protein |
| psiA | Compl. 96923-97642 | Plasmid SOS inhibition protein A |
| psiB | Compl. 97639-98073 | Plasmid SOS inhibition protein B |
| yfhA | Compl. 98128-100086 | Hypothetical protein |
| orf16 | 100145-100378 | Hypothetical protein |
| ssb | Compl. 100436-100963 | Single-stranded DNA-binding protein |
| orf17 | 101084-101557 | Hypothetical protein |
| yffA | Compl. 101735-101926 | Hypothetical protein |
| yfeC | Compl. 101926-102345 | Hypothetical protein |
| yfeB | Compl. 102392-102817 | Hypothetical protein |
| yfdA | Compl. 103234-104004 | Hypothetical protein |
| yfcB | Compl. 103961-104482 | Hypothetical protein |
| yfcA | Compl. 104496-104717 | Hypothetical protein |
| yfbB | Compl. 104714-105400 | Hypothetical protein |
| yfbA | Compl. 105474-105779 | Hypothetical protein |
| yfaB | Compl. 105783-106709 | Hypothetical protein |
| yfaA | Compl. 106723-106992 | Hypothetical protein |
| impC | 107104-107352 | UV protection |
| impA | 107349-107786 | UV protection |
| impB | 107786-109057 | UV protection |
| parA-like gene | 109258-110184 | ParA-like partitioning protein |
GenBank accession no. GU256641.
“Compl.” indicates that the gene is the reverse complement of the positions shown.
FIG. 1.
Overview of the IncB plasmid pR3521. The main regions as well as the indicative genes are shown in the outer circle (a complete list of genes can be found in Table 1). The next circle shows ORFs in the plus orientation and the 3rd-circle ORFs in the minus orientation. The innermost circle shows the G+C content plotted against the average G+C content of the entire plasmid sequence (52.6%). The inner circle plots the G+C skew. MDR, multidrug resistant.
Plasmid scaffold.
pR3521 possessed a single replication region of 1,368 bp (positions 5133 to 6500) identical to that of the IncB plasmid pMU707 (GenBank accession no. M93062) (23). It also exhibited significant similarities, ranging from 92 to 99%, with the replication regions of the IncI1 plasmids R64, pO113, pSERB1, and pEK204 (GenBank accession no. AP005147, AY258503, AY686591, and EU935740, respectively). The replication region of pR3521 included, apart from repA, two segments, RNAI and RNAII, that control copy number by inhibiting RepA translation through an antisense-RNA-mediated mechanism (27). The origin of replication (ori) located downstream of repA and the cis regulatory region between repA and ori were identical to those of pMU707 (22).
The relatively large transfer region of pR3521 (>50 kb; positions 6866 to 59455) was upstream of the RepA-encoding sequence and shared extensive structural as well as sequence similarities (71 to 98%) with the transfer regions of the IncI1 plasmids pO113, pSERB1, and pEK204 (7, 14, 28). The transfer region was organized in two blocks, tra and trb. tra (positions 6866 to 47247) included 12 pil genes (pilI and pilL to pilV), encoding thin pili required for liquid matings (11), as well as 22 additional genes, namely, traB, traC, traE, traF, traH to traK, and traL to traY, implicated in conjugal transfer (12). Between traC and pilI, the adhesin-encoding yqiJ and yqiK genes were identified (2). The transfer region also contained a sog gene upstream of traL. SogL is a SogS-regulated DNA primase suppressing dnaG mutations (16). trb (positions 54734 to 59455) comprised the trbABC operon, whose products are also involved in conjugation (9). In the region intervening between tra and trb, a segment including excA, analogous to the exc gene of IncI1 plasmids required for surface exclusion (8), along with pndA and pndC was identified. The products of the pnd genes contribute to plasmid maintenance (9). Also, 10 ORFs of unknown function were scattered throughout tra. The oriT operon, which included the origin of transfer and the nikA and nikB genes (whose products form a relaxation complex at the oriT site [10]), was distantly located from the trbABC operon due to insertion of a 26.4-kb acquired region in the sequence intervening between trbC and nikB. However, the oriT operon was apparently functional, as indicated by the self-transfer capability of pR3521 (18).
The plasmidic backbone also included the following: an ardA gene (positions 95367 to 95867), encoding an antirestriction protein; a psiBA operon (positions 96930 to 98080), whose products, PsiB and PsiA, inhibit the SOS response (1); and an ssb locus (positions 100443 to 100970), encoding Ssb, a single-stranded DNA protein (25). At positions 107104 to 109057, an impCAB operon, implicated in survival and induction of mutagenesis under UV irradiation, was found (24). pR3521 also possessed a parA-like gene, involved in segregational stability.
The close relationship of the rep regions of the complex I plasmids (IncI, -B, -K, and -Z) has been established in earlier studies (5, 17). The structure of the backbone of pR3521, being the first fully characterized IncB plasmid, extends this similarity to additional regions, such as tra, further supporting the common origin of these replicons.
Multidrug resistance region.
A 26.4-kb mosaic region was inserted into an ORF (here designated orfA) of the plasmidic backbone that corresponded to the LH0063 ORF of p0113 (GenBank accession no. AY258503) (Fig. 2). A remnant of the first 167 bp of orfA (orfA*1) was found at the boundary of the acquired region. A segment of 281 bp, exhibiting no significant homology with any known sequence, was adjacent to the remnant of orfA. This segment was followed by a noncoding sequence (302 bp), sul2, strA, and an strB gene with a truncation of the 3′ end (strB*1) similar to that described for plasmid RSF1010 (GenBank accession no. M28829). strAB could be a remnant of Tn5393, as suggested by the presence of an inverted repeat (IR) characteristic of this transposon (positions 64704 to 64758). The module comprising trbC, orfA*1, sul2, strA, and strB*1 has also been described to occur in other partially characterized IncB plasmids from E. coli (3).
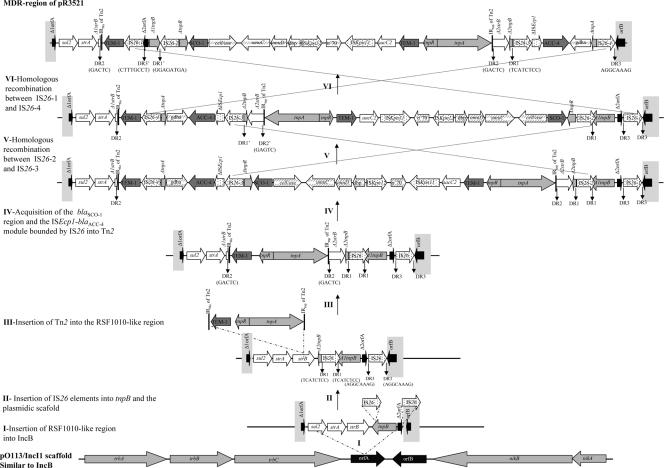
FIG. 2.
Schematic diagram of the hypothetical successive steps (I to VI) leading to the evolution of the multiresistant acquired region of pR3521 (top of the figure). Sequences are drawn to scale. ORFs are shown by arrows indicating direction of transcription. The sequence depicted at the bottom of the figure is from the IncB-related pO113/IncI plasmid. Target site duplications (TSD) generated by transposition are indicated as DR1, DR2, and DR3. The complement and reverse sequences of DR1, DR2, and DR3 are indicated as DR1′, DR2′, and DR3′, respectively.
A fragment of a Tn2-like transposon including a 38-bp inverted repeat (IRbla) and blaTEM-1 was adjacent to strB*1. IS26-1, along with the remaining part of orfA (orfA*2), was located upstream of blaTEM-1. However, orfA*2 was in an orientation opposite to that expected, indicating an inversion event. The bounding sequence contained ΔtnpB (also designated rcr2 in the IncB plasmid p838B-R; GenBank accession no. HM749967), IS26, ΔtnpR, blaSCO-1, and a glycosidase-like gene (18). Downstream (1,118 bp) of the glycosidase gene, two probably chromosomal genes, umuC and umuD (needed for maximal SOS mutagenesis [26]), as well as dbp, encoding a putative DNA-binding protein (15), were identified. A mosaic sequence comprising ISKpn12, ISKpn11, aacC2, and a Tn2 transposon was located upstream of dbp. This region was highly homologous to a sequence carried by K. pneumoniae 12836 (15). The IRtnp repeat of Tn2 was adjacent to the remaining part of strB (strB*2). Direct repeats (DRs) of 5 bp (GACTC) (DR2 in Fig. 2) were found within the coding sequences of strB*1 and strB*2 at the boundaries of IRs of Tn2. Downstream of strB*2, there was the remaining part of tnpB (tnpB*2) adjoining IS26-3. Notably, 8-bp reverse and complement sequences were located at the boundaries of tnpB*1 (tnpB with a truncation of the 3′ end) and IS26-2 (GGAGATGA) (DR1′ in Fig. 2) and tnpB*2 (tnpB with a truncation of the 5′ end) and IS26-3 (TCATCTCC) (DR1 in Fig. 2) as well as orfA*2 and IS26-1 (CTTTGCCT) (DR3′ in Fig. 2) and orfB and IS26-4 (AGGCAAAG) (DR3 in Fig. 2). The reverse and complement orientation of these 8-bp sequences suggested IS26-mediated inversions. Upstream of the IS26-4 sequence was an ACC-4-encoding segment that has been described previously (19). This segment comprised an ISEcp1 element that was truncated (due to insertion of IS26-3), a fragment that originated from the chromosome of H. alvei (blaACC-4 and gdhA), and an IS26-4 sequence in parallel orientation with IS26-3, therefore forming a class I composite transposon.
Hypotheses on the formation of the acquired region.
The multiresistant region apparently arose from multiple insertions and DNA rearrangements (Fig. 2). A possible initial event could be the insertion of the RSF1010-originated module comprising sul2, strA, strB, and tnpB within orfA of pR3521 (possible step I). This hypothesis is corroborated by sequencing data from other IncB plasmids, such as p99309, p99051, and p99171, showing insertion of similar modules in orfA-like genes (3). A subsequent event could be the insertion of two IS26 elements, one disrupting the 3′ end of the tnpB gene and another inserted into the IncB scaffold in the sequence intervening between orfA*2 and orfB, thus creating 8-bp DRs at the boundaries of the elements (DR1 and DR3, respectively) (possible step II). Truncation of strB was likely due to transposition of a Tn2-like element, as indicated by the target site duplications (TSD) (DR2) flanking the IRs of the transposon (possible step III). Of note, the structure resulting from steps I, II, and III was similar to a sequence identified in the IncB plasmid p838B-R. A SCO-1-encoding segment (comprising blaSCO-1, the glycosidase gene, umuC, and umuD) and a module comprising ISEcp1, blaACC-4, and gdhA, both associated with IS26 elements, could then have been inserted within Tn2 (possible step IV). It was not clear whether ISKpn11 and ISKpn12 had been inserted independently or recruited in a single event along with Tn2. However, the latter notion is supported by the existence of a similar sequence (comprising umuD, dbp, ISKpn12, the σ′70 gene, ISKpn11, Tn1000, and the Tn2* hybrid) in K. pneumoniae 12836 (15). Homologous recombination between IS26-2 and IS26-3 and then recombination between IS26-1 and IS26-3 could be proposed as the final events in the evolution of the acquired region (possible steps V and VI). This assumption is in line with the reverse and complement orientation of the 8-bp sequences found within tnpB*1 (DR1′) and orfA*2 (DR3′). The related sequences discussed by Doloy et al. (6) and Partridge (20) underscore the central role of IS26 in the formation of this structure type.
Nucleotide sequence accession number.
The complete nucleotide sequence of plasmid pR3521 has been assigned GenBank accession no. GU256641.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Althorpe, N. J., P. M. Chiley, A. T. Thomas, W. J. Brammar, and B. M. Wilkins. 1999. Transient transcriptional activation of the IncI1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol. Microbiol. 31:133-142. [DOI] [PubMed] [Google Scholar]
- 2.Antão, E.-M., L. H. Wieler, and C. Ewers. 2009. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean, D. C., D. M. Livermore, and L. M. C. Hall. 2009. Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob. Agents Chemother. 53:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carattoli, A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couturier, M., F. Bex, P. L. Bergquist, and W. K. Maas. 1988. Identification and classification of bacterial plasmids. Microbiol. Rev. 52:375-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doloy, A., C. Verdet, V. Gautier, D. Decré, E. Ronco, A. Hammani, A. Philippon, and G. Arlet. 2006. Genetic environment of acquired blaACC-1 β-lactamase gene in Enterobacteriaceae. Antimicrob. Agents Chemother. 50:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley, E. G., C. Abe, J. M. Ghigo, P. Latour-Lambert, J. C. Hormazabal, and J. P. Nataro. 2006. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect. Immun. 74:2102-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuya, N., and T. Komano. 1994. Surface exclusion gene of IncI1 plasmid R64: nucleotide sequence and analysis of deletion mutants. Plasmid 32:80-84. [DOI] [PubMed] [Google Scholar]
- 9.Furuya, N., and T. Komano. 1996. Nucleotide sequence and characterization of the trbABC region of the IncI1 plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 178:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuya, N., T. Nisioka, and T. Komano. 1991. Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J. Bacteriol. 173:2231-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komano, T., N. Funayama, S.-R. Kim, and T. Nisioka. 1990. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 172:2230-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komano, T., T. Yoshiba, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348-1359. [DOI] [PubMed] [Google Scholar]
- 13.Ktari, S., G. Arlet, C. Verdet, S. Jaoua, A. Kachrid, S. Ben Redjeb, F. Mahjoubi-Rhimi, and A. Hammami. 2009. Molecular epidemiology and genetic environment of acquired blaACC-1 in Salmonella enterica serotype Livingstone causing a large nosocomial outbreak in Tunisia. Microb. Drug Resist. 15:279-286. [DOI] [PubMed] [Google Scholar]
- 14.Leyton, D. L., J. Sloan, R. E. Hill, S. Doughty, and E. L. Hartland. 2003. Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter. Infect. Immun. 71:6307-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Márquez, C., M. Labbate, C. Raymondo, J. Fernadez, A. M. Gestal, M. Holley, G. Borthagaray, and H. W. Stokes. 2008. Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J. Clin. Microbiol. 46:3417-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merryweather, A., C. E. D. Rees, N. M. Smith, and B. M. Wilkins. 1986. Role of sog polypeptides specified by plasmid ColIb-P9 and their transfer between conjugating bacteria. EMBO J. 5:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikoletti, S., P. Bird, J. Praszkier, and J. Pittard. 1988. Analysis of the incompatibility determinants of I-complex plasmids. J. Bacteriol. 170:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papagiannitsis, C. C., A. Loli, L. S. Tzouvelekis, E. Tzelepi, G. Arlet, and V. Miriagou. 2007. SCO-1, a novel plasmid-mediated class A β-lactamase with carbenicillinase characteristics from Escherichia coli. Antimicrob. Agents Chemother. 51:2185-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papagiannitsis, C. C., L. S. Tzouvelekis, E. Tzelepi, and V. Miriagou. 2007. Plasmid-encoded ACC-4, an extended-spectrum cephalosporinase variant from Escherichia coli. Antimicrob. Agents Chemother. 51:3763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge, S. R. 2007. Genetic environment of ISEcp1 and blaACC-1. Antimicrob. Agents Chemother. 51:2658-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., S. Corvec, M. Rapoport, P. Mugnier, A. Petroni, F. Pasteran, D. Faccone, M. Galas, H. Drugeon, V. Cattoir, and P. Nordmann. 2007. Identification of the novel narrow-spectrum β-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 51:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Praszkier, J., S. Murthy, and J. Pittard. 2000. Effect of CIS on activity of the replication initiator protein of an IncB plasmid. J. Bacteriol. 182:3972-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Praszkier, J., T. Wei, K. Siemering, and J. Pittard. 1991. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J. Bacteriol. 173:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shereda, R. D., D. A. Bernstein, and J. L. Keck. 2007. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 282:19247-19258. [DOI] [PubMed] [Google Scholar]
- 26.Smith, C. M., and E. Eisenstadt. 1989. Identification of a umuDC locus in Salmonella typhimurium LT2. J. Bacteriol. 171:3860-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, I. W., J. Praszkier, and J. Pittard. 1994. Molecular analysis of RNAI control of repB translation in IncB plasmids. J. Bacteriol. 176:6497-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodford, N., A. Carattoli, E. Karisik, A. Underwood, M. J. Ellington, and D. M. Livermore. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53:4472-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]