Abstract
Amoxicillin is commonly used to treat Helicobacter pylori, a major cause of peptic ulcers, stomach cancer, and B-cell mucosa-associated lymphoid tissue lymphoma. Amoxicillin resistance in H. pylori is increasing steadily, especially in developing countries, leading to treatment failures. In this study, we characterize the mechanism of amoxicillin resistance in the U.S. clinical isolate B258. Transformation of amoxicillin-susceptible strain 26695 with the penicillin binding protein 1 gene (pbp1) from B258 increased the amoxicillin resistance of 26695 to equal that of B258, while studies using biotinylated amoxicillin showed a decrease in the binding of amoxicillin to the PBP1 of B258. Transformation with 4 pbp1 fragments, each encompassing several amino acid substitutions, combined with site-directed mutagenesis studies, identified 3 amino acid substitutions in PBP1 of B258 which affected amoxicillin susceptibility (Val 469 Met, Phe 473 Leu, and Ser 543 Arg). Homology modeling showed the spatial orientation of these specific amino acid changes in PBP1 from 26695 and B258. The results of these studies demonstrate that amoxicillin resistance in the clinical U.S. isolate B258 is due solely to an altered PBP1 protein with a lower binding affinity for amoxicillin. Homology modeling analyses using previously identified amino acid substitutions of amoxicillin-resistant PBP1s demonstrate the importance of specific amino acid substitutions in PBP1 that affect the binding of amoxicillin in the putative binding cleft, defining those substitutions deemed most important in amoxicillin resistance.
Helicobacter pylori infection is strongly associated with the development of numerous gastric pathologies, including chronic type B gastritis, gastric and duodenal ulcers, gastric adenocarcinoma, and B-cell mucosa-associated lymphoid tissue lymphoma (10). It is the most common cause of peptic ulcer disease (31) and is responsible for 5.5% of the cases of human cancer each year (27). There is also some evidence to suggest that H. pylori may infect other organs, including the liver (10), and may be associated with Alzheimer's disease (20). Currently, the rates of H. pylori infection in middle-aged adults are 20 to 50% in industrialized countries and 80% or more in many developing countries (11, 32), with higher prevalence rates seen in people of low socioeconomic status (11).
Effective treatment of H. pylori infection typically requires a combination of drugs, including two antibiotics and a proton pump inhibitor or a bismuth compound (14). First-line antibiotics include amoxicillin, clarithromycin, tetracycline, and metronidazole (14). While often effective, as many as 20 to 30% of these treatments fail, usually as a result of a lack of patient compliance or, increasingly, antibiotic resistance (14).
Antibiotic resistance of H. pylori to many of these antibiotics is increasing, with various levels of resistance noted (22). The rates of resistance are higher in developing countries than in developed countries, mainly due to the variation of prescribing patterns (17). Many strains of H. pylori are naturally competent, allowing it to take up resistance genes from other microbes, as well as naked DNA from the environment (25, 35). However, most of the antibiotic resistance mechanisms described to date are a result of point mutations on the bacterial chromosome (14), a consequence of a significantly high mutation rate in H. pylori (23).
Although the prevalence rates of amoxicillin resistance in H. pylori strains in the United States are generally still very low, resistance rates as high as 33% have been seen in other countries (17). Resistance to β-lactam drugs in Gram-negative bacteria is mostly due to β-lactamase activity (2). Although the H. pylori genome contains a β-lactamase-like gene which codes for the protein HcpA (24), this does not appear to confer amoxicillin resistance. Tseng et al. (34) recently described the presence of a TEM β-lactamase conveying high-level amoxicillin resistance in a clinical isolate from Taiwan; however, amoxicillin resistance in most clinical isolates of H. pylori is not due to β-lactamase activity. For these strains, resistance seems to be mainly mediated by alterations to penicillin binding proteins (PBPs). The role of amino acid mutations in PBP1 in amoxicillin resistance has been described in a number of studies, both in clinical isolates (12, 13, 19, 21, 26, 29) and in in vitro-derived amoxicillin-resistant strains (4, 9, 28). In some cases, this moderate to low-level amoxicillin resistance has been attributed to single amino acid substitutions (4, 13, 21), whereas other studies have reported multiple amino acid changes (12, 19, 26, 28, 29).
In this report, we have characterized the amoxicillin resistance of a clinical isolate identified in the United States (to our knowledge, the first U.S. strain to be characterized), identified the role of amino acid substitutions in PBP1 in resistance, and used homology modeling to clarify the role of these substitutions in affecting amoxicillin binding. We have also examined the previously published amino acid substitutions with structural and physicochemical analysis of the homology models in order to better understand their contributions to amoxicillin resistance. This approach has led to the hypothesis of a defined set of amino acid substitutions in PBP1 which affect amoxicillin binding and, consequently, reduce the susceptibility of these strains to the bactericidal effect of amoxicillin.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The H. pylori clinical isolate B258 was kindly provided by Richard M. Peek, Jr., Department of Gastroenterology, Vanderbilt University Medical Center. H. pylori ATCC strain 26695 was used as a reference strain and for creating transformants. Stock cultures of H. pylori B258 and 26695 were streaked for isolation on brucella agar (Becton-Dickinson Microbiology, Cockeysville, MD) supplemented with 5% defibrinated sheep blood (Colorado Serum Company, Denver, CO) and cultured at 37°C in a humidified 10% CO2 incubator. Liquid cultures were prepared by suspension of H. pylori colonies in brucella broth (Difco Laboratories, Detroit, MI) supplemented with 10% heat-inactivated fetal bovine serum (Gibco Bethesda Research Laboratories, Grand Island, NY) and 1% IsoVitaleX (Becton-Dickinson Microbiology). Cultures were routinely passed by dilution into fresh medium at 48-h intervals. Freezer stocks of cultures were prepared by resuspending 48-h cultures in 1% proteose peptone-20% glycerol, flash frozen in liquid nitrogen, and kept at −80°C.
General DNA techniques.
All DNA manipulations were performed according to standard protocols. The primers used in this study were created using the published sequence of pbp1 of 26695 and vector NTI (Invitrogen, Carlsbad, CA) and synthesized by Sigma-Aldrich (The Woodlands, TX). PCR was performed in an automated PTC100 thermal cycler (MJ Research, Ramsey, MN), using Choice-Taq Blue polymerase (Denville Scientific, Metuchen, NJ). PCR products were run on 1% agarose gel (Bio-Rad, Hercules, CA) and subsequently cut out and purified using the MinElute agarose purification system (Qiagen, Inc., Valencia, CA). Sequencing of purified DNA products was performed by the Genomics Core facility at the University of California, Riverside.
Transformation of H. pylori.
Transformations of the 26695 reference strain were done using ∼200 ng of purified PCR-amplified DNA and the electroporation protocol described by Wang et al. (36), using an ECM 630 electroporator (BTX, Hawthorne, NY). Transformation mixtures were plated on brucella agar supplemented with 5% defibrinated sheep blood; after 48 to 72 h, transformants were selected on brucella agar medium supplemented with 5% defibrinated sheep blood and 0.125 μg/ml amoxicillin.
Generation of 26695_T and other transformants.
The transpeptidase region of pbp1 from B258 was PCR amplified, purified, and subsequently used to transform strain 26695. Various fragments of the transpeptidase domain from B258 (as shown in Fig. 2a) were PCR amplified, purified, and used to transform 26695. Site-directed mutations were created for the Ser 543 Arg and Met 533 Ile loci using the QuikChange mutagenesis protocol (Stratagene, La Jolla, CA). Potential transformants were selected as indicated above, and pbp1 genes were amplified and sequenced to confirm transformation. The MICs of antibiotics were determined for B258 and all transformants using the broth microdilution method according to a previously published protocol (8).
Bio-Amox labeling of PBP1.
Biotinylated amoxicillin (bio-Amox) was prepared by the method of Dargis and Malouin (7). Whole cells were obtained from 48-h cultures from agar medium, suspended in 0.1 M sodium phosphate buffer, pH 7.4, and then adjusted for consistent protein concentrations using a Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA). Equal protein aliquots of approximately 100 μg were reacted with bio-Amox at concentrations of 4, 2, 1, 0.4, 0.2, 0.1, 0.05, and 0.02 μg/ml for 30 min at room temperature. Proteins were then separated using sodium dodecyl sulfate-14% polyacrylamide gel electrophoresis (SDS-14% PAGE). After electrophoresis, proteins were transferred to nitrocellulose and prepared for detection by chemiluminescence as described previously (8). Density readings were taken using Odyssey software (Li-Cor, Lincoln, NE).
Antibiotic competition experiment.
In antibiotic competition experiments, 26695 and 26695_T whole cells were obtained from 48-h agar cultures, resuspended into 0.1 M sodium phosphate buffer, pH 7.4, adjusted for consistent protein concentrations (∼100 μg), and incubated with penicillin G at 0.1, 1, and 10 μg/ml for 30 min at room temperature. These samples were then incubated with 2 μg/ml of bio-Amox for 30 min at room temperature. Bio-Amox-labeled proteins were detected by chemiluminescence, and density readings were taken as described above. The 50% inhibitory concentration (IC50) for penicillin G for PBP1 was estimated using densitometry tracing data.
Homology modeling.
Homology models of the transpeptidase region of PBP1 from H. pylori 26695 and B258 were created using as a template the crystal structure of PBP1A from Streptococcus pneumoniae (chain A, residues 267 to 650, PDB [Protein Data Bank] code 2C5W) (5). The sequence of Streptococcus pneumoniae PBP1A was the best match to the sequence of H. pylori among the deposited structures in the PDB at the time of modeling. We also examined the suitability of the structure of Streptococcus pneumoniae PBP1B for multiple template homology modeling; however, despite the overall similar fold, there are significant loop length differences among the three structures, rendering the structure of Streptococcus pneumoniae PBP1B nonoptimal for use as a template. The Swiss-Model server (http://swissmodel.expasy.org) (15) was used to perform homology modeling of the transpeptidase region (amino acids 262 to 628), using complete PBP1 protein sequences from H. pylori strains. The transpeptidase domain of the two PBP1 proteins is 24% identical and 38% similar to that of PBP1A from Streptococcus pneumoniae. The homology models were visualized using the program DeepView (15) and were inspected for van der Waals clashes, secondary structure quality, and disulfide bond correctness. Comparative analyses of hydrogen bonds, salt bridges, and charge and apparent pKa values were performed for the mutated amino acids in PBP1s from 26695 and B258.
Electrostatic calculations.
Electrostatic potential calculations were performed using the Adaptive Poisson-Boltzmann Solver (APBS) (1) implementation within the program VMD (18) and the PARSE force field (33). A dielectric coefficient of 4 was used for the protein, whereas a dielectric coefficient of 78.54 was used for the solvent in the absence of solvent counterions. The temperature was set to 298.15 K. The dielectric surface was defined as the protein contact surface using a sphere with probe radius of 1.4 Å, corresponding to that of a water molecule. The grid size was 129 by 129 by 129 points, and the box sizes were 105 Å by 127 Å by 111 Å for the coarse grid and 82 Å by 95 Å by 86 Å for the fine grid.
RESULTS
Characterization of H. pylori strain B258.
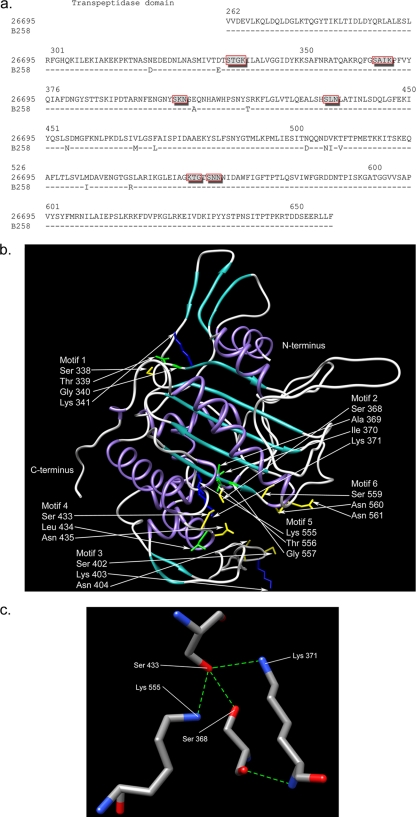
The MIC for clinical strain B258 is 1 μg/ml for amoxicillin; by comparison, our reference strain 26695 has a MIC of 0.06 μg/ml (Table 1). Strain B258 had similar MICs for ampicillin and penicillin but a MIC of 2 μg/ml for cefotaxime. Sequencing of the transpeptidase domain of pbp1 of strain B258 revealed 13 amino acid changes in comparison to the reference strain 26695, as shown in Fig. 1a. The transformation of 26695 with the transpeptidase region of pbp1 from B258, creating 26695_T, resulted in an increase in its MIC for amoxicillin to 1 μg/ml, equivalent to that of B258 (Table 1), suggesting that these changes in PBP1 account for the entire resistance to amoxicillin seen in B258. Strain 26695_T also had increased resistance for the other 3 β-lactams tested (Table 1).
TABLE 1.
MIC determinations of reference strain 26695, clinical isolate B258, and 26695 transformed with pbp1 from B258 (26695_T) for several β-lactam antibiotics
| β-Lactam | MIC (μg/ml) for strain: |
||
|---|---|---|---|
| 26695 | B258 | 26695_T | |
| Amoxicillin | 0.06 | 1 | 1 |
| Ampicillin | 0.125 | 1 | 2 |
| Penicillin G | 0.125 | 1 | 2 |
| Cefotaxime | 1 | 2 | 4 |
FIG. 1.
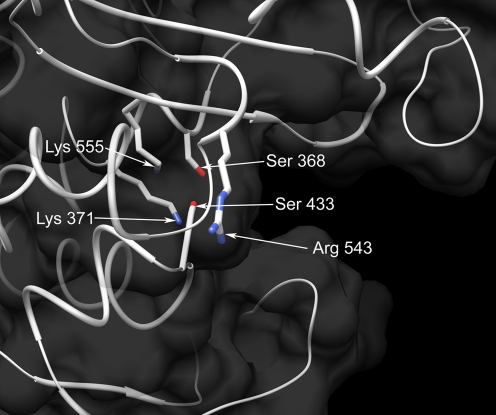
(a) Alignment of the PBP1 transpeptidase domains from 26695 and B258. The dashes indicate no change in B258 at these loci in comparison to the 26695 sequence. Specific amino acid substitutions are denoted by the letter of the changed amino acid. Boxed amino acids indicate putative penicillin binding motifs (PBMs). (b) Secondary structure of PBP1 transpeptidase region from 26695 depicted in ribbon representation (purple, alpha-helices; cyan, beta-sheets; gray, loops). The putative PBMs (boxed amino acids in panel a) are also shown, with amino acid side chains depicted in stick representation and colored by type (green, hydrophobic; yellow, polar; blue, polar positively charged). All other side chains have been deleted for clarity. (c) Hydrogen bond network within the putative binding cleft of the PBP1 transpeptidase region from 26695, involving the putative catalytic Ser 368 with Ser 433, Lys 371, and Lys 555 (blue, nitrogen; red, oxygen; grey, carbon; green dashed lines, hydrogen bonds). The putative catalytic Ser 368 is also amenable to weak hydrogen bonds with Lys 371 and Lys 555 (dashed lines not shown).
Effects of various amino acid substitutions in PBP1 on amoxicillin susceptibility.
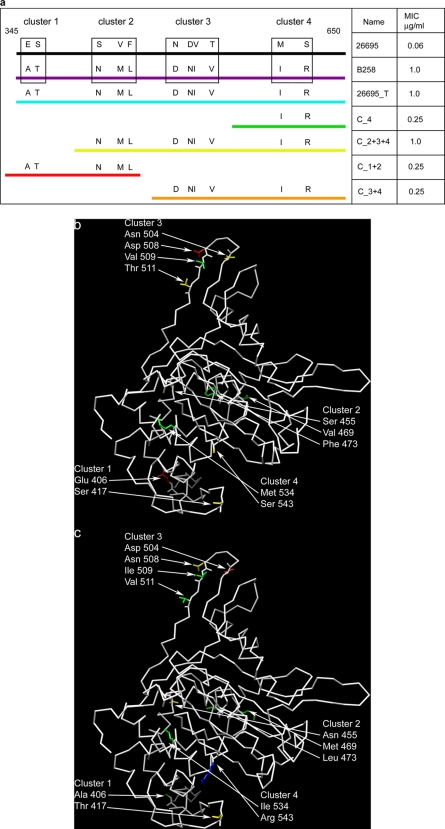
Strain 26695 was transformed with various segments of pbp1 from B258, representing clusters of amino acid substitutions (Fig. 2a), and the transformants were selected on amoxicillin-containing plates. Transformant C_2+3+4, which includes 9 of the observed amino acid substitutions, had a MIC of amoxicillin of 1 μg/ml, accounting for the entire resistance of B258. This suggests that the 4 upstream amino acid substitutions (Asn 322 Asp, Asp 336 Glu, Glu 406 Ala, and Ser 417 Thr, shown in Fig. 1a) do not play a significant role in conferring amoxicillin resistance. Consistent with this conclusion, we were unable to isolate a C_1 transformant (containing Glu 406 Ala and Ser 417 Thr) on amoxicillin-containing plates; these amino acids are located on the external face of the protein, close to but not within the putative binding cleft (Fig. 2b and c). However, transformant C_1+2 has a MIC of 0.25 μg/ml for amoxicillin, and based on the information above, this would suggest that amino acid substitutions Ser 455 Asn, Val 469 Met, and Phe 473 Leu are responsible for this increased resistance to amoxicillin. Transformant C_3+4 and transformant C_4 both have MICs of 0.25 μg/ml for amoxicillin, suggesting that the 4 amino acid changes seen in cluster 3 do not contribute to amoxicillin resistance; as seen in Fig. 2b and c, cluster 3 is located at a site removed from the putative binding cleft. In a separate experiment (data not shown here), a single point mutation, Ser 543 Arg, was created in 26695, resulting in a MIC of 0.25 μg/ml for amoxicillin, which suggests that Met 533 Ile does not contribute to amoxicillin resistance (which was confirmed by the inability of a 26695 strain transformed with a PBP1 containing only the Met 533 Ile substitution to demonstrate any increase in amoxicillin resistance; data not shown). A transformant containing Val 469 Met and Phe 473 Leu from cluster 2 in addition to Ser 543 Arg was created in 26695, and this construct had a MIC of 1.0 μg/ml, suggesting that these 3 sites were sufficient to account for all of the amoxicillin resistance of B258. These amino acids are located in the vicinity of the large putative binding cleft (Fig. 2b and c). Several efforts to select a Phe 473 Leu Ser 543 Arg double mutant, without Val 469 Met, have been unsuccessful.
FIG. 2.
(a) The amino acid substitutions seen in the PBP1 transpeptidase domain from B258 were divided into clusters based on their sequence proximity to each other. Strains 26695, B258, and 26695_T contained all clusters. Other transformants contained different combinations of clusters as indicated by the numbers in their names. (b) Alpha-carbon trace representation of transpeptidase region of PBP1 of 26695 with the side chains of amino acids changed to stick representation (all other amino acids are deleted for clarity). The color code is by amino acid type (green, hydrophobic; yellow, polar; blue, polar positively charged; red, polar negatively charged). (c) Alpha-carbon trace model of transpeptidase region of PBP1 of B258 showing amino acid substitutions as described for panel b.
PBP1 binding.
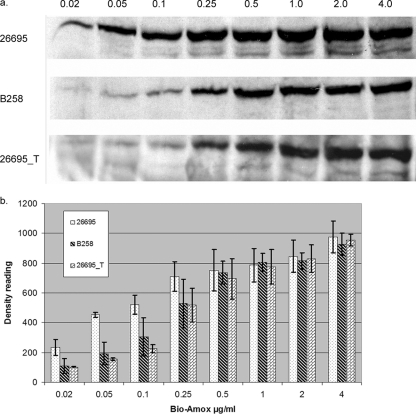
Whole cells from strains 26695, B258, and 26695_T were prepared and incubated with bio-Amox as described in Materials and Methods. The results of a representative experiment are shown in Fig. 3a, and the data from repeat experiments analyzed quantitatively are presented in Fig. 3b. Labeling of 26695_T and B258 was identical at each bio-Amox concentration and was lower than that of 26695 for all bio-Amox concentrations of <0.5 μg/ml. We also examined the affinity of the PBP1s from 26695 and 26695_T for amoxicillin using an antibiotic competition experiment. Each strain was preincubated with penicillin G at 0.1, 1, or 10 μg/ml for 30 min prior to incubation with 2 μg/ml of bio-Amox. The IC50 for penicillin G for PBP1 in 26695 was estimated to be 0.57 μg/ml, whereas it was 29.7 μg/ml for PBP1 in 26695_T.
FIG. 3.
(a) Bio-Amox-labeling profiles of PBP1 of H. pylori 26695, B258, and 26695_T. Numbers above the lanes indicate the amount of bio-Amox used in the experiment, in μg/ml. (b) Densitometric comparisons of PBP1 from the 26695, B258, and 26695_T strains labeled with various concentrations of bio-Amox (shown in μg/ml on the x axis). Bars represent the mean results ± standard deviations based on 3 separate experiments.
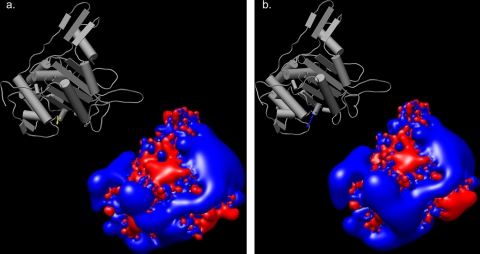
Homology modeling of PBP1 from H. pylori.
Protein modeling of the transpeptidase region placed 5 of the 6 putative penicillin binding motifs (PBMs) (16) within a putative binding cleft (Fig. 1b). The first motif, however, mapped away from the cleft. The Ser 543 Arg transformation results in an increase in protein net charge from +5 to +6. This charge increase results in a significant increase in the spatial distribution of positive electrostatic potential, which forms an electrostatic hot spot surrounding the putative binding cleft (Fig. 4a and b). The arginine at 543, although about 19 Å away from the putative catalytic Ser 368, is located in a loop in the vicinity of the large putative binding cleft and may interfere with the binding of the β-lactam antibiotic (Fig. 5). The arginine at 543 may be responsible for diverting the negatively charged β-lactam antibiotic from the site of catalysis, either through nonspecific long-range electrostatic recognition or through specific short-range charge-charge interaction or both. This type of electrostatic steering leading to recognition and binding has been discussed in the case of protein-protein interactions (3, 37-39). The remaining substitutions in the PBP1 of B258 that had an effect on the MIC, Val 469 Met and Phe 473 Leu, are about 20 Å away from the catalytic Ser 368, but they are located in a loop enclosing the putative binding cleft.
FIG. 4.
Isopotential contour surfaces depicting the spatial distributions of electrostatic potentials of the transpeptidase region of PBP1. Blue and red represent positive and negative electrostatic potentials, respectively, at ±2kBT/e units (kB, Boltzmann constant; T, temperature; e, electron charge). (a) Transpeptidase region of PBP1 from 26695. Inset shows Ser 543 in yellow. (b) Transpeptidase region of PBP1 from B258. Inset shows Arg 543 in blue.
FIG. 5.
Surface representation of the cleft showing amino acids within 4 Å of the putative catalytic serine (Ser 368) of the PBP1 transpeptidase region from 26695. In addition, although Arg 543 from the superimposed structure B258 is located about 19 Å away from the putative catalytic Ser 368, it potentially interferes with ligand entry. Blue, nitrogen; red, oxygen; white, carbon.
Our homology modeling (or the crystallographic structure of the template, for that matter) does not depict protein dynamics, which are more pronounced in solvent-exposed (>20% solvent accessibility) flexible loop regions. Substitutions Val 469 Met and Phe 473 Leu belong to the 459-to-476 loop. Met 469 and Leu 473 have solvent accessibilities of 43% and 11%, respectively, in the modeled loop conformation (out of many possible conformations due to inherent loop flexibility). The substituted amino acids, Val 469 and Phe 473 in the case of PBP1 of 26695, have solvent accessibilities of 30% and 17%. Substitution Ser 543 Arg belongs to the 540-to-552 loop. Arg 543 has a solvent accessibility of 40%. The substituted Ser 543 in the case of PBP1 of 26695 has a solvent accessibility of 25%. (Solvent accessibilities were calculated in the presence of the hydrogen atoms in the model structures.) Higher solvent accessibility values correspond to amino acids in highly flexible portions of the loops. The flexible loop on which Ser 543 is located is likely to play an important role in the dynamics of protein-ligand binding. Therefore, the significant change of Ser 543 Arg, which modifies both the steric bulk and the charge of this loop, may conceivably affect the binding kinetics, thermodynamics, or both. The Phe 473 Leu change may result in the loss of a very weak π-cation interaction between the aromatic ring of Phe 473 and Lys 464. The steric hindrance argument applies to substitution Ser 543 Arg, which occurs in the solvent-exposed mobile loop, in addition to the electrostatic arguments described above. Most of the other amino acid substitutions in the PBP1 of B258 were not predicted to make any significant change to the putative binding cleft. The putative catalytic serine at position 368 (5) is in the middle of the cleft (Fig. 1b) and participates in a complex hydrogen bond network (Fig. 1c). Although the putative catalytic Ser 368 is well anchored by hydrogen bonds, the mobility of the surrounding loops of the putative binding cleft may be responsible for controlling ligand entry and binding, depending on the type of loop mutations. Molecular dynamics simulations are planned to study the effects of loop flexibility and mobility on the formation of the putative binding cleft.
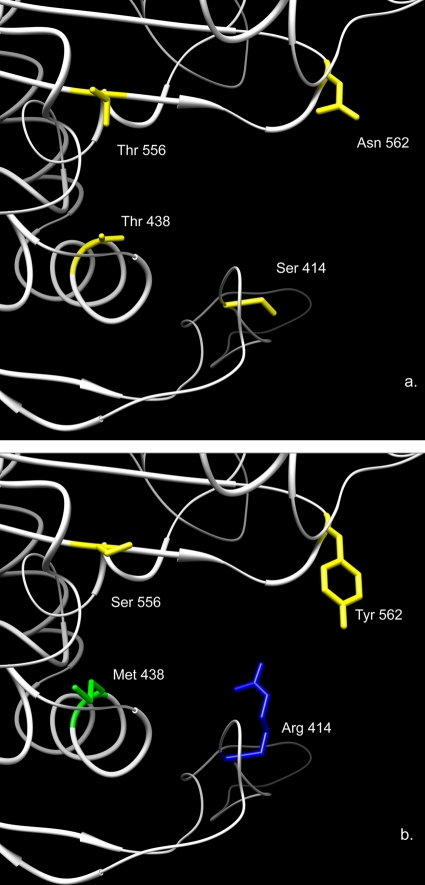
Previously published data identified several mutations in PBP1 which confer resistance on H. pylori (4, 12, 13, 19, 21, 26, 28, 29, 34), and we chose to visualize the putative effect of three of the most commonly reported mutations, namely, Ser 414 Arg, Thr 556 Ser, and Asn 562 Tyr, as well as the Thr 438 Met mutation we previously reported in an in vitro-derived amoxicillin-resistant strain (4). The mutation Ser 414 Arg results in an increased net positive charge comparable to that seen with Ser 543 Arg. The mutations of these two serines to arginines perhaps share the same mechanism of interaction with the β-lactam antibiotic, although Ser 414 Arg is located right at the entrance of the binding cleft and may be more prone to specific charge-charge interaction with the β-lactam antibiotic (Fig. 6a and b). The Thr 438 M and Asn 562 Tyr mutations result in replacement with an amino acid of a significantly larger size, likely causing steric hindrance within the putative binding cleft (Fig. 6a and b). Thr 556 Ser is not a major amino acid change; however, it is adjacent to Lys 555 and in proximity to putative catalytic serine, possibly altering the hydrogen bonding patterns of Ser 368. Other amino acid substitutions did not contribute to resistance regardless of their proximity to the PBMs or the putative binding cleft (e.g., Glu 406 Ala or Ser 417 Thr).
FIG. 6.
(a) The localization and stick structure representation of 4 key amino acids within the putative binding cleft in strain 26695 (amoxicillin susceptible). (b) The predicted stick structure representation of unique amino acid substitutions for these 4 amino acids which have been previously reported to increase amoxicillin resistance in various clinical strains. Amino acid side chains depicted in stick representations are colored by type (green, hydrophobic; yellow, polar; blue, positively charged).
DISCUSSION
Our initial studies demonstrated that the MICs of H. pylori clinical isolate B258 to ampicillin, amoxicillin, and penicillin G were significantly increased (8-fold or greater) in comparison to those of the amoxicillin-susceptible reference strain 26695. On the other hand, the MIC to cefotaxime was only modestly increased. PCR amplification and sequencing of the pbp1 gene from B258 identified 13 amino acid substitutions in comparison to the pbp1 gene from 26695. Replacement of the transpeptidase domain (amino acids 600 to 1850) of the pbp1 gene in 26695 with the pbp1 gene from B258 resulted in the transformant 26695_T, with a MIC for amoxicillin identical to that of B258. The results of this experiment demonstrated that the amoxicillin resistance of strain B258 is due entirely to amino acid changes in PBP1. It should be noted that besides PBP1 changes, there have been a few reports describing changes in outer membrane permeability which have affected amoxicillin susceptibility (4, 19), whereas in other reports, only part of the amoxicillin resistance has been attributed to PBP1 changes and the other resistance mechanisms remain uncharacterized (12, 13, 26, 28, 29). Finally, Rimbara et al. (30) reported that amino acid mutations in PBP2 and PBP3, in concert with although of less consequence than amino acid changes in PBP1, increase amoxicillin resistance.
In our studies using bio-Amox labeling, the transformation of 26695 with the transpeptidase domain of PBP1 from B258 demonstrates that the increased amoxicillin resistance of B258 is due to decreased binding of amoxicillin to the transpeptidase domain of PBP1. Since penicillin G, ampicillin, and amoxicillin are structurally similar, it is not surprising that the altered PBP1 in B258 would bind less effectively to each of these antibiotics, resulting in an increased MIC for each of these antibiotics. Using antibiotic competition experiments, we calculated the IC50 for penicillin to be more than 50 times higher for the 26695_T PBP1 than for the PBP1 of the amoxicillin-susceptible strain 26695. While other studies have reported that amino acid substitutions in PBP1 affect amoxicillin resistance, this study is one of the few to both demonstrate reduced amoxicillin binding and calculate the IC50 for the altered PBP1.
Using chimeric PBP1 transformants, we were able to eliminate from consideration or establish the minimal importance of most of the amino acid substitutions in PBP1 in strain B258 in conferring amoxicillin resistance, suggesting 3 amino acid sites as critical. Although the percent identity between the homology model and its template was relatively low (24% identity and 38% similarity), the experimental data from chimeric transformants supported our results. We confirmed the importance of the Ser 543 Arg mutation by using site-directed mutagenesis of the pbp1 gene in 26695, resulting in a 4-fold increase in amoxicillin resistance. This same site has also been reported to be important for amoxicillin resistance by Kwon et al. (19).
As seen in the projected model, penicillin binding motifs 3 and 6 are located at the entrance points of the putative binding cleft, perhaps as gatekeepers for antibiotic entry (Fig. 1b). Lys 371 (motif 2), Ser 433 (motif 4), and Lys 555 (motif 5) are projected to form hydrogen bonding interactions with the putative catalytic Ser 368 (Fig. 1c). The replacement of Ser 543 with Arg 543 increases the overall charge by one unit and renders a stronger positive electrostatic field to the protein, which may interfere with the recognition and binding of the negatively charged β-lactam antibiotic (Fig. 4). The increase and change in the directionality of the positive electrostatic field caused by the Ser 543 Arg mutation may increase the recognition ability for β-lactam and divert its entry path away from the catalytic site. Another or an additional mechanism involves possible local Coulombic attraction between Arg 543+ and COO− of β-lactam, which may cause inaccessibility to the catalytic site.
On the other hand, the finding of a mutation at the Phe 473 Leu site is novel and may be significant, because this change results in the loss of a very weak π-cation interaction which previously occurred between Phe 473 and Lys 464. Interestingly, we were unable to isolate a Phe 473 Leu and Ser 543 Arg double mutant without a corresponding Val 469 Met amino acid substitution. It may be that Val 469 Met is a compensatory mutation needed to stabilize the PBP1 protein with the Phe 473 Leu mutation.
Our data suggest that the mobility of solvent-exposed loops surrounding the putative binding cleft may control ligand entry toward the catalytic Ser 368, which is anchored by a hydrogen bond network in the base of the cleft (Fig. 1b and c). Certain mutations in these loops that produce antibiotic resistance may mediate steric and electrostatic effects which hinder antibiotic access to the catalytic Ser 368. Such “active site breathing” has recently been proposed as being a possible mechanism for β-lactam resistance in pneumococci (6).
A careful review of the amino acid changes in PBP1 in amoxicillin-resistant strains in previously published studies has led to our hypothesis that only a few amino acid substitutions can alter amoxicillin binding—Ser 414 Arg (reported in references 12, 13, 28, and 29), Thr 438 Met (4), Phe 473 Leu (this paper), Ser 543 Arg (19; also this paper), Thr 556 Ser (12, 19, 21, 26, 34), and Asn 562 Tyr (12, 19, 26, 29). These amino acid substitutions have appeared as single changes with corresponding increased amoxicillin resistance (4, 13, 21) or as parts of clusters of changes (12, 19, 26, 28, 29, 34; also this paper). The homology modeling demonstrates how these individual changes affect the putative binding cleft, apparently impeding amoxicillin docking. It is also clear that combinations of these specific substitutions, as seen in this paper, can additively increase the level of amoxicillin resistance found. However, the myriad of other amino acid substitutions sometimes found in PBP1 proteins from amoxicillin-resistant strains are unlikely to add to the effects seen for these specific amino acid changes.
These studies reveal that it is not the proximity of amino acid changes to PBMs per se that determines whether a substitution will confer resistance; for example, the Glu 406 Ala substitution is very close to the SKN PBM but does not affect amoxicillin susceptibility. Rather, it is the impact of the specific amino acid change within the putative binding cleft which appears to be more important. Our studies also suggest that changes which have a significant effect on the structure of the protein, particularly around the putative binding cleft, can have an effect on amoxicillin binding regardless of their lack of proximity to the PBMs (for example, Ser 414 Arg and Phe 473 Leu). On the other hand, Thr 556 Ser and Asn 562 Tyr are in or adjacent to a PBM, respectively, and clearly affect amoxicillin susceptibility.
Amoxicillin resistance in H. pylori is slowly increasing in the United States. A better understanding of its resistance mechanisms will allow changes in treatment strategies to prevent the occurrence of treatment failures. Furthermore, rapid screening of resistant isolates could be performed on clinical isolates using molecular techniques if the absolute number of amino acid mutations conferring resistance could be determined. In this study, we suggest that currently, only 6 sites seem critical, and of these 6, only three substitutions—Ser 414 Arg, Thr 556 Ser, and Asn 562—have been reported in multiple clinical isolates, suggesting that these are the most common amino acid changes in PBP1 connected to amoxicillin resistance.
Acknowledgments
We thank Richard Peek from Vanderbilt University for providing H. pylori strain B258 and Brenton Bauer for his assistance with some of the experiments described.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush, K. 2001. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, A., C. Kieslich, J. Yang, and D. Morikis. 2010. Solvation effects in calculated electrostatic association free energies for the C3d-CR2 complex and comparison with experimental data. Biopolymers 93:509-519. [DOI] [PubMed] [Google Scholar]
- 4.Co, E.-M. A., and N. L. Schiller. 2006. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 50:4174-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras-Martel, C., V. Job, A. M. Di Guilmi, T. Vernet, O. Dideberg, and A. Dessen. 2006. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in beta-lactam resistance in Streptococcus pneumoniae. J. Mol. Biol. 355:684-696. [DOI] [PubMed] [Google Scholar]
- 6.Contreras-Martel, C., C. Dahout-Gonzalez, A. Dos Santos Martins, M. Kotnik, and A. Dessen. 2009. PBP active site flexibility as the key mechanism for β-lactam resistance in pneumococci. J. Mol. Biol. 387:899-909. [DOI] [PubMed] [Google Scholar]
- 7.Dargis, M., and F. Malouin. 1994. Use of biotinylated beta-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob. Agents Chemother. 38:973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLoney, C. R., and N. L. Schiller. 1999. Competition of various beta-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: antibacterial activity and effects on bacterial morphology. Antimicrob. Agents Chemother. 43:2702-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLoney, C. R., and N. L. Schiller. 2000. Characterization of an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3368-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frenck, R. W., Jr., and J. Clemens. 2003. Helicobacter pylori in the developing world. Microbes Infect. 5:705-713. [DOI] [PubMed] [Google Scholar]
- 12.Gerrits, M. M., A. P. O. Godoy, E. J. Kuipers, M. L. Ribeiro, J. Stoof, S. Mendonca, A. H. M. van Vliet, J. Pedrazzoli, Jr., and J. G. Kusters. 2006. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter 11:181-187. [DOI] [PubMed] [Google Scholar]
- 13.Gerrits, M. M., D. Schuijffel, A. A. Van Zwet, E. J. Kuipers, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2002. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2229-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerrits, M. M., A. H. M. van Vliet, E. J. Kuipers, and J. G. Kusters. 2006. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect. Dis. 6:699-709. [DOI] [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Harris, A. G., S. L. Hazell, and A. G. Netting. 2000. Use of digoxigenin-labelled ampicillin in the identification of penicillin-binding proteins in Helicobacter pylori. J. Antimicrob. Chemother. 45:591-598. [DOI] [PubMed] [Google Scholar]
- 17.Hu, C.-T., C.-C. Wu, C.-Y. Lin, C.-C. Cheng, S.-C. Su, Y.-H. Tseng, and N.-T. Lin. 2007. Resistance rate to antibiotics of Helicobacter pylori isolates in eastern Taiwan. J. Gastroenterol. Hepatol. 22:720-723. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: Visual Molecular Dynamics. J. Mol. Graphics 14:33-38. [DOI] [PubMed] [Google Scholar]
- 19.Kwon, D. H., M. P. Dore, J. J. Kim, M. Kato, M. Lee, J. Y. Wu, and D. Y. Graham. 2003. High-level β-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malaguarnera, M., R. Bella, G. Algona, R. Ferri, A. Carnemolla, and G. Pennisi. 2004. Helicobacter pylori and Alzheimer's disease: a possible link. Eur. J. Intern. Med. 15:381-386. [DOI] [PubMed] [Google Scholar]
- 21.Matteo, M. J., G. Granados, M. Olmos, A. Wonaga, and M. Catalano. 2008. Helicobacter pylori amoxicillin heteroresistance due to point mutations in PBP-1A in isogenic isolates. J. Antimicrob. Chemother. 61:474-477. [DOI] [PubMed] [Google Scholar]
- 22.Megraud, F. 2004. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megraud, F., and P. Lehours. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 20:280-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittl, P. R. E., L. Luthy, P. Hunzinger, and M. G. Grutter. 2000. The cysteine-rich protein A from Helicobacter pylori is a β-lactamase. J. Biol. Chem. 275:17693-17699. [DOI] [PubMed] [Google Scholar]
- 25.Nedenskov-Sorensen, P., G. Bukholm, and K. Bovre. 1990. Natural competence for genetic transformation of Campylobacter pylori. J. Infect. Dis. 161:365-366. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto, T., H. Yoshiyama, T. Nakazawa, I.-D. Park, M.-W. Chang, H. Yanai, K. Okita, and M. Shirai. 2002. A change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J. Antimicrob. Chemother. 50:849-856. [DOI] [PubMed] [Google Scholar]
- 27.Parkin, D. M. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030-3044. [DOI] [PubMed] [Google Scholar]
- 28.Paul, R., S. Postius, K. Melchers, and K. P. Schafer. 2001. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob. Agents Chemother. 45:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rimbara, E., N. Noguchi, T. Kawai, and M. Sasatsu. 2007. Correlation between substitutions in penicillin-binding protein 1 and amoxicillin resistance in Helicobacter pylori. Microbiol. Immunol. 51:939-944. [DOI] [PubMed] [Google Scholar]
- 30.Rimbara, E., N. Noguchi, T. Kawai, and M. Sasatsu. 2008. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 61:995-998. [DOI] [PubMed] [Google Scholar]
- 31.Rosenstock, S., T. Jorgensen, O. Bonnevie, and L. Andersen. 2003. Risk factors for peptic ulcer disease: a population based prospective cohort study comprising 2416 Danish adults. Gut 52:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothenbacher, D., and H. Brenner. 2003. Burden of Helicobacter pylori and H. pylori-related disease in the developed countries: recent developments and future implications. Microbes Infect. 5:693-704. [DOI] [PubMed] [Google Scholar]
- 33.Sitkoff, D., K. A. Sharp, and B. Honig. 1994. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 98:1978-1988. [Google Scholar]
- 34.Tseng, Y.-S., D.-C. Wu, C.-Y. Chang, C.-H. Kuo, Y.-C. Yang, C.-M. Jan, Y.-C. Su, F.-C. Kuo, and L.-L. Chang. 2009. Amoxicillin resistance with β-lactamase production in Helicobacter pylori. Eur. J. Clin. Invest. 39:807-812. [DOI] [PubMed] [Google Scholar]
- 35.Tsuda, M., M. Karita, and T. Nakazawa. 1993. Genetic transformation in Helicobacter pylori. Microbiol. Immunol. 37:85-89. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 37.Wu, J., and D. Morikis. 2006. Molecular thermodynamics for charged biomacromolecules. Fluid Phase Equilibria 241:317-333. [Google Scholar]
- 38.Zhang, L., and D. Morikis. 2006. Immunophysical properties and prediction of activities for vaccinia virus complement control protein and smallpox inhibitor of complement enzymes using molecular dynamics and electrostatics. Biophys. J. 90:3106-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, L., B. Mallik, and D. Morikis. 2007. Immunophysical exploration of C3d-CR2(CCP1-2) association using molecular dynamics and electrostatics. J. Mol. Biol. 369:567-583. [DOI] [PubMed] [Google Scholar]