Abstract
Ammonia oxidation—the microbial oxidation of ammonia to nitrite and the first step in nitrification—plays a central role in nitrogen cycling in coastal and estuarine systems. Nevertheless, questions remain regarding the connection between this biogeochemical process and the diversity and abundance of the mediating microbial community. In this study, we measured nutrient fluxes and rates of sediment nitrification in conjunction with the diversity and abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing betaproteobacteria (β-AOB). Sediments were examined from four sites in Elkhorn Slough, a small agriculturally impacted coastal California estuary that opens into Monterey Bay. Using an intact sediment core flowthrough incubation system, we observed significant correlations among NO3−, NO2−, NH4+, and PO43+ fluxes, indicating a tight coupling of sediment biogeochemical processes. 15N-based measurements of nitrification rates revealed higher rates at the less impacted, lower-nutrient sites than at the more heavily impacted, nutrient-rich sites. Quantitative PCR analyses revealed that β-AOB amoA (encoding ammonia monooxygenase subunit A) gene copies outnumbered AOA amoA gene copies by factors ranging from 2- to 236-fold across the four sites. Sites with high nitrification rates primarily contained marine/estuarine Nitrosospira-like bacterial amoA sequences and phylogenetically diverse archaeal amoA sequences. Sites with low nitrification rates were dominated by estuarine Nitrosomonas-like amoA sequences and archaeal amoA sequences similar to those previously described in soils. This is the first report measuring AOA and β-AOB amoA abundance in conjunction with 15N-based nitrification rates in estuary sediments.
Nitrification, the microbially mediated oxidation of ammonia (NH3) to nitrite (NO2−) and nitrate (NO3−), represents a key process in the global cycling of nitrogen and plays a critical role in facilitating the removal of nitrogen, particularly in coastal and estuarine ecosystems. The first step in this chemoautotrophic process (the oxidation of NH3 to NO2−) has been known for decades to be carried out by ammonia-oxidizing bacteria (AOB) belonging to the beta- and gammaproteobacteria lineages. In fact, AOB were among the first chemoautotrophs ever grown in culture (63). However, recent cultivation and molecular ecological studies have revealed that many crenarchaea may also be capable of ammonia oxidation (ammonia-oxidizing archaea [AOA]) (14, 16, 19, 25, 43, 57, 60). Indeed, AOA and ammonia-oxidizing betaproteobacterial (β-AOB) genes putatively encoding the α subunit of ammonia monooxygenase (amoA), responsible for catalyzing the first and rate-limiting step of nitrification, are present in a wide variety of marine and terrestrial environments (16, 20, 27, 33, 36, 64).
Very little is known about the distribution, environmental tolerances, and relative contribution of β-AOB and AOA to the process of nitrification. Emerging studies comparing the impacts of environmental factors (e.g., salinity, temperature, and pH) on AOA and β-AOB community composition (20, 54, 56) and relative abundances (9, 31, 34, 36, 51) are only beginning to sort out the primary controls on the distribution and activity of these two groups of ammonia oxidizers. Generally, an overwhelming dominance of archaeal amoA abundance over bacterial amoA has been observed in open-ocean studies (4, 19, 33, 64) and soils (20, 27, 54). However, increasing evidence indicates that many coastal and estuarine sediments have a higher relative abundance of β-AOB amoA genes than AOA amoA genes (9, 31, 34, 51). Factors proposed to explain this pattern include salinity (31, 34, 51) and sediment C/N ratio (34). In addition, it has recently been demonstrated that the cultivated ammonia-oxidizing crenarchaeote Nitrosopumilus maritimus SCM1 has a far greater affinity (lower Km) for NH4+ than many well-known AOB, suggesting that NH4+ availability may also play a major role in governing the relative distribution and niche partitioning of AOA versus AOB in the environment (32).
In addition to assessing the relative abundances of AOA and β-AOB communities, measures of amoA functional gene diversity within each of these groups may help shed light on their relative ecological importance. In light of the immense microbial diversity observed in natural environments, the question remains whether diversity (and, more specifically, functional diversity, e.g., amoA) plays a role in controlling rates of biogeochemical processes, such as nitrification. Indeed, the relationship between functional gene diversity and N transformation rates regulated by the corresponding enzyme remains poorly understood (10). In this study, we explore whether differences in the rates of nitrification correspond to the diversity and/or relative abundance of AOA and β-AOB communities responsible for catalyzing this process in the agriculturally impacted Elkhorn Slough estuary, California.
MATERIALS AND METHODS
Site descriptions.
Elkhorn Slough is a small estuary extending approximately 11.4 km inland from the Pacific Ocean (Monterey Bay) with a mean tidal range of approximately 1.7 m. While the majority of the watershed is rural residential, approximately 26% of the watershed is agricultural, supporting such row crops as strawberries, artichokes, flowers and brussels sprouts.
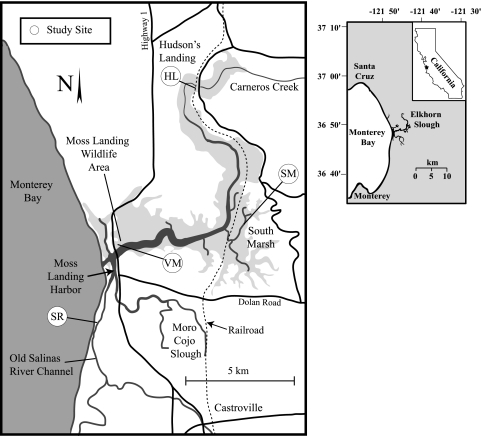
Four sites representative of the diverse environmental conditions found in Elkhorn Slough were chosen for this study (Fig. 1). Site characteristics were defined on the basis of a 15-year database of water quality sampling (Table 1). Sediments from all sites consisted of very-fine-grained, clay-rich mud (e.g., low hydraulic conductivity) high in organic matter content and quite sulfidic (Table 1). Two sites, Hudson Landing (HL) and Salinas River Marsh (SR), receive heavy amounts of agricultural runoff from nearby farms within the Carneros Creek (HL) and the old Salinas River, Tembladero, and Moro Cojo watersheds (SR). HL and SR are regularly exposed to high concentrations of NO3−; mean annual NO3− concentrations are 166.5 ± 323.3 μM at HL and 602.9 ± 602.6 μM at SR. The high variability in terrestrially derived nutrient concentrations results from both extensive tidal exchange and the episodic nature of storm-derived terrestrial runoff during the wet season (winter). Concentrations of water column NH4+ at these sites are also somewhat elevated; mean annual NH4+ concentrations are 11.8 ± 11.8 μM at HL and 12.9 ± 11.7 μM at SR. Agricultural practices in the area vary considerably, but N-based fertilizer is commonly used throughout the watershed (28). Additionally, irrigation practices are supplemented through the use of reclaimed wastewater from Monterey County (44).
FIG. 1.
Map of the Elkhorn Slough estuary illustrating the four sites where sediments were collected for intact core incubation experiments.
TABLE 1.
Site water column and sediment characteristicsa
| Parameter | Impacted |
Unimpacted |
||
|---|---|---|---|---|
| HL | SR | SM | VM | |
| Water | ||||
| Temp (°C) | 18.1 ± 4.3 | 16.2 ± 3.1 | 16.1 ± 3.4 | 15.3 ± 2.6 |
| Salinity (psu) | 26.8 ± 10.0 | 17.4 ± 10.9 | 29.8 ± 7.5 | 29.9 ± 6.6 |
| NO3− concn (μM) | 166.5 ± 323.3 | 602.9 ± 602.6 | 41.2 ± 75.7 | 8.13b ± 8.3 |
| NH4+ concn (μM) | 11.8 ± 11.8 | 12.9 ± 11.7 | 8.6 ± 7.5 | 12.6b ± 27.6 |
| PO43− concn (μM) | 6.2 ± 6.2 | 11.4 ± 9.7 | 4.3 ± 4.2 | 1.38b ± 0.8 |
| Dissolved O2 concn (mg/liter) | 8.0 ± 3.0 | 8.8 ± 3.1 | 7.7 ± 2.3 | 7.7b ± 2.5 |
| Dissolved O2 (% saturated) | 101.8 ± 38.1 | 95.9 ± 25.0 | 90.4 ± 21.9 | 96.3b ± 37.2 |
| Turbidity (NTUc) | 22.5 ± 26.3 | 44.7 ± 60.9 | 10.9 ± 15.0 | |
| pH | 8.2 ± 0.6 | 8.2 ± 0.5 | 8.1 ± 0.5 | 8.2 ± 0.4 |
| No. of samples | 97 | 110 | 109 | 104 (16b) |
| Sediment | ||||
| Organic N (%) | 0.20 ± 0.13 | 0.20 ± 0.10 | 0.26 ± 0.07 | 0.15 ± 0.07 |
| Organic C (%) | 1.81 ± 1.21 | 1.86 ± 0.85 | 3.17 ± 0.93 | 1.67 ± 0.96 |
| C/N (atomic) | 11.4 ± 2.7 | 11.1 ± 1.6 | 14.4 ± 1.8 | 12.2 ± 3.6 |
| δ15N (‰ vs N2) | 6.9 ± 1.9 | 7.1 ± 2.4 | 4.4 ± 1.5 | 3.8 ± 2.4 |
| δ13C (‰ vs VPDBd) | −24.1 ± 0.7 | −25.6 ± 0.9 | −25.3 ± 0.7 | −24.6 ± 0.8 |
| Porositye (%) | 37.2 ± 0.5 | 58.9 ± 0.2 | 47.1 ± 0.7 | 36.9 ± 2.3 |
| Specific density (g/ml) | 1.65 ± 0.02 | 1.42 ± 0.00 | 1.35 ± 0.01 | 1.61 ± 0.07 |
Data are from the Elkhorn Slough National Estuarine Research Reserve (NERR) Water Quality Database (www.nerr.gov), unless otherwise noted.
From field data collected at the Moss Landing Wildlife Area (VM), near site 1 of the Elkhorn Slough NERR Water Quality Monitoring Database.
NTU, nephelometric turbidity unit.
VPDB, Vienna Pee Dee Belemnite.
Measured as water loss upon drying.
Two other sites (Fig. 1), Moss Landing Wildlife Area (VM) and Elkhorn Slough Reserve South Marsh (SM), typically receive only limited direct terrestrial runoff and are primarily flushed by water from the main channel of Elkhorn Slough, in which 70% of the water is exchanged diurnally by tides (7). Thus, in contrast to HL and SR, these sites are generally less exposed to nutrient-rich agricultural runoff (i.e., “unimpacted”), as evidenced by lower mean NO3− concentrations (8.1 ± 8.3 μM at VM and 41.2 ± 75.7 μM at SM) (Table 1). While we acknowledge that these sites also have somewhat elevated nutrient concentrations compared to more pristine estuarine environments, we choose the term “unimpacted” for ease of comparison with the agriculturally “impacted” sites in the following discussion.
Flowthrough core incubation system.
A flowthrough core incubation system (see Fig. S1 in the supplemental material) was constructed for measurement of steady-state N transformation rates as described previously (38, 39, 50, 59). Flowthrough incubation has several advantages over bottle sediment-slurry batch-type incubation, including minimization of sediment disturbance (and associated physical/chemical changes), the ability to directly measure and calculate aerial N transformation rates, maintenance of steady-state conditions, and the ability to monitor many processes simultaneously.
Clear acrylic core tubes (diameter, 7.5 cm; length, 30 cm) were used to collect intact sediment cores from each of the sites during January 2005. Sediments were cored to depths of approximately 10 cm, keeping the overlying 20 cm filled with site water during transport to minimize disruption of the sediments. Cores were kept in the dark and on ice during transport back to the laboratory, where they were immediately incorporated into the flowthrough incubation system. Two cores were collected and analyzed from each site. Once the cores were back at the lab, the overlying water was gently poured off and was replaced in all cores with seawater from Monterey Bay (e.g., representative of water delivered to the sites during a flood tide).
During the experiment, the system was set up in a dark, temperature-controlled (15°C) room and consisted of a large aerated reservoir of low-nutrient seawater (salinity, 33.4 practical salinity units [psu]; from Monterey Bay) which was pumped using an 8-channel peristaltic pump (flow rate, 3.0 ml/min) into the overlying water of each core. The effluent of each core was routed into collection flasks during sampling. To allow for initial equilibration of the sediments and overlying seawater, cores were preincubated for approximately 22 h before measurements commenced. Samples were collected approximately every 8 h for 7 days, and steady-state conditions (based on a lack of change in nutrient concentration in the overlying water column) were observed after 4 days. To evaluate the processes within the N cycle and their rates, 100 μM isotopically labeled (95% 15N) KNO3− was added to the cores, and sediment fluxes and nitrogen transformation rates were calculated from the steady-state changes in nutrient concentration and isotopic composition of N2, NH4+, and NO3− (50). Finally, as a means for inducing additional biogeochemical variability, the overlying water column was gently stirred in one set of incubated cores (“stirred”), while the overlying water column in the other set was not (“unstirred”). This approach was designed to represent a range of water column mixing conditions which might be routinely experienced on estuarine intertidal mudflats within Elkhorn Slough.
Flux measurements and transformation rates.
Aerial fluxes and transformation rates were calculated using the difference in concentrations and/or isotopic compositions between the inflow and the outflow solutions and multiplying by flow rate and sediment surface area. Positive nutrient fluxes are indicated by outflow concentrations that are higher than inflow concentrations, implying net production occurring within the sediment. Although several studies of coastal shelf sediments have found that pore water advection can influence sediment-based nitrogen transformations (18, 22, 45), given the high clay content and low hydraulic conductivity of the sediments in this study, pore water advection is unlikely to play a prominent role in sediment nitrogen fluxes and/or transformations and is therefore not considered in the analysis.
Rates of denitrification (both that supported by water column NO3− [Dw] and that coupled to nitrification [Dn]) were calculated using the isotope pairing method by measuring the difference in production rates of 29N2 and 30N2 (37, 50, 55). Additionally, rates of gross nitrate uptake (U) and efflux of unlabeled NO3− (R) were calculated using the isotope dilution method by measuring changes in the isotopic composition of 15NO3− (24, 39, 50). Nitrification rates were calculated as the production rate of unlabeled NO3− plus that considered to be directly coupled to denitrification by the rate of production of 29N2 and 30N2 (see below). We did not include explicit consideration of anaerobic ammonium oxidation (anammox), which has been shown to be less important than denitrification in estuarine sediments (26, 46, 47, 58) and which was undetectable in previous slurry incubations of Elkhorn Slough sediments (61).
Isotopic measurements. (i) 15N content of NO3−.
A KNO3 standard solution was mixed with a known volume of sample water containing 15NO3−, yielding an approximately 20:1 dilution. Samples were then freeze-dried into tin boats, crimped, and analyzed using a conventional elemental analyzer combustion coupled to a GVI IsoPrime isotope ratio mass spectrometer (IRMS). A simple mass balance calculation was then used to determine the 15N content of the sample. Calibration of the δ15N was made using international nitrate isotopic standards USGS 32 (δ15N = 180.0‰) and USGS 34 (δ15N = −1.9). Analytical precision was assessed by multiple measurements of known 95% 15N-enriched KNO3 (ICON Isotopes) over a dilution series with a stock natural-abundance KNO3 and was better than ±0.5%.
(ii) 15N content of NH4+.
The percent 15N content of the effluent NH4+ was measured using the ammonium diffusion method (21), whereby NH4+ in approximately 100 ml of sample water is converted to NH3 (by increasing the pH with MgO), which is then trapped on an acidified filter disk encased in a Teflon membrane. Filters are later combusted and analyzed for percent 15N via conventional EA-IRMS analysis. A similar isotope dilution series was used to evaluate analytical precision, which also was better than ±0.5%.
(iii) 15N content of N2.
Using a 5-ml syringe, 3 ml of sample water was drawn directly from the effluent end of each core without entraining air (any bubbles in the syringe sample were gently expelled from the syringe). The sample was immediately transferred underwater (to minimize contamination by air N2) through a blue butyl septum into a 10-ml serum vial (preflushed with ultra-high-purity helium and containing 100 μl of saturated ZnCl2 solution to stop biologic activity). Samples were stored at room temperature until analysis for isotopic abundance of 29N2 and 30N2.
Samples were analyzed on a GVI IsoPrime isotope ratio mass spectrometer, using a customized sample prep system (AS 1200), allowing direct injection of a 30-μl headspace sample. A headspace sample was drawn from the sample vial using a Hamilton gas-tight syringe. N2 was separated chromatographically from O2 using a 1 m 1/16-in. packed molecular sieve (pore size, 5 Å) column. Using empty preflushed vials or 3 ml deionized water equilibrated with air, as well as blank injections, we were able to account for the potential influence of accidentally entrained air during sample injection and/or collection.
Nitrification rates.
Rates of nitrification were measured via isotope dilution (24, 39, 50), taking advantage of the fact that any NO3− produced from NH4+ via nitrification will not be labeled and will isotopically dilute the originally labeled 15NO3− pool. Using this approach, the efflux of unlabeled NO3− (R) (from nitrification) was estimated with the following equation (50):
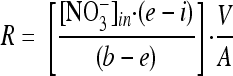 |
(1) |
where R is the net efflux of unlabeled NO3− out of the sediment; e and i are the fraction of 15N-labeled NO3− in the effluent and influent water, respectively; b is the percent 15N content of the nitrified NH4+ (based on the measured 15N content of effluent NH4+); V is the flow rate; and A is the sediment surface area. The nitrification rate is then calculated as the sum of R and Dn, where Dn is coupled nitrification-denitrification. The process of dissimilatory nitrate reduction to ammonium (DNRA) was estimated as follows:
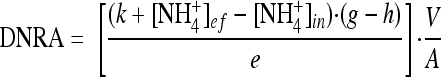 |
(2) |
where ef and in refer to effluent and influent concentrations of NH4+, respectively; g and h are the percent 15N content of the NH4+ in the effluent and influent, respectively; e is the percent 15N in the NO3− pool; and k is the nitrification rate expressed in terms of the concentration of NH4+ removed by nitrification. The inclusion of the influence of nitrification on the calculation of DNRA is a slight modification of the approach, outlined by others, meant to include the effect of NH4+ consumption by nitrification (50). Finally, remineralization rates (REMIN) reflect the difference between the sum of sediment efflux and nitrification and the sum of influx and DNRA:
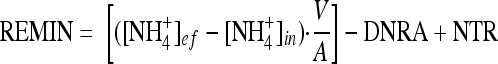 |
(3) |
where NTR is nitrification. For more details on the isotope pairing method and calculations of coupled nitrification-denitrification, in addition to DNRA and remineralization, see references 37 and 55.
The approach outlined above is designed for estimating an aerial rate of nitrification occurring at the sediment surface. However, it must be noted that we cannot account for nitrification occurring in the oxic water column overlying the sediment. Calculations are made using the difference between the inflow and outflow, a reasonable approach for estimating processes occurring at the quasi-two-dimensional oxic-anoxic interface. However, considering that NH4+ was generally present in the overlying water column at concentrations ranging from 0.2 to 11 μM, it is likely that nitrification was not entirely restricted to this oxic-anoxic interface. Hence, rates derived from this experiment are more appropriately considered in terms of moles of NO3− produced per core system. In this study, rates of nitrification can be considered to reflect a sediment/water system proportional to a sediment surface area of 1 m2 and a 20-cm-deep overlying water column. Rates thus include both sediment-based nitrification (where there is far greater microbial biomass and, presumably, activity) and any water column nitrification occurring within the system.
Statistical analyses.
All regression analyses and statistical comparisons were carried out using SPSS statistics software (version 17.0).
DNA extraction and PCR amplification.
At the termination of the 8-day experiment, surface sediment (∼1.5 cm) was collected from each core and immediately frozen on dry ice and stored at −80°C until DNA extraction. DNA was extracted from surface sediment (∼0.5 g) using a FastDNA spin kit for soil (MP Biomedicals) with a FastPrep instrument, according to the manufacturer's instructions. The following reaction chemistry was used for amplification of β-AOB amoA: 20 μl 2× PCR premix E (Epicentre), a 1 μM concentration of each primer (49), and 1.25 U AmpliTaq LD Taq polymerase (Applied Biosystems, Inc., Foster City, CA). Since gammaproteobacterial AOB have not been shown to be important in marine sediments (40), they were not specifically examined via PCR in this study. For amplification of AOA amoA, the following reaction chemistry was used: 12.5 μl 2× PCR premix F (Epicentre), a 0.2 μM concentration of each primer (16), 3 μl MgCl2, and 1.25 U AmpliTaq LD Taq polymerase (Applied Biosystems, Inc.). Triplicate PCR products were pooled, gel purified, and cloned into the pCR2.1 vector using a TOPO-TA cloning kit (Invitrogen).
Sequencing and phylogenetic analysis.
Gene sequencing was performed using an ABI 3730xl capillary sequencer (PE Applied Biosystems). Nucleotide sequences were edited in the Sequencher program (version 4.8; GeneCodes Corporation) and then aligned with GenBank sequences using the MacClade program (version 4.08) (30). A 579-bp region of the sequence alignment was used for archaeal amoA phylogeny, and a 444-bp region was used for bacterial amoA phylogeny. Neighbor-joining phylogenetic trees (based on Jukes-Cantor correction with 300 bootstrap replicates) were constructed in ARB software (29).
Richness and diversity analyses.
Jukes-Cantor distance matrices were generated in the PAUP program (version 4.0b10; Sinnauer Associates). Operational taxonomic units (OTUs), diversity indices (Shannon and Simpson), and nonparametric richness estimates (e.g., Chao1) were determined using the furthest-neighbor algorithm in the DOTUR program (52). OTUs were defined as amoA clones sharing ≥95% nucleotide sequence identity. Abundance-based Sørensen-type (Labd), Jaccard-type (Jabd), and θ (65) similarity indices for comparing communities among sites and treatments were calculated using the SONS program (53).
Quantification of amoA gene abundance.
Quantitative PCR (qPCR) was used to estimate the number of archaeal and bacterial amoA copies in sediments as previously described (34), except that 0.4 μM each bacterial amoA primer was used. Template DNA (0.5 to 8 ng) was added to each reaction mixture. Standard curves spanned a range from 5.3 to 1.1 × 105 amoA copies per μl for the AOA assay and from 16.2 to 1.6 × 105 amoA copies per μl for the AOB assay. Sediment DNA and standard DNA were quantified using Quant-iT high-sensitivity and broad range DNA assays, respectively, with a Qubit fluorometer (Invitrogen). All sample and standard reactions were performed in triplicate, and an average value was calculated. An outlier was removed from some triplicate measurements to maintain a standard deviation of less than 10% for each sample. The standard deviation for site HL2 was 11.7% for the archaeal amoA quantification. Melt curves were generated after each assay to check the specificity of amplification. PCR efficiencies were 91 to 93% for archaeal amoA and 97% for bacterial amoA. Correlation coefficients (r2) were greater than 0.99 for all runs. The ratio of AOA amoA to β-AOB amoA was calculated using copy numbers normalized to ng DNA and then log10 transformed.
Nucleotide sequence accession numbers.
The sequences reported in this study have been deposited in GenBank under accession numbers HM363789 to HM364038 for archaeal amoA and HM364039 to HM364274 for bacterial amoA.
RESULTS AND DISCUSSION
Nitrogen transformation rates.
Nutrient fluxes and nitrogen transformation rates in sediments from the Elkhorn Slough estuary were measured using intact core incubations. Nitrification rates ranged from 7.1 to 32.8 mmol NO3− m−2 day−1 (Table 2), calculated as aerial rates, though they include the rates in the oxic 20-cm water column overlying the sediment surface. Because the measurement of nitrification is based on the production of unlabeled 14NO3−, these values are a direct measure of ammonia oxidation (and not simply NO3− production) and exclude interference by reoxidation of NO2− produced by NO3− reduction or denitrification. The highest nitrification rate (32.8 mmol NO3− m−2 day−1) corresponded to core SM2, although the rate in the parallel stirred core, SM1, was somewhat lower (15.0 mmol NO3− m−2 day−1).
TABLE 2.
Steady-state rates of nitrogen transformation from intact core incubations
| Site identifier | Site name | Treatment | Type | Rate (μmol N m−2 day−1) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Nitrification | Denitrification | Coupled N-Da | Remineralization | Gross NO3− uptake | DNRA | ||||
| VM1 | Elkhorn Slough protection area | Stirred | Unimpacted | 22,930 | 1,595 | 74 | 16,179 | 32,542 | 944 |
| SM1 | South Marsh | Stirred | Unimpacted | 14,976 | 2,086 | 510 | 7,787b | 20,487 | NAc |
| SR1 | Salinas River | Stirred | Impacted | 7,149 | 4,592 | 387 | 7,183 | 22,352 | 240 |
| HL1 | Hudson Landing | Stirred | Impacted | 11,835 | 12,961 | 388 | 18,809 | 47,741 | 4,775 |
| VM2 | Elkhorn Slough protection area | Unstirred | Unimpacted | 27,417 | 39 | NDd | 19,230 | 39,057 | 328 |
| SM2 | South Marsh | Unstirred | Unimpacted | 32,796 | 1,824 | 534 | 24,498b | 47,876 | NA |
| SR2 | Salinas River | Unstirred | Impacted | 23,642 | 4,706 | 413 | 24,458 | 37,096 | 1,241 |
| HL2 | Hudson Landing | Unstirred | Impacted | 27,588 | 2,464 | 64 | 19,431 | 40,897 | 1,224 |
N-D, nitrification-denitrification.
In the absence of any constraint on rates of DNRA; the calculated remineralization rate includes DNRA as a source of NH4+.
NA, not available, samples lost.
ND, not detected.
Nitrification rates within the unstirred cores were significantly higher than those in the stirred cores (P < 0.01) (Fig. 2; Table 2). Despite differences in the magnitude of the nitrification rates between the stirred and unstirred cores, similar overall trends in nitrification rates were observed across the four sites. Higher rates occurred at the more well-flushed sites (VM and SM), and lower rates occurred at the more poorly flushed, agriculturally impacted sites (HL and SR). In fact, averaging the nitrification rates within each site yields the same trend, and the average rates range from ∼15 to 25 mmol NO3− m−2 day−1, generally toward the high end of the values observed by others using similar techniques (50, 59).
FIG. 2.
Correlations among nutrient fluxes and 15N-based nitrification rates. Empty symbols denote experiments in which the overlying water column was gently stirred throughout the incubation, while symbols with internal circles denote an unstirred water column.
There are a number of possible explanations for the higher rates of nitrification in the unstirred cores. First, more extensive water column nitrification may have occurred above the sediments in the unstirred cores than in the stirred cores. Notably, it was recently demonstrated that the growth of N. maritimus SCMI is impaired by agitation (32), which may have implications for our stirred and unstirred core incubations. Second, in addition to reducing water column activity, the stirring may have physically perturbed/disrupted the narrow nitrification zone associated with the surface sediments of the stirred cores, relative to the unstirred cores. It is also possible that a thin nitrifying biofilm may have formed at the sediment-water interface (or on the inner walls of the core system itself) in the unstirred cores. Given that both core setups were subjected to the same flow rate and source of seawater, the unstirred core system may simply have offered conditions that were more favorable for nitrification.
Overall, denitrification rates were generally lower than nitrification rates, ranging from 0.04 to 12.9 mmol N m−2 day−1, and rates of DNRA (0.3 to 4.8 mmol N m−2 day−1) were generally lower than denitrification rates and accounted for <10% of NO3− consumption (Table 2). The relative amount of nitrification that was attributable to coupled nitrification-denitrification (i.e., the amount of NH3 oxidation that is immediately converted to N2 via denitrification) was also small (<10%), implying spatial decoupling of these processes in our reactor columns. Because light can either enhance or interfere with the coupling of nitrification-denitrification, e.g., through stimulation of the microphyotobenthos (2, 47, 48), our reactor system was operated in the dark. The low relative rates of coupled nitrification-denitrification observed in our reactors are consistent with observations by others during dark incubations (23, 50).
Nutrient fluxes.
Nutrient fluxes were variable both between parallel cores (stirred/unstirred) and among the four distinct sites within the estuary (Table 3). Stirred and unstirred cores from the same site often exhibited considerable differences, highlighting potential heterogeneity within a particular site, as well as the influence of environmental conditions (e.g., mixing) on biogeochemical fluxes and processes. Fluxes of inorganic PO43− were negative across all sites and cores, ranging from −0.9 to −1.5 mmol PO43− m−2 day−1, indicating net uptake of PO43− by the sediments. Fluxes of NO3− were also always negative, ranging from −5.7 to −35.9 mmol NO3− m−2 day−1, with the highest (most negative) fluxes corresponding to sites impacted by agricultural runoff (SR2 and HL1), suggesting a high capacity of these sediments to take up and transform NO3−. Nitrite (NO2−) fluxes were generally small, although variable in direction, ranging from 0.9 to −1.7 mmol m−2 day−1. The two cores with positive NO2− fluxes (SR1 and HL1) correspond to agriculturally impacted sites. NH4+ fluxes were variable, ranging from −8.3 to 11.7 mmol NH4+ m−2 day−1. As with the NO2− fluxes, the three cores with positive NH4+ fluxes (SR1, SR2, and HL1) corresponded to agriculturally impacted sites.
TABLE 3.
Steady-state fluxes of nutrients from intact core incubations
| Site identifier | Site name | Treatment | Type | Steady-state flux (μmol m−2 day−1) |
|||
|---|---|---|---|---|---|---|---|
| NO2− | NO3− | PO43− | NH4+ | ||||
| VM1 | Elkhorn Slough protection area | Stirred | Unimpacted | −54 ± 328 | −9,320 ± 1,573 | −1,352 ± 34 | −5,807 ± 334 |
| SM1 | South Marsh | Stirred | Unimpacted | −790 ± 272 | −5,655 ± 1,764 | −1,132 ± 58 | −7,189 ± 835 |
| SR1 | Salinas River | Stirred | Impacted | 906 ± 216 | −15,224 ± 1,327 | −888 ± 70 | 730 ± 1,273 |
| HL1 | Hudson Landing | Stirred | Impacted | 514 ± 203 | −35,929 ± 2,351 | −872 ± 41 | 11,750 ± 1,330 |
| VM2 | Elkhorn Slough protection area | Unstirred | Unimpacted | −1,700 ± 386 | −10,093 ± 1,658 | −1,430 ± 76 | −7,859 ± 623 |
| SM2 | South Marsh | Unstirred | Unimpacted | −1,452 ± 351 | −14,033 ± 3,160 | −1,399 ± 78 | −8,298 ± 845 |
| SR2 | Salinas River | Unstirred | Impacted | −676 ± 290 | −28,003 ± 2,577 | −1,349 ± 40 | 1,419 ± 1,967 |
| HL2 | Hudson Landing | Unstirred | Impacted | −1,284 ± 360 | −11,489 ± 211 | −1,452 ± 100 | −6,933 ± 990 |
Fluxes of various nutrients viewed simultaneously often provide insight into important processes occurring at the sediment-water interface (Fig. 2). For example, NH4+ and PO43− fluxes were positively and significantly correlated (r2 = 0.55, P = 0.03), pointing to the role of organic matter remineralization in the production of bioavailable forms of N and P from organic-rich sediments. In addition, NO2− fluxes were positively correlated with PO43− fluxes (r2 = 0.73, P < 0.01). While NO2− is a product of both NH3 oxidation and NO3− reduction, this correlation suggests that organic remineralization is an important process for delivering NH3/NH4+ as a substrate to the ammonia-oxidizing microbial community. Furthermore, NH4+ and NO2− fluxes were also significantly and positively correlated (r2 = 0.52, P = 0.04), consistent with the NH4+ supply playing a role in regulating rates of NO2− production in this environment. All of these lines of evidence indicate a coupling of remineralization of organic matter in estuarine sediments to rates of ammonia oxidation by microbial communities inhabiting the oxic sediment-water interface, as described previously.
In addition to the trends observed among nutrient fluxes, nitrification rates showed a strong negative correlation with NO2− fluxes (r2 = 0.76, P < 0.01; Fig. 2), consistent with the observation that NO2− oxidation is not the rate-limiting step in nitrification and that the two steps (NH3 and NO2− oxidation) are tightly coupled. Thus, when ammonia oxidation rates are highest, the resulting NO2− is oxidized to NO3− more quickly than when ammonia oxidation rates are low. This suggests that, where conditions are conducive to nitrification, nitrifier communities (both ammonia-oxidizing and nitrite-oxidizing microorganisms) are well poised for carrying out the two-step process. Notably, NH4+ fluxes were not correlated with nitrification rates (Fig. 2), probably due to a larger, more variable influence of remineralization on the sediment NH4+ pool. However, we did observe a significant negative correlation between NH4+ and NO3− fluxes (r2 = 0.83, P < 0.01), consistent with the relatively high rates of nitrification, which would have the effect of lessening the observed uptake of NO3− by the sediments (although a positive correlation between nitrification and NO3− fluxes was not found). Additionally, this observation suggests that denitrification, driven largely by a heterotrophic microbial community in organic-rich estuarine sediments, could play an important role in the NH4+ dynamics by contributing to the remineralization of nitrogenous organic matter and release of NH4+.
Agriculturally impacted versus unimpacted sites.
Agriculturally impacted sites exhibited greater net uptake of NO3− and greater net efflux of NH4+ than unimpacted sites (P < 0.05) (Table 3). Furthermore, average rates of nitrification for the impacted sites were lower than those for the unimpacted sites for both the stirred and unstirred reactor conditions (Table 2). This is consistent with previously observed nitrate dual stable isotope patterns, which show that while sediments were important for both denitrification and nitrification, the shallow, unimpacted marsh regions of Elkhorn Slough exhibited a particularly strong influence by nitrification (62). Because the production of NO3− by nitrification acts to decrease the apparent net microbial uptake of NO3− by the sediments, use of the gross NO3− uptake rate may be more appropriate for gauging differences among sites. Nonetheless, gross NO3− uptake rates showed no difference between impacted and unimpacted sites. Thus, the differences in NO3− flux observed between impacted and unimpacted sites were primarily driven by differences in rates of nitrification between the two types of sites; higher rates of nitrification at unimpacted sites acted to lessen the net uptake of NO3−, effectively diminishing the capacity for sediments to remove NO3− from the estuary.
Though it has been observed that eutrophic estuarine sediments can have a higher relative capacity for NO3− uptake (13, 41), our findings suggest that, while this may be true, the mechanism responsible does not necessarily involve increased denitrification or DNRA (e.g., consumption of NO3−). Rather, we observed that agriculturally impacted sites had lower nitrification rates, which gave rise to higher net uptake of NO3−. In this way (and under these experimental conditions), the nitrifier community is influencing the capacity for the sediments to remove NO3− from the overlying water column. While the observation that agriculturally impacted sediments have a higher capacity for removing N (via denitrification and N2O or N2 loss) (13, 41) is consistent with our results, our data suggest a substantial involvement of the nitrifying community in the process. This underscores the unique ecological role of nitrifiers, often touted as important because they provide a substrate for denitrification and aid in the elimination of excess N from aquatic ecosystems. However, as observed in our experiments, when spatially uncoupled from denitrification, nitrifiers also serve as significant producers of NO3−, ultimately contributing to N release and transport. Finally, this emphasizes the benefit of intact core incubations: steady-state intact core incubations allow simultaneous measurement of many processes such that interactions among processes are more easily revealed.
AOA and β-AOB abundance.
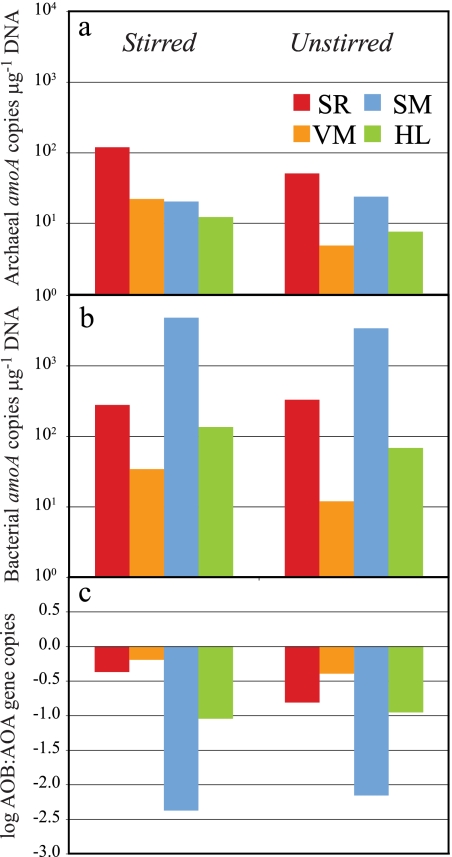
With the observed patterns among N transformation and flux data and the notable differences between the agriculturally impacted and unimpacted sites, we sought to determine the abundance and diversity of the ammonia-oxidizing microbial communities responsible for the biogeochemical activity observed under these experimental conditions. The abundance of archaeal and betaproteobacterial amoA genes was measured in each of the eight incubation systems from surface sediment samples collected at the end of the incubation (after ∼8 days). The AOA amoA gene copy numbers ranged from 4.9 × 103 to 1.2 × 105 copies μg−1 DNA (Fig. 3a). These estimates are within the range observed in other estuarine systems, such as San Francisco Bay in California (34) and coastal beach aquifer sediments (51). β-AOB amoA gene copy numbers, ranging from 1.2 × 104 to 4.8 × 106 copies μg−1 DNA, were substantially higher than AOA amoA gene copy numbers (Fig. 3b). These values are typical of the range observed in several other studies (12, 31, 34, 51).
FIG. 3.
Results from quantitative PCR analysis of archaeal amoA gene abundance (a), betaproteobacterial amoA gene abundance (b), and the log ratio of AOA to β-AOB amoA gene abundance (c), showing dominance of β-AOB at all sites and with all treatments.
β-AOB amoA genes were most abundant in sediments cores from SM and least abundant at VM, both sites that were considered less impacted by agricultural runoff (Fig. 3b). Relative to β-AOB, AOA amoA gene copy numbers were less variable across the sites. AOA amoA genes were most abundant in sediment cores from SR, a heavily impacted nutrient-rich site that routinely receives high flows (>20 m3 s−1) of agricultural runoff containing large amounts of suspended sediment from nearby fields. There were no significant differences in β-AOB or AOA amoA gene abundance between the stirred and unstirred treatments for any given site (Fig. 3, middle panel), suggesting that the difference in physical mixing of water overlying the sediments did not affect the abundance of either β-AOB or AOA (with the exception of a lower AOA amoA abundance in the unstirred VM core).
In every incubation, β-AOB amoA gene abundance was greater than AOA amoA gene abundance by a factor of 2 to 236 (Fig. 3). The relative ratio of AOA to β-AOB (Fig. 3c) varied substantially among the four estuarine sites. Numerous studies based on qPCR analysis of amoA genes have shown AOA to greatly outnumber AOB, calling into question the biogeochemical role of β-AOB in many systems (4, 20, 27, 33, 35, 54). However, mounting evidence from various coastal and estuarine studies (9, 31, 34, 51), including this study, suggests that β-AOB amoA gene abundance may actually be greater than AOA amoA gene abundance in certain regions of estuaries. For example, a study within the San Francisco Bay estuary showed greater AOA amoA gene abundance only at sites with low salinity and high sediment C/N ratios (34), with β-AOB amoA genes predominating at sites with salinities of 22 to 31 and low C/N ratios. A similar study across a groundwater-seawater beach interface also revealed that β-AOB amoA abundance exceeded AOA amoA abundance with proximity to the ocean and higher salinity (51).
While salinity appears to play a role in the relative distribution of AOA and β-AOB communities in larger estuaries and/or across stable salinity gradients (34, 51), Elkhorn Slough is a small, tidally flushed estuary and thus does not have the stable salinity gradient often found in larger estuaries. We conducted our experiments under a constant salinity of ∼33 psu, typical of the salinity of seawater that each site generally experiences twice daily. Thus, overall, the predominance of β-AOB amoA genes over AOA amoA genes in experimental sediment cores from Elkhorn Slough is entirely consistent with the trends observed in saline regions of a large estuary (34), within a coastal aquifer (51), and within sandy estuarine sediments (31). Additionally, the relatively high organic matter content (Table 1) and pore water NH4+ (8) of these sediments, together with the higher NH4+ affinity of AOA versus β-AOB (32), may also explain a large degree of the dominance of β-AOB in Elkhorn Slough.
AOA and AOB richness, phylogenetic diversity, and community comparisons.
A total of 411 cloned amoA gene fragments from β-AOB and AOA from surface sediments of all eight core incubations were sequenced. On the basis of a 5% OTU cutoff, 3 to 8 OTUs were observed within each β-AOB amoA clone library (Table 4), comparable to the results of many other studies of bacterial amoA in estuarine environments (3, 5, 6, 15, 34). AOA amoA richness was much greater for all eight libraries, with 10 to 24 OTUs occurring in each library. Total richness estimates for β-AOB amoA genes (6 to 12 OTUs per library and 21 OTUs for the entire data set, predicted by Chao1; Table 4) were in the same range as those from other estuarine studies (3, 11, 12, 15, 34). In contrast, Chao1 predictions of AOA amoA richness (14 to 81 OTUs per library; 52 for the entire library; Table 4) were always higher than predictions of bacterial amoA richness. On average, the number of observed AOA amoA OTUs was 64% of that predicted by the Chao1 index, while 71% of the observed β-AOB amoA OTUs were predicted. The Shannon diversity index for AOA was also consistently higher (2.4 to 3.1) than that for β-AOB (1.2 to 2.2) (Table 4). In summary, as in San Francisco Bay (34) and Huntington Beach (51), AOA amoA richness and diversity estimates were consistently higher than those for β-AOB in Elkhorn Slough (Fig. 4).
TABLE 4.
DOTUR-based indices of amoA diversity of nitrifying communities
| Core identifier | No. of clones | No. of OTUs | No. of singletons | Chao1 (95% CIa) | Simpson index | Shannon index |
|---|---|---|---|---|---|---|
| Archaeal | ||||||
| VM1 | 27 | 10 | 3 | 17.2 (13.8, 35.3) | 0.12 | 2.6 |
| SM1 | 34 | 17 | 8 | 23.3 (15.9, 59) | 0.10 | 2.6 |
| SR1 | 32 | 21 | 16 | 37.5 (23.2, 90.9) | 0.10 | 2.9 |
| HL1 | 34 | 16 | 7 | 22.6 (18.2, 44.1) | 0.05 | 2.9 |
| VM2 | 29 | 11 | 6 | 19.5 (13.3, 54.5) | 0.11 | 2.5 |
| SM2 | 35 | 24 | 19 | 81 (37.7, 237.2) | 0.05 | 3.1 |
| SR2 | 32 | 13 | 5 | 14 (12.3, 26) | 0.18 | 2.4 |
| HL2 | 27 | 11 | 4 | 14 (12.3, 26) | 0.10 | 2.6 |
| All | 250 | 52 | 14 | 62.5 (55.1, 87.5) | 0.04 | 3.5 |
| Bacterial | ||||||
| VM1 | 20 | 7 | 2 | 12 (9.4, 32) | 0.15 | 2.2 |
| SM1 | 21 | 5 | 2 | 6 (5.1, 18.5) | 0.32 | 1.5 |
| SR1 | 17 | 7 | 4 | 11.3 (8.5, 30.1) | 0.47 | 1.7 |
| HL1 | 16 | 5 | 1 | 5 (5, 5) | 0.24 | 1.8 |
| VM2 | 27 | 8 | 2 | 8.3 (8, 14) | 0.16 | 2.1 |
| SM2 | 24 | 5 | 1 | 8.8 (8.1, 16.5) | 0.32 | 1.8 |
| SR2 | 17 | 3 | 1 | 6.5 (5.1, 20.1) | 0.62 | 1.2 |
| HL2 | 19 | 6 | 3 | 7.5 (6.1, 21.1) | 0.36 | 1.5 |
| All | 136 | 21 | 4 | 21.9 (21.1, 29.0) | 0.15 | 2.2 |
CI, confidence interval.
FIG. 4.
Rarefaction curves illustrating higher amoA diversity of AOA than β-AOB. OTUs were defined on the basis of a 5% nucleotide identity cutoff.
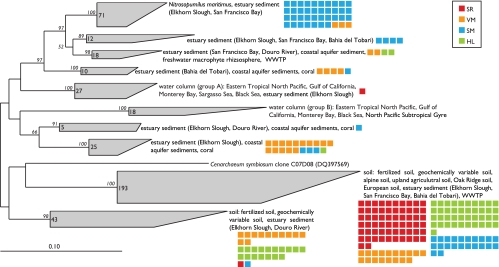
Archaeal amoA sequences were phylogenetically diverse and grouped within the previously described (16) sediment and soil/sediment clusters (Fig. 5). Sequences from site VM were diverse, with sequences falling throughout the phylogenetic tree. A majority of the site SM sequences fell into the sediment cluster comprised almost entirely of coastal/estuarine sequences, along with N. maritimus (25). Over 95% of the site HL and SR sequences fell into the soil/sediment clusters which include the group 1.1b kingdom Crenarchaeota fosmid clone 54d9 (57). The dominance of soil-like AOA amoA phylotypes at these sites may be a result of seeding of the population with terrestrial soil runoff. This is consistent with the heavy agricultural influence observed at these sites and was also noted at internal sites in the Bahía del Tóbari estuary in Mexico that received high agricultural runoff (3).
FIG. 5.
Neighbor-joining phylogenetic tree showing the affiliation of archaeal amoA sequences (579-bp fragment) from Elkhorn Slough core incubations and GenBank sequences from other environments. Colored squares represent individual sequences from each site (see legend). Significant bootstrap values (>50) are shown in italics at branch nodes.
Bacterial amoA sequences from Elkhorn Slough showed substantial overlap with those from adjacent Monterey Bay (42), as well as the San Francisco Bay, Chesapeake Bay, Plum Island Sound, and Bahía del Tóbari estuaries (3, 5, 15, 34) (Fig. 6). The majority of the sequences fell into the estuarine/marine Nitrosospira-like cluster and the estuarine/marine Nitrosomonas-like cluster, and site-specific phylotypes were evident. Interestingly, most of the SM and VM sequences fell into the estuarine/marine Nitrosospira-like cluster, while the SR and HL sequences generally fell into the estuarine/marine Nitrosomonas-like cluster. Because the salinity at each of these sites is generally polyhaline (20 to 30 psu) and because all the incubations were carried out at a constant salinity, the stark differences in β-AOB community composition are more likely due to other site-specific environmental conditions. In addition to slightly lower and more variable mean salinities, sites HL and SR also exhibit higher concentrations of agriculturally associated nutrients (Table 1). Thus, we suggest that prolonged exposure of these sediments to high concentrations of nutrients (e.g., NO3−, NH4+, PO43+, and dissolved organic carbon) and/or any other potentially agriculturally associated pollutants likely plays a more important role than salinity alone, in contrast to observations in other estuaries (5, 6, 15, 17).
FIG. 6.
Neighbor-joining phylogenetic tree showing the affiliation of bacterial amoA sequences (444-bp fragment) from Elkhorn Slough core incubations and GenBank sequences from other environments. Colored squares represent individual sequences from each site (see legend). Jasper Ridge is in California. Significant bootstrap values (>50) are shown in italics at branch nodes.
The archaeal amoA sequences had considerable compositional overlap between the stirred and unstirred cores at all of the Elkhorn Slough sites, except HL. This was confirmed with the abundance-based Jaccard index of similarity (Jabund) and θ, a nonparametric maximum-likelihood estimate of similarity calculated using SONS (Table 5) (53). High Jabund values indicate that the most abundant members are shared between two communities (i.e., they have a high degree of overlap), whereas high values of θ indicate a high degree of similarity of community structure by accounting for similarities in the relative abundances among shared OTUs. Jabund values indicated that >50% of the communities were shared between stirred and unstirred treatments (with the exception of site HL, where Jabund was 0.40; Table 5). When the relative abundances are considered, values of θ show an even higher similarity (>0.67). As with the AOA communities, there was considerable overlap between AOB communities between the stirred and unstirred cores at all sites, with Jabund values being >0.63 for all sites (except site HL, for which Jabund was 0.33) and θ values being >0.72 at all sites (Table 5).
TABLE 5.
Abundance-based microbial community analysis by SONSa
| Community analyzed by SONS and parameter | Stirred vs unstirred |
Impacted vs unimpacted |
||||
|---|---|---|---|---|---|---|
| HL | SM | SR | VM | Stirred | Unstirred | |
| Archaeal | ||||||
| Shared Chaob | 6.6 | 37.7 | 29.0 | 10.9 | 47.0 | 15.3 |
| ThetaYCc | 0.15 | 0.71 | 0.72 | 0.67 | 0.15 | 0.22 |
| Jaccard indexd | 0.40 | 0.78 | 0.53 | 0.63 | 0.74 | 0.35 |
| Sorensen indexd | 0.57 | 0.88 | 0.69 | 0.77 | 0.85 | 0.52 |
| Bacterial | ||||||
| Shared Chao | 3.5 | 7.5 | 20.8 | 19.0 | 5.8 | 10.5 |
| ThetaYC | 0.87 | 0.90 | 0.95 | 0.72 | 0.08 | 0.09 |
| Jaccard index | 0.33 | 0.63 | 0.98 | 0.89 | 0.58 | 0.77 |
| Sorensen index | 0.50 | 0.77 | 0.99 | 0.94 | 0.74 | 0.87 |
Community similarities are compared at each site between treatments as well as between agriculturally impacted versus unimpacted sites for each treatment.
Estimate of the number of OTUs that are shared between the two communities.
Nonparametric maximum-likelihood estimate of similarity (65).
The probability that a randomly selected OTU is found in both communities (given that it is in at least one of the communities).
Both AOA and AOB communities show a very strong spatial segregation between agriculturally impacted and unimpacted sites (Fig. 5 and 6 and Table 5). AOA community composition at agriculturally impacted sites (both HL and SR combined) shared only 15 to 22% similarity (based on θ values) with unimpacted sites (SM and VM). For β-AOB communities, θ values indicated only 8 to 9% similarity between agriculturally impacted sites and unimpacted sites. Thus, this would suggest again that site-specific environmental conditions (e.g., impacts from agricultural runoff) play an important role in dictating the ammonia oxidizer community composition found in estuarine sediments.
Relationship between nitrifier community and nitrification rates.
Despite the strong site specificity observed among nitrification rates and microbial community composition, we found no significant correlation (P > 0.05) between nitrification rates and the abundance of either AOA or β-AOB amoA genes. The abundance of amoA transcripts or AMO proteins (instead of gene copy numbers measured here) may have a stronger correlation with nitrification rates. In addition, different AOA and β-AOB phylotypes at the impacted and unimpacted sites in the estuary may have distinct physiologies (e.g., enzyme kinetics) and tolerances that affect nitrification rates in ways not reflected by total amoA gene abundance.
To date, only a few other estuarine studies have measured both potential nitrification rates, unlike the 15N-based rates measured here, and amoA abundance (AOA and β-AOB) (1, 9, 31). Caffrey et al. (9) showed a significant relationship between nitrification potential and AOA amoA abundance in two out of six estuaries but no relationship with β-AOB amoA abundance and concluded that AOA may play an important role in nitrification within certain estuaries. However, Magalhães et al. (31) saw no correlation between β-AOB or AOA amoA abundance and nitrification potential or NH4+ flux in the intertidal sediments of the Douro River estuary in Portugal. They concluded that β-AOB were the major contributors to nitrification, largely on the basis of the relative dominance of β-AOB amoA gene copies over AOA amoA gene copies, which may also be the case in this study of Elkhorn Slough.
Conclusions.
In this study, we observed higher 15N-based nitrification rates at less impacted, lower-nutrient sites in comparison to those at more heavily impacted, nutrient-rich sites. In addition, both the betaproteobacterial and archaeal ammonia-oxidizing communities were strongly spatially segregated. Sites with high nitrification rates primarily contained marine/estuarine Nitrosospira-like bacterial amoA sequences and phylogenetically diverse archaeal amoA sequences, while sites with low nitrification rates were dominated by estuarine Nitrosomonas-like amoA sequences and archaeal amoA sequences from previously described soil groups. Statistical community analyses confirmed these findings, showing only 8 to 9% overlap between impacted and unimpacted sites for β-AOB and only 15 to 22% overlap for AOA. Despite this spatial trend in nitrification rates and community composition, quantitative PCR revealed that the relative abundances of AOA and AOB communities did not vary significantly by site or agricultural impact. β-AOB amoA gene copies outnumbered AOA amoA gene copies at all sites and treatments. Agriculturally associated nutrients (and possibly pollutants) may play an important role in dictating the spatial distribution and activity of these estuarine ammonia-oxidizing communities.
Supplementary Material
Acknowledgments
We thank Alyson E. Santoro for invaluable field support and Katie Roberts for tireless assistance both in the field and throughout the core incubations. Helpful comments were also provided by Jason Smith. We also acknowledge support from the staff of the Elkhorn Slough National Estuarine Research Reserve, especially Kerstin Wasson and John Haskins.
Support for S.D.W. was provided by a NOAA National Estuarine Research Reserve Graduate Fellowship at the Elkhorn Slough National Estuarine Research Reserve. Additional support for S.D.W. and A.P. was provided in part by NSF grant ECS-0308070. A.C.M. was supported in part by an EPA-STAR Graduate Research Fellowship. This work was also supported in part by NSF grant MCB-0604270 to C.A.F.
Footnotes
Published ahead of print on 5 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abell, G. C., A. T. Revill, C. Smith, A. P. Bissett, J. K. Volkman, and S. S. Robert. 2010. Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 4:286-300. [DOI] [PubMed] [Google Scholar]
- 2.An, S., and S. B. Joye. 2001. Enhancement of coupled nitrification-denitrification by benthic photosynthesis in shallow estuarine sediments. Limnol. Oceanogr. 46:62-74. [Google Scholar]
- 3.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing Archaea and Bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beman, J. M., B. N. Popp, and C. A. Francis. 2008. Molecular and biogeochemical evidence of ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429-441. [DOI] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., J. Tucker, A. E. Giblin, and D. A. Stahl. 2007. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ. Microbiol. 9:1439-1447. [DOI] [PubMed] [Google Scholar]
- 7.Broenkow, W. W., and L. C. Breaker. 2005. A 30-year history of tide and current measurements in Elkhorn Slough, California. Scripps Institution of Oceanography Library, San Diego, CA.
- 8.Caffrey, J., N. Harrington, and B. B. Ward. 2002. Biogeochemical processes in a small California estuary 1: benthic fluxes and porewater constituents reflect high nutrient freshwater inputs. Mar. Ecol. Prog. Ser. 233:39-53. [Google Scholar]
- 9.Caffrey, J. M., N. Bano, K. Kalanetra, and J. T. Hollibaugh. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660-662. [DOI] [PubMed] [Google Scholar]
- 10.Casciotti, K. L., D. M. Sigman, and B. B. Ward. 2003. Linking diversity and stable isotope fractionation in ammonia-oxidizing bacteria. Geomicrobiol. J. 20:335-353. [Google Scholar]
- 11.Cebron, A., T. Berthe, and J. Garnier. 2003. Nitrification and nitrifying bacteria in the Lower Seine River and Estuary (France). Appl. Environ. Microbiol. 69:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dollhopf, S. L., J.-H. Hyun, A. C. Smith, H. J. Adams, S. O'Brien, and J. E. Kostka. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl. Environ. Microbiol. 71:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, L. F., D. B. Nedwell, and A. Stott. 2006. Sources of nitrogen used for denitrification and nitrous oxide formation in sediments of the hypernutrified Colne, the nutrified Humber, and the oligotrophic Conwy estuaries, United Kingdom. Limnol. Oceanogr. 51:545-557. [Google Scholar]
- 14.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 15.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 16.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag, T. E., L. Chang, and J. I. Prosser. 2005. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684-696. [DOI] [PubMed] [Google Scholar]
- 18.Gihring, T. M., A. Canion, A. Riggs, M. Huettel, and J. E. Kostka. 2010. Denitrification in shallow, sublittoral Gulf of Mexico permeable sediments. Limnol. Oceanogr. 54:43-54. [Google Scholar]
- 19.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, P. M. Richardson, and E. F. DeLong. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, J.-Z., J.-P. Shen, L.-M. Zhang, Y.-G. Zhu, Y.-M. Zheng, M.-G. Xu, and H. Di. 2007. Quantitative analyses of the abundance and compostion of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9:2364-2374. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, R. M., J. W. McClelland, D. M. Sigman, B. Fry, and B. J. Peterson. 1998. Measuring 15N-NH4+ in marine, estuarine and fresh waters: an adaptation of the ammonia diffusion method for samples with low ammonium concentrations. Mar. Chem. 60:235-243. [Google Scholar]
- 22.Hunter, E. M., H. J. Mills, and J. E. Kostka. 2006. Microbial community diversity associated with carbon and nitrogen cycling in permeable shelf sediments. Appl. Environ. Microbiol. 72:5689-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, K., M. Jensen, and E. Kristensen. 1996. Nitrification and denitrification in Wadden Sea sediments (Konigshafen, Island of Sylt, Germany) as measured by nitrogen isotope pairing and isotope dilution. Aquat. Microb. Ecol. 11:181-191. [Google Scholar]
- 24.Koike, I., and A. Hattori. 1978. Simultaneous determinations of nitrification and nitrate reduction in coastal sediments by a 15N dilution technique. Appl. Environ. Microbiol. 35:853-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Könneke, M., A. E. Bernhard, J. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia oxidizing marine archeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 26.Koop-Jackobsen, K., and A. E. Giblin. 2009. Anammox in Tidal marsh sediments: the role of salinity, nitrogen loading, and marsh vegetation. Estuaries Coasts 32:238-245. [Google Scholar]
- 27.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 28.Los Huertos, M., L. Gentry, and C. Shenan. 2001. Land use and stream nitrogen concentrations in agricultural watersheds along the central coast of California. Sci. World 1:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, R. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Rieichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schliefer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddison, D. R., and W. P. Maddison. 2000. MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland, MA. [DOI] [PubMed]
- 31.Magalhães, C. M., A. Machado, and A. A. Bordalo. 2009. Temporal variability in the abundance of ammonia-oxidizing bacteria vs. archaea in sandy sediments of the Douro River estuary, Portugal. Aquatic Microb. Ecol. 56:13-23. [Google Scholar]
- 32.Martens-Habbena, W., P. M. Berube, H. Urakawa, J. de la Torre, and D. A. Stahl. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976-979. [DOI] [PubMed] [Google Scholar]
- 33.Mincer, T. J., M. J. Church, L. T. Taylor, C. M. Preston, D. M. Karl, and E. F. DeLong. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162-1175. [DOI] [PubMed] [Google Scholar]
- 34.Mosier, A. C., and C. A. Francis. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002-3016. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa, T., K. Mori, C. Kato, R. Takahashi, and T. Tokuyama. 2007. Distribution of cold-adapted ammonia-oxidizing microorganisms in the deep-ocean of the north eastern Japan Sea. Microbes Environ. 22:365-372. [Google Scholar]
- 36.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia-oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen, L. P. 1992. Denitrification in sediment determined from nitrogen isotope pairing. FEMS Microbiol. Ecol. 86:357-362. [Google Scholar]
- 38.Nishio, T., I. Koike, and A. Hattori. 1982. Denitrification, nitrate reduction, and oxygen consumption in coastal and estuarine sediments. Appl. Environ. Microbiol. 43:648-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishio, T., I. Koike, and A. Hattori. 1983. Estimates of denitrification and nitrification in coastal and estuarine sediments. Appl. Environ. Microbiol. 45:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogilvie, B. G., D. B. Nedwell, R. M. Harrison, A. D. Robinson, and A. Sage. 1997. High nitrate, muddy estuaries as nitrogen sinks: the nitrogen budget of the River Colne estuary (UK). Mar. Ecol. Prog. Ser. 150:217-228. [Google Scholar]
- 42.O'Mullan, G. D., and B. B. Ward. 2005. Comparison of temporal and spatial variability of ammonia-oxidizing bacteria to nitrification rates in Monterey Bay, California. Appl. Environ. Microbiol. 71:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prosser, J. I., and G. W. Nicol. 2008. Relative contributions of archaea and bacteria to aerobic ammonium oxidation in the environment. Environ. Microbiol. 10:2931-2941. [DOI] [PubMed] [Google Scholar]
- 44.PVWMA. 2010. Surface water monitoring. Pajaro Valley Water Management Agency, Watsonville, CA. http://www.pvwma.dst.ca.us.
- 45.Rao, A., J. J. McCarthy, W. S. Gardner, and R. A. Jahnke. 2008. Respiration and denitrifcation in permeable continental shelf deposits on the South Atlantic Bight: N2:Ar and isotope pairing measurements in sediment column experiments. Continental Shelf Res. 28:602-613. [Google Scholar]
- 46.Rich, J. J., O. R. Dale, B. Song, and B. B. Ward. 2008. Anaerobic ammonium oxidation (anammox) in Chesapeake Bay sediments. Microb. Ecol. 55:311-320. [DOI] [PubMed] [Google Scholar]
- 47.Risgaard-Petersen, N., R. L. Meyer, M. Schmid, M. S. M. Jetten, A. Enrich-Prast, S. Rysgaard, and N. P. Revsbech. 2004. Anaerobic ammonium oxidation in an estuarine sediment. Aquatic Microb. Ecol. 36:293-304. [Google Scholar]
- 48.Risgaard-Petersen, N., L. P. Nielsen, S. Rysgaard, T. Dalsgaard, and R. L. Meyer. 2003. Application of the isotope pairing technique in sediments where anammox and denitrification coexist. Limnol. Oceanogr. Methods 1:63-73. [Google Scholar]
- 49.Rotthauwe, J., K. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rysgaard, S., N. Risgaard-Petersen, L. P. Nielsen, and N. P. Revsbech. 1993. Nitrification and denitrification in lake and estuarine sediments measured by the 15N dilution technique and isotope pairing. Appl. Environ. Microbiol. 59:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santoro, A. E., C. A. Francis, N. R. de Sieyes, and A. B. Boehm. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068-1079. [DOI] [PubMed] [Google Scholar]
- 52.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, J.-P., L.-M. Zhang, Y.-G. Zhu, J.-B. Zhang, and J.-Z. He. 2008. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 10:1601-1611. [DOI] [PubMed] [Google Scholar]
- 55.Steingruber, S. M., J. Friedrich, R. Gächter, and B. Wehrli. 2001. Measurement of denitrification in sediments with the 15N isotope pairing technique. Appl. Environ. Microbiol. 67:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tourna, M., T. E. Freitag, G. W. Nicol, and J. I. Prosser. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357-1364. [DOI] [PubMed] [Google Scholar]
- 57.Treusch, A., S. Leininger, A. Kletzin, S. C. Schuster, H.-P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 58.Trimmer, M., J. C. Nicholls, and B. Deflandre. 2003. Anaerobic ammonium oxidation measured in sediments along the Thames Estuary, United Kingdom. Appl. Environ. Microbiol. 69:6447-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Usui, T., I. Koike, and N. Ogura. 2001. N2O production, nitrification and denitrification in an estuarine sediment. Estuarine Coastal Shelf Sci. 52:769-781. [Google Scholar]
- 60.Venter, J. C., K. Remington, J. F. Heidelbery, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Pualsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 61.Wankel, S. D. Nitrogen sources and cycling in coastal ecosytems: insights from a nitrogen and oxygen stable isotope approach. Stanford University, Stanford, CA.
- 62.Wankel, S. D., C. Kendall, and A. Paytan. 2009. Using nitrate dual isotopic composition (δ15N and δ18O) as a tool for exploring sources and cycling of nitrate in an estuarine system: Elkhorn Slough, California. J. Geophys. Res. 114:G01011. doi: 10.1029/2008JG000729. [DOI] [Google Scholar]
- 63.Winogradsky, S. N. 1890. Sur les organismes de la nitrification. C. R. Acad. Sci. 110:1013-1016. [Google Scholar]
- 64.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. S. Damste. 2006. Archael nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yue, J. C., and M. K. Clayton. 2005. A similarity measure based on species proportions. Commun. Stat. Theor. Methods 34:2123-2131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.