Abstract
The 13 Shiga-toxigenic Escherichia coli (STEC) strains isolated from wholesale spinach and lettuce consisted mostly of serotypes that have not been implicated in illness. Among these strains, however, were two O113:H21 that carried virulence genes common to this pathogenic serotype (stx2, ehxA, saa, and subAB), suggesting that their presence in ready-to-eat produce may be of health concern.
Consumption of fresh produce has been implicated in several outbreaks of food-borne illness, thus raising concerns about the microbial safety of these products. The Microbiological Data Program (MDP) of the USDA's Agricultural Marketing Service (http://www.ams.usda.gov/AMSv1.0/mdp) was initiated in 2001 to conduct microbiological surveillance of fresh produce samples (including domestic, imported, and organically grown) collected from terminal markets and wholesale distribution centers. The samples are tested for the presence of Salmonella, enterotoxigenic Escherichia coli, E. coli serotype O157:H7, and other Shiga-toxigenic E. coli (STEC) strains using methods that are described on that website. In the MDP surveys performed thus far, no E. coli O157:H7 strains have been found and the small number of STEC strains found, about 2 to 5 per year from 2002 to 2008 (see MDP Reports on their website), came mainly from samples of lettuce or sprouts and were of serotypes that have not been implicated in human illness or that have seldom been found in isolates from cattle or the environment (5). In the 2009 survey, MDP tested 15,354 samples that included alfalfa sprouts, lettuce, spinach, hot peppers, green onions, cantaloupe, and cilantro. From these, 12 STEC strains were isolated from the analysis of 2,328 spinach samples and 1 STEC strain was isolated from the 2,336 lettuce samples tested—all from domestically grown products. The 13 STEC strains, serotyped by the E. coli Reference Center at Penn State University, were primarily of rare serotypes that are seldom isolated; however, a few serotypes that may potentially be of health concern were found. In this study, we further characterized the 13 STEC strains isolated from produce to determine whether they carried other virulence genes and may be potentially pathogenic.
All strains were plated on MacConkey agar with ColiComplete (BioControl, Belleview, WA) (see reference 20) and were determined to exhibit β-galactosidase and β-glucuronidase activities, consistent with E. coli strains (Table 1). The somatic (O) and flagellar (H) serotypes were determined for most strains (Table 1), but the three that could not be serotyped (strains 09-00002, 09-00031, and 09-00043) were subjected to genetic O serotyping by several PCR assays (12, 16). The O types of these strains remained undetermined but were confirmed not to be O157, O111, O26, O121, O145, O45, or O113.
TABLE 1.
Summary of analysis results of STEC strains isolated from produce
| Strain | Serotypea | Source | Results of analysis |
|||||
|---|---|---|---|---|---|---|---|---|
| PCRb | Stx2c | EhxAd | saae | subABf | sabg | |||
| 09-00002 | NA | Spinach | stx2, ehxA | + | + | + | + | NAh |
| 09-00009 | O172:H2 | Spinach | stx2 | − | NA | − | − | NA |
| 09-00024 | O116:H21 | Spinach | stx2, ehxA | + | + | + | + | NA |
| 09-00025 | O116:H21 | Spinachi | stx2, ehxA | + | + | + | + | NA |
| 09-00026 | O116:H21 | Spinachi | stx2 | + | NA | − | − | NA |
| 09-00027 | O113:H? | Spinach | stx2, ehxA | + | + | + | + | − |
| 09-00028 | O8:H− | Spinach | stx2, ehxA | − | Weak + | − | − | NA |
| 09-00029 | O11:H15 | Spinach | stx2? | − | NA | − | − | NA |
| 09-00031 | O−:H19 | Spinach | stx2, ehxA | + | + | + | + | NA |
| 09-00039 | O168:H8 | Spinach | stx2, ehxA | + | − | − | − | NA |
| 09-00043 | O−:H21 | Spinach | stx2 | − | NA | − | − | NA |
| 09-00047 | O113:H21 | Spinach | stx2, ehxA | + | + | + | + | − |
| 09-00049 | O168:H− | Lettuce | stx2, ehxA | + | − | − | − | NA |
O and H serotyping results from the E. coli Reference Center, Penn State University.
Various PCRs were used to test for stx1 and stx2, ehxA (enterohemolysin), γ-eae, and other eae alleles, and for O antigen genes for O157, O26, O111, O121, O145, and O103 serotypes.
Serological analysis was done to test for Stx2 using VTEC-RPLA (Denka Seiken, Japan).
Enterohemolysis was done on blood agar plates with Ca2+ to test for EhxA.
PCR was used to test for the Saa gene.
PCR was used to test for the subtilase cytotoxin genes.
PCR was used to test for the gene encoding the STEC autotransporter protein.
NA, not applicable.
Organic product.
All 13 isolates were found to be STEC by a preliminary screening multiplex PCR (13), which used a single primer pair to detect both Shiga toxin (stx) genes 1 and 2. To determine the specific Stx types carried by the strains, another multiplex PCR was used (3); this PCR differentiates stx1 and stx2 and detects other virulence and trait markers, including, γ-eae (intimin), ehxA (enterohemolysin), and the uidA (β-glucuronidase) single-nucleotide polymorphism at position +93 in O157:H7 strains. The γ-eae allele is found mostly in O157:H7 strains, but since there are ∼30 known eae alleles that are carried by STEC strains of other serotypes, the isolates were also tested by another PCR for the presence of other eae alleles (12).
All the isolates were confirmed to be STEC carrying only stx2 (Table 1). An exception was strain 09-00029, which was initially believed to be stx positive but was subsequently found not to carry either stx1 or stx2. There are several genetic alleles of stx2, and sequence analysis showed that the stx-specific primers used in the different PCR assays varied in their abilities to detect stx2 alleles. Hence, it is plausible that strain 09-00029 is carrying a stx2 allele that was detected only by the initial screening PCR assay. Serological analysis for Stx using the VTEC-RPLA (verotoxin-producing E. coli reverse passive latex agglutination) assay (Denka Seiken, Japan) showed that 9 of 13 strains produced Stx2 (Table 1), but 4 strains that carried stx2, including 09-00029, were Stx2 negative. The VTEC-RPLA assay does not detect products of all stx2 alleles; hence, these results are consistent with our speculation that strain 09-00029, and perhaps the other 3 strains as well, are carrying a stx2 allele. Enterohemolysin, encoded by the ehxA gene, is a putative STEC virulence factor and was found in 9 of 13 strains (Table 1). However, phenotypic analysis showed that the two O168 serotype strains (09-00039 and 09-00049) that had ehxA did not exhibit enterohemolysis on blood agar plates supplemented with 10 mM CaCl2 (Table 1).
PCR analysis showed that none of the 13 STEC strains carried eae alleles that encode intimin, a protein that enables bacterial attachment to intestinal epithelial cells. Intimin is a known virulence factor of STEC and enteropathogenic E. coli, but STEC strains devoid of eae have also caused severe illnesses, including hemolytic-uremic syndrome (HUS) (19), suggesting that these pathogens have other means for cellular attachment (2). Characterization of eae-negative STEC strains have identified Saa (STEC autoagglutinating adhesin) as a candidate adherence protein (18) but have also found other putative virulence factors, like subtilase cytotoxin (encoded by subAB) (14), that are also predominant in eae-negative STEC strains. Since these STEC isolates from produce also did not carry eae, they were tested by PCR for the presence of saa (17) and subAB (14) genes. Results showed that only six STEC strains carried the saa gene but that all six also had subAB, stx2, and ehxA and expressed both Stx2 and EhxA (Table 1). This is consistent with the findings from the analysis of a large panel of STEC isolates in Brazil, which showed that strains that carry saa and subAB often also have stx2 and ehxA (7). Two of the six STEC strains from produce that had all four of these virulence genes were of the O116:H21 serotype. There was a third O116:H21 strain (09-00026), but it did not carry ehxA, saa, or subAB (Table 1), suggesting that there is diversity within the serotype. STEC strains of serotype O116:H21 have been isolated from animals, the environment (21), and ground beef samples in various countries (10, 15) but have seldom been isolated from humans or caused illness (5, 21). However, an O116:H21 strain has been isolated from a HUS patient (11); hence, the pathogenicity of O116:H21 strains, including these produce isolates, remains uncertain.
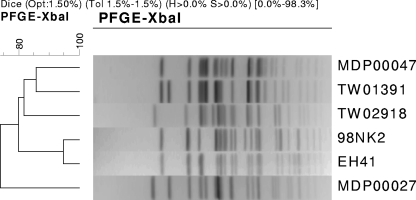
Of the other four STEC strains from produce that carried stx2, ehxA, saa, and subAB, two strains (09-00002 and 09-00031) could not be, or were only partially, serotyped. The other two strains were of the O113 serotype, and PCR (16) confirmed that both strains had the O113 antigen gene. Strain 09-00047 was serologically typed to be O113:H21, while strain 09-00027 had an undetermined H type (Table 1). The latter strain was therefore subjected to genetic H serotyping by PCR amplifying and sequencing the fliC gene that encodes flagellin (9). Analysis for sequence homologies in GenBank by BLAST analysis showed that strain 09-00027 had the H21 fliC sequence; hence, it is also an O113:H21 strain. STEC strains of the O113:H21 serotype do not carry eae; they were first implicated in HUS in 1983 (8) and also caused a cluster of HUS cases in Australia in 1998 (19). Analysis of the pathogenic O113:H21 strains from Australia showed that they carried stx2, ehxA, saa, and subAB (17, 18, 19) but also had the sab gene that encodes an outer membrane autotransporter protein (4) that enhances biofilm formation and is thought to be a putative virulence factor. PCR analysis for the sab gene (4) showed that it was absent in both O113:H21 strains that were isolated from produce (Table 1) but present in the two Australian O113:H21 strains that caused HUS (EH41 and 98NK2) that were used as positive controls (data not shown). The sab gene is reported to be carried by eae-negative STEC strains and found mostly in strains of O113:H21 and a few other selected serotypes (4). However, it appears that sab may not be present in all O113:H21 strains, as TW01391 and TW02918, both O113:H21 strains, isolated from HUS and diarrhea patients, respectively, and obtained from the STEC Center at Michigan State University, also did not have sab (data not shown). Pulsed-field gel electrophoresis analysis of the XbaI-digested genomic DNA showed that the profile of the two produce O113:H21 isolates (MDP 09-00027 and MDP 09-00047) did not resemble the profiles of either the Australian O113:H21 strains (EH41 and 98NK2) or those of TW01391 and TW02918 (Fig. 1).
FIG. 1.
PFGE profiles of XbaI-digested genomic DNA of O113:H21 strains and a dendrogram comparing profile similarities. The strain designations are shown at the right of the profiles. The cluster tree was constructed using the BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) software package.
Serotype O113:H21 strains have been isolated from foods, animals, and the environment in several countries (1, 6, 15). An analysis of 34 O113:H21 strains isolated from healthy cattle in Brazil (10) showed that many carried stx2, ehxA, saa, and subAB. Since these are virulence factors that are common in the pathogenic O113:H21 strains that have caused HUS, the authors cautioned that these cattle isolates may also pose a health risk. In prior years of MDP analysis, STEC strains of the O113:H21 serotype had not been encountered. However, the isolation of two O113:H21 strains from produce in the 2009 MDP study presents a situation analogous to the Brazilian study (10), as both produce strains carried virulence factors identical to those of the pathogenic O113:H21 strains and, therefore, may potentially be of health concern.
Lastly, it is interesting to note that 12 of the 13 STEC isolates from produce in 2009 came from spinach samples. MDP initiated spinach testing in 2008 and, in that year, 5 of the 7 STEC isolates also came from spinach, while the remaining 2 were isolated from bagged lettuce. At present, it may be premature to speculate on the high rate of STEC isolation from spinach; however, it will be interesting to monitor future survey data to see if this trend continues and whether any correlations exist.
Acknowledgments
We thank L. Hartland and A. Paton as well as the STEC Center at Michigan State University for providing the O113:H21 strains used as controls in this study.
Footnotes
Published ahead of print on 5 November 2010.
REFERENCES
- 1.dos Santos, L. F., K. Irino, T. M. I. Vaz, and B. E. C. Guth. 2010. Set of virulence genes and genetic relatedness of O113:H21 Escherichia coli strains isolated from the animal reservoir and human infections in Brazil. J. Med. Microbiol. 59:634-640. [DOI] [PubMed] [Google Scholar]
- 2.Dytoc, M. T., A. Ismaili, D. J. Philpott, R. Soni, J. L. Brunton, and P. M. Sherman. 1994. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect. Immun. 62:3494-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng, P., and S. R. Monday. 2000. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic Escherichia coli serotypes. Mol. Cell. Probes 14:333-337. [DOI] [PubMed] [Google Scholar]
- 4.Herold, S., J. C. Paton, and A. W. Paton. 2009. Sab, a novel autotransporter of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation. Infect. Immun. 77:3234-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussein, H. S. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85:E63-E72. [DOI] [PubMed] [Google Scholar]
- 6.Irino, K., M. A. M. F. Kato, T. M. I. Vaz, I. I. Ramos, M. A. C. Souza, A. S. Cruz, T. A. Gomes, M. A. M. Vieira, and B. E. C. Guth. 2005. Serotypes and virulence markers of Shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in Sao Paulo State, Brazil. Vet. Microbiol. 105:29-36. [DOI] [PubMed] [Google Scholar]
- 7.Irino, K., M. A. Vieira, T. A. Gomes, B. E. Guth, Z. V. Naves, M. G. Oliveira, L. F. dos Santos, M. Guirro, C. D. Timm, C. P. Pigatto, S. M. Farah, and T. M. Vaz. 2010. Subtilase cytotoxin-encoding subAB operon found exclusively among Shiga toxin-producing Escherichia coli strains. J. Clin. Microbiol. 48:988-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verocytotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 9.Lacher, D. W., H. Steinland, T. E. Blank, M. S. Donnenberg, and T. S. Whittam. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189:342-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchesi, P. M. A., A. Kruger, and A. E. Parma. 2006. Distribution of saa variants in verocytotoxigenic Escherichia coli isolated from cattle and food. Res. Microbiol. 157:263-266. [DOI] [PubMed] [Google Scholar]
- 11.Luck, S. N., V. Bennett-Wood, R. Poon, R. M. Robins-Browne, and E. L. Hartland. 2005. Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 73:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monday, S. R., A. Beisaw, and P. C. H. Feng. 2007. Identification of Shiga toxigenic E. coli seropathotypes A and B by multiplex PCR. Mol. Cell. Probes 21:308-311. [DOI] [PubMed] [Google Scholar]
- 13.Monday, S. R., C. Keys, P. Hansen, Y. Shen, T. S. Whittam, and P. Feng. 2006. Produce isolates of Escherichia coli Ont:H52 serotype that carry both Shiga toxin 1 and stable toxin genes. Appl. Environ. Microbiol. 72:3062-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton, H. J., J. Sloan, D. M. Bulach, T. Seemann, C. C. Allison, M. Tauschek, R. M. Robins-Browne, J. C. Paton, T. S. Whittam, A. W. Paton, and E. L. Hartland. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parma, A. E., M. E. Sanz, J. E. Blanco, J. Blanco, M. R. Viñas, M. Blanco, N. L. Padola, and A. I. Echeverria. 2000. Virulence genotypes and serotypes of verotoxigenic Escherichia coli isolated from cattle and foods in Argentina. Eur. J. Epidemiol. 16:757-762. [DOI] [PubMed] [Google Scholar]
- 16.Paton, A. W., and J. C. Paton. 1999. Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR. J. Clin. Microbiol. 37:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli strains by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. Food and Drug Administration. July 2009. Bacteriological analytical manual, chapter 4a. Diarrheagenic Escherichia coli, section P.1.e. http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/UCM070080.
- 21.WHO. 1998. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a WHO Scientific Working Group Meeting. World Health Organization, Geneva, Switzerland. http://whqlibdoc.who.int/hq/1998/WHO_CSR_APH_98.8.pdf.