Abstract
Visible-wavelength Raman spectroscopy was used to investigate the uptake and metabolism of the five-carbon sugar alcohol xylitol by Gram-positive viridans group streptococcus and the two extensively used strains of Gram-negative Escherichia coli, E. coli C and E. coli K-12. E. coli C, but not E. coli K-12, contains a complete xylitol operon, and the viridans group streptococcus contains an incomplete xylitol operon used to metabolize the xylitol. Raman spectra from xylitol-exposed viridans group streptococcus exhibited significant changes that persisted even in progeny grown from the xylitol-exposed mother cells in a xylitol-free medium for 24 h. This behavior was not observed in the E. coli K-12. In both viridans group streptococcus and the E. coli C derivative HF4714, the metabolic intermediates are stably formed to create an anomaly in bacterial normal survival. The uptake of xylitol by Gram-positive and Gram-negative pathogens occurs even in the presence of other high-calorie sugars, and its stable integration within the bacterial cell wall may discontinue bacterial multiplication. This could be a contributing factor for the known efficacy of xylitol when taken as a prophylactic measure to prevent or reduce occurrences of persistent infection. Specifically, these bacteria are causative agents for several important diseases of children such as pneumonia, otitis media, meningitis, and dental caries. If properly explored, such an inexpensive and harmless sugar-alcohol, alone or used in conjunction with fluoride, would pave the way to an alternative preventive therapy for these childhood diseases when the causative pathogens have become resistant to modern medicines such as antibiotics and vaccine immunotherapy.
Although the exact mechanism of the efficacy of xylitol on pathogenic bacteria is not known, there is an impressive collection of clinical reports claiming its preventive action in a number of pediatric diseases. Xylitol is a natural five-carbon-sugar alcohol that can be safely applied as a preventive measure for diseases such as pneumonia, acute otitis media (AOM), dental caries, and meningitis (11, 16, 21, 38, 41, 45, 47). The causative pathogens of pneumonia in children are Gram-positive Streptococcus pneumoniae and Gram-negative Klebsiella pneumoniae with polysaccharide capsules. Both classes of bacteria are morphologically diplococcic (33). The pathogens causing AOM (middle-ear infection of children) are Gram-positive S. pneumoniae (30% of cases), Gram-negative bacteria Haemophilus influenzae (25% of cases), and Moraxella catarrhalis (20% of cases) (52). They are also diplococcic (7). It remains to be seen whether all of these pathogenic organisms contain xylitol operons and whether they are capable of utilizing xylitol even in the presence of other sugars or carbon sources.
One likely explanation for xylitol efficacy involves the phosphotransferase system of streptococci and its ability to take in and metabolize different sugars depending on the current sugar environment (48). It has been hypothesized that xylitol is metabolized to xylitol-5-phosphate in a “futile cycle,” which mutans group streptococci cannot utilize further and which may even be toxic to bacteria (44). It has also been observed that exposure to low concentrations of xylitol results in a reduction of cell adherence to epithelial cells (17), perhaps due to a disruption of polysaccharide production in the xylitol-exposed bacteria (39). There is also evidence for significant ultrastructure alteration in xylitol-exposed S. mutans (46) and S. pneumoniae (41). Careful Raman spectroscopic studies of the molecular alteration induced in xylitol-exposed streptococci may shed new light on one or more of these biomolecular processes.
The four structurally related known pentitols (or C5 polyol epimers) are xylitol, ribitol, d-arabitol, and l-arabitol. Only d-arabitol and ribitol are abundant in nature (2). d-Arabitol, which forms 10% of the dry weight of certain mushrooms, is capable of binding to capsules and teichoic acids of several Gram-positive bacteria. Ribitol is also present in free form in some higher plants (1, 10). Most of the previous work on catabolism of these compounds was done with various Aerobacter aerogenes strains, which are now classified as Klebsiella or Enterobacter species (32, 50).
Reiner and Scangos have characterized the catabolism of ribitol (rtl+) and d-arabitol (atl+) in E. coli C and the disadvantages of a constitutive catabolite pathway (30, 34). Briefly, each operon contains genes for a dehydrogenase and a kinase and is under negative control. They have also shown that constitutive production of ribitol catabolic enzymes selected in response to xylitol render the same mutant more sensitive to certain other five-carbon sugar alcohols. The ribitol operon is inducible by ribitol, and the d-arabitol operon is inducible by d-arabitol. Significantly, these two operons are very closely linked and located in the E. coli C chromosome between the metG and his (histidine) genes in a mirror-image arrangement, rtlB-rtlA-rtlC-atlC-atlA-atlB (30). The rtl and atl operons are well-characterized in E. coli C and Klebsiella aerogenes (25). Proteus, Klebsiella, and Enterobacter species are common urinary tract pathogens. However, the two other extensively used E. coli strains, E. coli K-12 and E. coli B, are genetically very close to E. coli C but, surprisingly, they lack the DNA region or xylitol operon (∼2.5 kb) loci involved in the utilization of ribitol (rtl) and d-arabitol (atl). The xylitol operon is located between his and metG loci and is genetically linked to the bacteriophage P2 attachment site (49).
In the present study we report on the use of visible wavelength Raman spectroscopy to analyze the uptake and retention of xylitol in E. coli and viridans group streptococcus. Raman spectroscopy is a versatile molecular vibrational technique that has only recently been widely applied for identification and characterization purposes in microbiology (22, 28). In these applications, a laser beam is nondestructively incident upon a bacterial target, and the inelastically scattered light is carefully dispersed. Shifts in the scattered photon energy corresponding to vibrational modes in the molecules of the target are then measured to determine molecular composition. A Raman spectrum is therefore a map where every peak can be associated with a particular bond in a specific molecule. Although the locations (and thus the identities) of Raman peaks from specific bonds in specific molecules can be predicted by ab initio calculations, this is seldom done in such complex and diverse systems as biological targets. Instead, it is more common to utilize the extensive body of published experimental Raman literature from the last 30 years of research to identify observed Raman peaks. For example, the identities of the Raman peaks observed in the spectra of xylitol and the other common sugars and polyols have been determined previously (5), as have the peaks in bacterial spectra such as S. aureus (50) and E. coli (13) and certain amino acids (37). While the peaks in homogenous, pure samples can therefore be identified with some certainty, in highly heterogeneous molecular systems such as bacterial cells the incredible diversity of molecules often reduces peak identification to accurate generalizations such as “lipid” or “protein.” Nonetheless, the Raman technique has been effectively used to biochemically characterize bacterial slurries (8), to probe bacterial colonies (3), to identify and discriminate many medically important microbes (including E. coli, Klebsiella spp., Enterococcus spp., Proteus mirabilis, Staphylococcus aureus, Candida strains, and Pseudomonas spp.) (13, 22), and even to probe individual microbial cells (9, 12).
MATERIALS AND METHODS
Bacterial strain selection and growth conditions.
We compared the uptake of xylitol by the four well-characterized strains of E. coli—E. coli K-12, JW1881-1, E. coli C, and E. coli HF4714—and an ATCC strain of viridans group streptococcus. E. coli K-12 and its flagellum- and pilus-negative mutant strain JW1881-1 lacking the genes to ferment xylitol were used as xylitol-nonfermenting Gram-negative controls. E. coli C was chosen because it possesses the xylitol operon in a repressed state, and E. coli HF4714 was chosen because it possesses the xylitol operon in a derepressed state (operator constitutive). HF4714, as constructed by a genetic recombination between the E. coli K-12 donor and the E. coli C recipient, contains the xylitol operon in a functional state (Howard Flanders, unpublished data). E. coli C is sensitive to the single-stranded DNA-containing semitemperate bacteriophage φX174 but produces very turbid plaques to its amber mutant of φX174am3, while HF4714 is equally sensitive to both wild-type φX174 and φXam3. E. coli K-12 is known to be resistant to these phages but sensitive to another temperate phage P2. The attachment site of this phage is closely linked to the ribitol or xylitol operon in E. coli C. Some of these characteristics are extremely useful to routinely test the genetic purity of all of the E. coli strains used in our experiments.
The selection of a well-characterized viridans group streptococcus strain for the present study was predicated on several factors. S. pneumoniae, on which the action of xylitol has previously been studied, is highly pathogenic and its inclusion here was not suggested for this reason. The dental pathogen S. mutans is a better candidate to study; however, it has a well-established growth problem in common laboratory nutrition medium since it is multiply auxotrophic with numerous nutritional requirements for normal growth (18). Viridans group streptococcus has several characteristics in common with S. mutans and S. pneumoniae, both of which have already been shown be susceptible to xylitol (41), and is known to cause a variety of infections commonly associated with dental caries (26). Using PCR technology with primers of 16S rRNA gene sequence, we verified that viridans group streptococcus and S. mutans do not differ in DNA sequences (our unpublished data).
The viridans group streptococcus strain and the above-mentioned E. coli strains were grown in brain heart infusion (BHI) or tryptic soy broth (TSB) (rich liquid medium) containing a minimal amount of dextrose together with xylitol (0 to 2%). Considerable dilution of the overnight bacterial culture prior to growth in xylitol was very important. The titer was significantly reduced from 108/ml to 104/ml to make sure the population consisted only of planktonic cells before they were exposed to xylitol. This low titer increased the chance for the entire bacterial population to grow through their log-phase in the presence of xylitol. After growth for 24 h, both aerobically and anaerobically at 37°C, these bacterial cultures were examined by Gram staining. Then, the Gram-positive viridans group streptococcus strain and the Gram-negative E. coli were spread, respectively, on two selective media, colistin-nalidixic acid (CNA) and BHI, for a comparison of their purity and titer. BHI with 1.5% agar and tryptic soy agar (TSA) are rich nutrient agar media, and the CNA is a differential blood-agar medium that contains two antibiotics, colistin (polymyxin B) and a quinolone drug (nalidixic acid). Viridans group streptococcus was selectively grown on the CNA medium, while the Gram-negative isolates were selected on bile-containing MacConkey-lactose media. The blood agar medium without antibiotics was used to distinguish the alpha, beta, and gamma hemolysis. Viridans group streptococcus demonstrates gamma hemolysis.
To study the retention of xylitol as a function of time, specimens of viridans group streptococcus and E. coli obtained from the xylitol-containing broth cultures (grown in a CO2 jar) were spread on xylitol-free TSA plates and incubated for 24 h both aerobically and anaerobically at 37°C. Colonies were collected from these plates, suspended in 50 μl of sterile-distilled water, centrifuged, and the pellet was used for Raman spectroscopy. Next, 10 μl of the moist pellet was mounted on a low-fluorescence quartz microscope slide and allowed to dry in air prior to Raman spectroscopy. In all cases, a final bacterial titer of ∼108 cells was utilized.
Raman spectroscopy.
Raman spectra were obtained by using a Jobin-Yvon Horiba Triax 550 spectrometer equipped with a liquid-nitrogen-cooled charge-coupled device. A Stellar-Pro-L continuous-wavelength argon-ion laser with a 514-nm wavelength was used to excite the sample with 8 mW of power. An Olympus model BX41 microscope equipped with a ×100 objective lens was used to focus the laser light on the sample and collect the back-scattered Raman-shifted light. A notch filter removed the elastically scattered light, and the Raman scattered light was dispersed using a 1,200-line/mm grating, focused on the detector, and recorded by LabSpec software that also controlled the experimental parameters, including the exposure time and the number of accumulations. Raman measurements were recorded in the spectral region between 600 and 2,000 cm−1 with an exposure time of 10 s and three accumulations. All Raman spectra were calibrated using an Si wafer as a standard. A Matlab program utilizing an adaptive min-max method was used to process the data by subtracting the broadband fluorescence background and normalizing each spectrum by setting the maximum intensity of the channel to one. Powdered dry xylitol was pressed and compacted to form a flat solid surface, and Raman measurements were recorded and used for comparison purposes.
Bacterial Raman spectra were obtained from dense pellets obtained from multiple cultures grown on different days, and these spectra were then averaged together to create “average” spectra. The spectra averaged in this way were always 100% representative of the changes being measured. No differences between spectra from bacteria prepared in a specific manner were ever observed. This was confirmed in hundreds of spectra acquired from multiple aliquots. Therefore, the averaging of spectra, while not necessary, was a highly effective way to reduce noise in the Raman spectrum while accentuating real changes in the spectrum. Typically, 40 to 50 spectra from a single quartz slide were averaged. Spectra were acquired from the bacteria prepared in three different ways as described in Materials and Methods: control (non-xylitol exposed), xylitol exposed (0 to 2% for 24 to 30 h), and post-xylitol exposure (incubated for 24 h in a xylitol-free medium subsequent to growth in xylitol). To highlight and accentuate differences in the Raman spectra, the differences of the normalized averaged spectra were calculated so that deviations from zero (which would represent no significant change) could be more easily identified. In all of the Raman studies described here, we did not attempt to directly compare the absolute intensity of Raman signals by varying the bacterial titer. Attempts were made to illuminate approximately equal numbers of cells for all bacterial specimens and species.
RESULTS
Differential Gram staining.
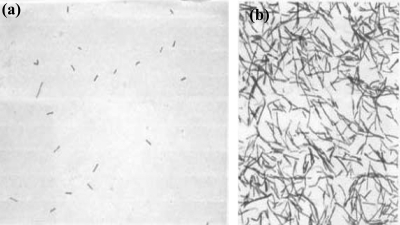
Microscopy of viridans group streptococcus after xylitol-exposure revealed that xylitol-exposed viridans group streptococcus behaved differently under differential Gram-staining than the nonexposed control. Fig. 1 a shows the low titer of the viridans group streptococcus population after 10,000-fold dilution in BHI broth prior to growth in xylitol. Figure 1b shows the effect of xylitol on the viridans group streptococcus after growth in the presence of xylitol (∼13 generations). The viridans group streptococcus population grown in BHI without xylitol (control) stained purple, as expected (not shown). Growth studies of viridans group streptococcus comparing the CFU after 24 h of growth at 37°C under anaerobic conditions in BHI medium with or without xylitol confirm the observations of others that xylitol affected the normal growth patterns. Differential Gram-staining showed that xylitol disturbed the retention of the crystal violet in viridans group streptococcus. In rich-growth medium (BHI or TSB), viridans group streptococcus normally appeared purple and in a population of diplococci, while the xylitol-exposed bacterial population demonstrated a remarkable change: 30 to 50% of them were in long chains and stained pink (appearing like Gram-negative or dead bacteria) instead of purple diplococci (Fig. 1). As our control, after an acetone alcohol wash, the Gram-positive bacteria retained the crystal violet-iodine complex (blue or purple), but the Gram-negative organisms usually lose this, and the counterstain safranin adds its pink color.
FIG. 1.
Action of 2% xylitol on viridans group streptococcus ultrastructure as revealed by differential Gram-staining imaged with the same magnification. (a) Planktonic viridans group streptococcus cells cultured in BHI medium and 0% xylitol anaerobically for 24 h prior to growth in xylitol. The titer has been reduced 10,000-fold to ensure that all bacteria attain log-phase growth in the presence of the xylitol. (b) Gram-stained viridans group streptococci cultured in BHI medium and 2% xylitol anaerobically for 24 h. The xylitol-exposed (2% or higher) bacterial population demonstrated a remarkable change; 50% of them stained pink, appearing like Gram-negative or dead bacteria instead of purple.
Raman spectroscopy of the uptake of xylitol by E. coli K-12 (xylitol operon deficient), its pilus- and flagellum-deficient derivative E. coli JW1881-1, and E. coli C (xylitol operon positive, but in a repressed state).
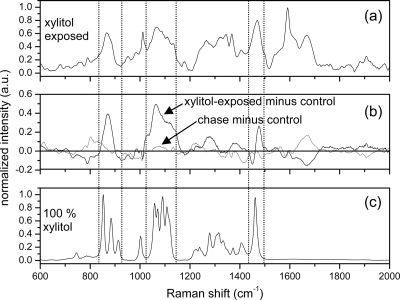
Raman spectra from 600 to 2,000 cm−1 obtained from E. coli K-12 which does not possess the xylitol operon are shown in Fig. 2. Figure 2a shows the averaged Raman spectrum from the xylitol-exposed E. coli K-12. Figure 2b shows two spectra, one is the difference of the xylitol-exposed and the control E. coli K-12 spectra (black), and the other is the difference of the postexposure chase and the control E. coli K-12 spectra (gray). Deviations from the zero-line indicate alterations in the Raman spectrum due to xylitol exposure. Several significant new peaks appear in spectral regions corresponding to strong Raman bands in xylitol (which is shown in Fig. 2c for comparison). Specifically, a new broad Raman feature (not a narrow isolated peak) centered at 861 cm−1 was observed corresponding to the three xylitol Raman bands at 863, 893, and 922 cm−1. A second very broad new feature between 1,030 and 1,140 cm−1 was observed in E. coli corresponding to the multiple Raman-peak band of xylitol between 1,030 and 1,140 cm−1 and a fairly sharp new feature at 1,470 cm−1 was correlated with a very sharp isolated Raman peak in xylitol at 1440 cm−1. Taken together, we interpret these new features as direct observation of the xylitol molecule in a semiliquid/liquid environment that allows the xylitol to retain its molecular stoichiometry and structure but significantly reduces the signal-to-noise ratio and broadens Raman-active bands. The spectrum in Fig. 2c was obtained from 100% dried, crushed, and powdered xylitol; therefore, it was not unexpected that the Raman spectrum from xylitol within the bacterial body would appear significantly altered.
FIG. 2.
Raman spectra of E. coli K-12 and xylitol. (a) Averaged Raman spectrum from xylitol-exposed E. coli K-12. (b) Difference in the xylitol-exposed spectra and the control E. coli K-12 (black) and difference in the postexposure chase spectra and the control E. coli K-12 (gray). Deviations from zero denote changes from the control bacteria and are observed strongly in the spectral regions located between the dashed lines in the xylitol-exposed minus control spectra. (c) Raman spectrum from 100% dried, powdered, and compacted xylitol.
Conversely, the averaged spectrum of the E. coli K-12 grown for 24 h in a xylitol-free medium exhibited no Raman bands that are indicative of the presence of xylitol. These data confirm a passive (unregulated) entry of xylitol into E. coli K-12 (black) and a silent exit (gray) from the cell without affecting bacterial growth or metabolism. However, we would conclude that the xylitol molecule is quite stable in the growing bacterial cell for the 24 h of their growth, i.e., the early log phase to the stationary phase. Significantly, the xylitol pool of these bacteria is immediately depleted when they are regrown in BHI agar medium without xylitol (chase) for ∼10 generations.
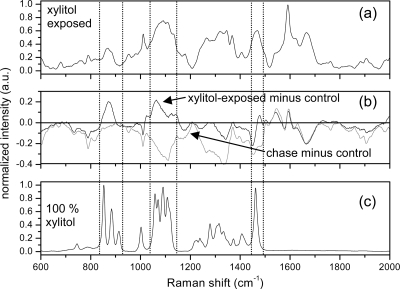
The Raman spectra of the E. coli C which carries the xylitol operon in a repressed state is shown in Fig. 3. Comparison of the spectra between 600 to 2,000 cm−1 of the E. coli C with that of E. coli K-12 shows that these two strains of E. coli exhibited nearly identical behavior both in their uptake of xylitol and in their inability to retain it during postexposure growth. No significant reproducible differences were observed in the Raman spectra obtained from the E. coli K-12 and E. coli C strains, which thus indicated the passive uptake, stable presence, and passive exit of xylitol without any participation in bacterial metabolism.
FIG. 3.
Raman spectra of E. coli C and xylitol. (a) Averaged Raman spectrum from xylitol-exposed E. coli C. (b) Difference in the xylitol-exposed spectra and the control E. coli C (black) and difference in the postexposure chase spectra and the control E. coli C (gray). Deviations from zero denote changes from the control bacteria and are observed strongly in the spectral regions located between the dashed lines in the xylitol-exposed minus control spectra. (c) Raman spectrum from 100% dried, powdered, and compacted xylitol.
Identical experiments were carried out with the pilus- and flagellum-negative E. coli mutant JW 1881-1. The major peaks of xylitol were again observed in the Raman spectra, and the intensity of these peaks showed no significant difference from the other strains. The spectra of the JW1881-1 were very similar to those of K-12 and E. coli C and therefore are not shown. These data lead us to the important conclusion that the flagella or common pili (motility) apparently play no role either in the passive entry of xylitol into the bacterial cells or in the exit of xylitol from the bacterial pool.
Xylitol uptake and stability in E. coli HF4714.
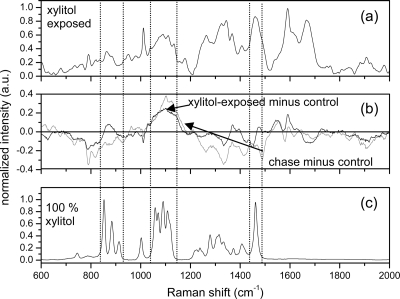
Experiments identical to those performed on the other E. coli strains were performed on E. coli HF4714 which contains the derepressed xylitol operon. The results are shown in Fig. 4. In contrast to the other E. coli strains, the peaks observed in E. coli HF4714 due to xylitol did not disappear from the progeny when chased in the absence of xylitol. Specifically, the very broad feature between 1,030 and 1,140 cm−1 was observed in both the xylitol-exposed mother cells and the progeny E. coli HF4714 with comparable intensity. This band corresponds to the multiple Raman-peak pattern observed in xylitol between 1,030 and 1,140 cm−1. The retention of this Raman feature attributed to xylitol is broadly indicative of the stable accumulation of xylitol in this E. coli strain with minimal or no catabolism of the xylitol.
FIG. 4.
Raman spectra of E. coli HF4714 and xylitol. (a) Averaged Raman spectrum from xylitol-exposed E. coli HF4714. (b) Difference in the xylitol-exposed spectra and the control E. coli HF4714 (black) and difference in the postexposure spectra and the control E. coli HF4714 (gray). Deviations from zero denote changes from the control bacteria and are observed consistently in the spectral regions located between the dashed lines in the xylitol-exposed minus control spectrum and also in the chase minus control spectrum. (c) Raman spectrum from dried, powdered, and compacted xylitol.
Xylitol metabolism by Gram-positive viridans group streptococcus.
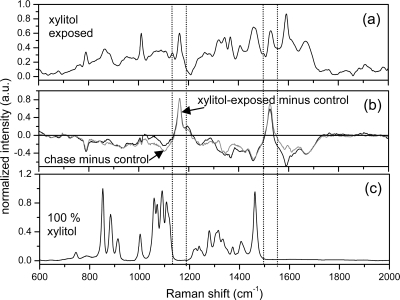
To characterize the molecular changes responsible for the observed Gram-staining change observed in viridans group streptococcus as shown in Fig. 1, Raman spectroscopy was performed on aliquots of the same bacterial samples. Figure 5 shows the Raman spectra obtained from the viridans group streptococcus (control), from the viridans group streptococcus grown in 2% xylitol for 24 h, and from the same viridans group streptococcus washed of xylitol and regrown in BHI-agar medium for an additional 24 h without xylitol. Comparison of the Raman spectra as presented in Fig. 4 and 5 shows that the Raman spectra of Gram-positive viridans group streptococcus are completely different from those of the Gram-negative E. coli. Two new Raman features were observed in both the xylitol-exposed mother cells and the postexposure progeny cells. These two new features at 1,165 cm−1 and at 1,527 cm−1 are not present at all in the xylitol spectrum or the spectrum of the control viridans group streptococcus strain and could be assigned to a COO stretch vibration in sucrose (5) and a protein NH/CH bend or C=C stretch vibration (36), respectively. Biochemically, the xylitol is partially metabolized by such streptococci, resulting in insoluble intermediates. Definitive assignment of these peaks at 1,165 cm−1 and at 1,527 cm−1 awaits further study and possibly the chemical purification of these xylitol intermediates and analysis by Raman spectroscopy. Nonetheless, it is apparent that these peaks are evidence of the use of xylitol by viridans group streptococci. These peaks either originate from smaller sugar by-product molecules resulting from xylitol catabolism or cellular proteins formed as a result of the xylitol catabolism. In future experiments, further growth of viridans group streptococcus in a medium containing only xylitol as a sole carbon source (after the complete utilization of dextrose) would confirm the relationship of these peaks at 1,165 cm−1 and at 1,527 cm−1 with xylitol catabolism.
FIG. 5.
Raman spectra of viridans group streptococcus and xylitol. (a) Averaged Raman spectrum from xylitol-exposed viridans group streptococcus. (b) Difference in the xylitol-exposed spectra and the control viridans group streptococcus strain (black) and difference in the postexposure spectra and the control viridans group streptococcus strain (gray). Deviations from zero denote changes from the control bacteria. New Raman peaks not observed at all in xylitol or the control viridans group streptococcus strain spectra are observed at 1,165 cm−1 and at 1,527 cm−1 in both spectra. (c) Raman spectrum from dried, powdered, and compacted xylitol. No xylitol Raman peaks appear in the spectral regions located between the dashed lines.
DISCUSSION
We compared the uptake of xylitol by four genetically well-defined strains of E. coli—E. coli K-12, its pilus- and flagellum-deficient derivative JW1881-1, E. coli C, and an E. coli C derivative HF4714—as well as a Gram-positive strain of viridans group streptococcus (ATCC 19950). It is known that the E. coli C contains a xylitol operon in a repressed state, apparently to avoid the disadvantages of a constitutive catabolic pathway (toxicity to galactitol and arabitol). Moreover, the flagella and pili responsible for motility are conspicuously absent on the surface of the E. coli C compared to E. coli HF4714 when visualized by scanning electron microscopy (S. Palchaudhuri, unpublished data). Thus, E. coli C may have avoided a fatal pathway to chemoattraction, but this is not a very unusual occurrence in an aquatic environment. These E. coli strains show comparable levels of xylitol uptake, despite their genetic differences (Fig. 2 and 3).
Significantly, the E. coli HF4714 which contains the xylitol operon in a derepressed state is capable of xylitol metabolism whenever the other carbon sources are exhausted. The preferred carbon source, dextrose in BHI-agar growth medium, is first utilized before the metabolic use of xylitol begins. Our experiments were conducted with a BHI medium containing a minimal amount of dextrose and 2% xylitol. Because of the availability of dextrose, the HF4714 may have developed a metabolic imbalance, resulting in an accumulation of xylitol that far exceeds the rate of its utilization (Fig. 4). After growth in BHI-agar medium for 2 days, the E. coli C and E. coli K-12 stopped growing, as evidenced by the colony size and bacterial titer, whereas the colonies of strain HF4714 continued to increase in size (Palchaudhuri, unpublished).
In a similar manner, viridans group streptococcus, which has an incomplete metabolic pathway of xylitol, cannot grow into larger colonies in the presence of 2% xylitol. The Gram staining of these colonies picked up from CNA or BHI-agar plates with 2% xylitol demonstrates the elongated shapes or the diplococcic bacteria in chains, an index of abnormality in bacterial septum formation or abnormality during stages of cell division or cytokinesis (Fig. 1).
It is known that pneumonia results in a considerable number of deaths worldwide in the very young and the elderly, especially given the rise of antibiotic- and/or immunoresistant organisms (19). It seems logical to determine the efficacy of xylitol alone or in combination with other agents, such as fluoride, on such drug-resistant pathogens. Our data support the work of others that it could be the anti-adherence effect of nasally administered xylitol which might lead to a significant reduction in the occurrence of AOM and pneumonia in children (15). Alteration of surface structures after exposure to xylitol and a lessening of the bacterial adhesion to both biotic and abiotic surfaces have been reported (17, 24, 27), as has a lessening of the bacterial adhesion of E. coli to uroepithelial cells subsequent to cranberry juice exposure (29), and this could probably minimize persistent urinary tract infections. Significantly, it has been reported that xylitol can damage the ultrastructure of pneumococci, compared to the effect of fructose or sucrose (42). Xylitol is a five-carbon sugar, and the Gram-positive diplococci are permeable to such sugars, except that there seems to be an antagonistic competition between fructose and xylitol at the initial stages of biochemical pathways (40). The molecular basis of such competition has not been properly investigated.
The observations of surface ultrastructure modification and reduction in adhesion are intriguing. The majority of the Gram-negative bacteria have grown flagella and pili (or fimbriae) emerging out of the cell envelope, but our data show that the presence of these fimbriae does not seem to affect xylitol uptake. It remains to be seen whether an adhesion change is measurable in xylitol-exposed pathogens and whether this adhesion change is pilus motivated (35, 43). The specific interaction of type 4 pili with xylitol has not been studied (23). Experiments are under way in our laboratory utilizing atomic force microscopy (AFM) to determine the change in the adhesion of xylitol-exposed viridans group streptococci to abiotic surfaces and other bacteria (6, 29). We hope to correlate the appearance of the new Raman molecular peaks observed in Fig. 5 with an alteration of surface ultrastructures responsible for adhesion changes. As well as measuring surface adhesion strengths, AFM can be used for direct visualization of bacterial surface ultrastructures (4), which may further illuminate these relationships. Subsequent experiments will examine in detail the effect that xylitol concentration and exposure time has on the Raman spectra and the adhesion of viridans group streptococci.
A remarkable coincidence is that the pathogens interacting with xylitol are all morphologically diplococcic organisms, regardless of their Gram-staining classification (33). All possible structures of bacteria are well characterized, but their specific interactions with different sugar and sugar-alcohol molecules have not been considered. We believe that the morphological shapes of these bacteria may play a critical role in their effective interactions with specific structures of sugar molecules (five-carbon, six-carbon, or sugar alcohols) and subsequent uptake. Since Gram-positive S. pneumoniae cross-reacts with the capsular Gram-negative Klebsiella pneumoniae and both are morphologically diplococcic, the xylitol seems to be equally accessible to these pathogens (33). In our work, the Gram-positive pathogen viridans group streptococcus is not fully capable of metabolizing the five-carbon sugar alcohol xylitol, but intermediate chemical compounds are formed and continue to exist in the bacterial metabolic pool. Their presence may adversely affect the thickness of the peptidoglycan walls of bacteria. This in turn probably weakens the stable stacking of crystal violet, and therefore a confusion may arise in the identification of Gram-positive bacteria by Gram staining (31).
It is already known that the use of fluoride is effective in minimizing dental decay (11). What is more, the simultaneous application of xylitol and fluoride produces a synergistic effect, but the precise mechanism of synergism is unknown (20). Analyses of intracellular glycolytic intermediates have shown that the fluoride inhibits the lower part of the glycolytic pathway and that xylitol affects the upper pathway. It is not known whether fluoride affects the biofilm formation and or the growth of diplococci in chains. Reversion of chains into single diplococci populations may allow a higher percentage of the bacterial population (or their receptors) to effectively interact with the xylitol molecules. Thus, the probability of success of the preventive therapy would highly increase and thus save many young lives. This would provide us with a molecular basis for the successful use of the synergistic approach toward a preventative therapy.
Acknowledgments
This study was supported in part by a generous donation by Richard Barber (S.J.R. and K.H.).
One of us (S.P.) is also grateful to A. Palchaudhuri for her valuable comments on the concept of synergistic effects in medicine and Gail Pecherski for sharing her data from bacterial growth curves.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Brimacombe, J. S., and J. M. Webber. 1972. Alditols and derivatives, p. 479-519. In W. Pigman and D. Horto (ed.), The carbohydrates, vol. IA, 2nd ed. Academic Press, London, England.
- 2.Charnetzky, W. T., and R. P. Mortlock. 1974. Close genetic linkage of the determinants of the ribitol and d-arabitol catabolic pathways in Klebsiella aerogenes. J. Bacteriol. 119:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo-Smith, L.-P., K. Maquelin, T. Van Vreeswijk, H. A. Bruining, G. J. Puppels, N. A. Ngo Thi, C. Kirschner, D. Naumann, D. Ami, A. M. Villa, F. Orsini, S. M. Doglia, H. Lamfarraj, G. D. Sockalingum, M. Manfait, P. Allouch, and H. P. Endtz. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross, S. E., J. Kreth, L. Zhu, F. Qi, A. E. Pelling, W. Shi, and J. K. Gimzewski. 2006. Atomic force microscopy study of the structure-function relationships of the biofilm-forming bacterium Streptococcus mutans. Nanotech 17:S1-S7. [DOI] [PubMed] [Google Scholar]
- 5.de Veij, M., P. Vandenabeele, T. De Beer, J. P. Remon, and L. Moens. 2009. Reference database of Raman spectra of pharmaceutical excipients. J. Raman Spectrosc. 40:297-307. [Google Scholar]
- 6.Dorobantu, L. S., S. Bhattacharjee, J. M. Foght, and M. R. Gray. 2009. Analysis of force interactions between AFM tips and hydrophobic bacteria using DLVO theory. Langmuir 25:6968-6976. [DOI] [PubMed] [Google Scholar]
- 7.Gladwin, M., and B. Trattler. 2008. Clinical microbiology, 4th ed. MedMaster, Miami, FL.
- 8.Goodacre, R., E. M. Timmins, R. Burton, N. Kaderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology (Reading, Engl.) 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 9.Harz, M., P. Rosch, K.-D. Peschke, O. Ronneberger, H. Burkhardt, and J. Popp. 2005. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 130:1543-1550. [DOI] [PubMed] [Google Scholar]
- 10.Hattori, K., and T. Suzuki. 1974. Microbial production of d-arabitol by n-alkane-grown Candida tropicalis. Agric. Biol. Chem. 38:1203-1208. [Google Scholar]
- 11.Honkala, E., S. Honkala, M. Shyama, and S. A. Al-Mutawa. 2006. Field trial on caries prevention with xylitol candies among disabled school students. Caries Res. 40:508-513. [DOI] [PubMed] [Google Scholar]
- 12.Huang, W. E., R. I. Griffiths, I. P. Thompson, M. J. Bailey, and A. S. Whiteley. 2004. Raman microscopic analysis of single microbial cells. Anal. Chem. 76:4452-4458. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis, R. M., A. Brooker, and R. Goodacre. 2004. Surface-enhanced Raman spectroscopy for bacterial discrimination utilizing a scanning electron microscope with a Raman spectroscopy interface. Anal. Chem. 76:5198-5202. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis, R. M., and R. Goodacre. 2004. Ultra-violet resonance Raman spectroscopy for the rapid discrimination of urinary tract infection bacteria. FEMS Microbiol. Lett. 232:127-132. [DOI] [PubMed] [Google Scholar]
- 15.Jones, A. 2003. The next step in infectious disease: taming bacteria. Med. Hypotheses 60:171-174. [DOI] [PubMed] [Google Scholar]
- 16.Kontiokari, T., M. Uhari, and M. Kosela. 1995. Effect of xylitol on growth of nasopharyngeal bacteria in vitro. Antimicrob. Agents Chemother. 39:1820-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontiokari, T., M. Uhari, and M. Koskela. 1998. Antiadhesive effects of xylitol on otopathogenic bacteria. J. Antimicrob. Chem. 41:563-565. [DOI] [PubMed] [Google Scholar]
- 18.Lamont, R. J., M. Meila, Q. Xia, and M. Hackett. 2006. Mass spectrometry-based proteomics and its application to studies of Porphyromonas gingivalis invasion and pathogenicity. Infect. Disord. Drug Targets 6:311-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, O. S., K. L. O'Brien, M. Knoll, R. A. Adegbola, S. Black, T. Cherian, R. Dagan, D. Goldblatt, A. Grange, B. Greenwood, T. Hennessy, K. P. Klugman, S. A. Madhi, K. Mulholland, H. Nohynek, M. Santosham, S. K. Saha, J. A. Scott, S. Sow, C. G. Whitney, and F. Cutts. 2006. Pneumococcal vaccination in developing countries. Lancet 367:1880-1882. [DOI] [PubMed] [Google Scholar]
- 20.Maehara, H., Y. Iwami, H. Mayanagi, and N. Takahashi. 2005. Synergistic inhibition by combinations of fluoride and xylitol on glycolysis of mutans streptococci and its biochemical mechanism. Caries Res. 34:231-234. [DOI] [PubMed] [Google Scholar]
- 21.Mäkinen, K., P. Alanen, P. Isokangas, K. Isotupa, E. Söderling, P.-L. Mäkinen, W. Wenhui, W. Weijian, C. Xiaohi, W. Yi, and Z. Boxue. 2008. Thirty-nine month xylitol chewing gum program in initially 8-year-old school children: a feasibility study focusing on mutans streptococci and lactobacilli. Int. Dent. J. 58:41-50. [DOI] [PubMed] [Google Scholar]
- 22.Maquelin, K., C. Kirschner, L.-P. Choo-Smith, N. Van den Braak, H. Ph. Endtz, D. Naumann, and G. J. Puppels. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255-271. [DOI] [PubMed] [Google Scholar]
- 23.Mattick, J. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 24.Modesto, A., and D. Drake. 2006. Multiple exposures to chlorhexadine and xylitol: adhesion and biofilm formation by Streptococcus mutans. Curr. Microbiol. 52:418-423. [DOI] [PubMed] [Google Scholar]
- 25.Mortlock, R. P. 1976. Catabolism of unnatural carbohydrates by micro-organisms. Adv. Microb. Physiol. 13:1-53. [DOI] [PubMed] [Google Scholar]
- 26.Murray, P. R., W. L. Drew, G. S. Kobayashi, and J. H. Thompson. 1990. Medical microbiology. CV Mosby Company, St. Louis, MO.
- 27.Naaber, P., E. Lehto, S. Salminen, and M. Mikelsaar. 1996. Inhibition of adhesion of Clostridium difficile to Caco-2 cells. FEMS Immunol. Med. Microbiol. 14:205-209. [DOI] [PubMed] [Google Scholar]
- 28.Navratil, M., G. A. Mabbott, and E. A. Arriaga. 2006. Chemical microscopy applied to biological systems. Anal. Chem. 78:4005-4019. [DOI] [PubMed] [Google Scholar]
- 29.Pinzón-Arango, P. A., Y. Liu, and T. A. Camesano. 2009. Role of cranberry on bacterial adhesion forces and implications for Escherichia coli: uroepithelial cell attachment. J. Med. Food 12:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Reiner, A. M. 1975. Genes for ribitol and d-arabitol catabolism in Escherichia coli: their loci in C strains and absence in K-12 and B strains. J. Bacteriol. 123:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes. Chapman & Hall, New York, NY.
- 32.Saint Martin, E. J., and R. P. Mortlock. 1976. Natural and altered induction of the l-fructose catabolic enzymes in Klebsiella aerogenes. J. Bacteriol. 127:91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salyers, A. A., and D. D. Whitt. 2002. Bacterial pathogenesis: a molecular approach, 2nd ed. ASM Press, Washington, DC.
- 34.Scangos, G. A., and A. M. Reiner. 1978. Ribitol and d-arabitol catabolism in Escherichia coli. J. Bacteriol. 134:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, J. R., and D. Zahner. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320-330. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta, A., M. L. Laucks, and E. J. Davis. 2005. Surface-enhanced Raman spectroscopy of bacteria and pollen. Appl. Spectrosc. 59:1016-1023. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta, A., M. L. Laucks, N. Dildine, E. Drapala, and E. J. Davis. 2005. Bioaerosol characterization by surface-enhanced Raman spectroscopy (SERS). J. Aerosol Sci. 36:651-664. [Google Scholar]
- 38.Söderling, E., T. Ekman, and T. Taipale. 2008. Growth inhibition of Streptococcus mutans with low xylitol concentrations. Curr. Microbiol. 56:382-385. [DOI] [PubMed] [Google Scholar]
- 39.Söderling, E., L. Alaräisänen, A. Scheinin, and K. K. Mäkinen. 1987. Effect of xylitol and sorbitol on polysaccharide production by and adhesive properties of Streptococcus mutans. Caries Res. 21:109-116. [DOI] [PubMed] [Google Scholar]
- 40.Tapiainen, T., T. Kontiokari, L. Sammalkivi, I. Ikaheimo, M. Koskela, and M. Uhari. 2001. Effect of xylitol on growth of the Streptococcus pneumoniae in the presence of fructose and sorbitol. Antimicrob. Agents Chemother. 45:166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapiainen, T. 2002. Microbiological effects and clinical uses of xylitol in preventing acute otitis media. Ph.D. thesis. University of Oulu, Oulu, Finland.
- 42.Tapiainen, T., R. Sormunen, T. Karjalainen, T. Kontiokari, I. Ikaheimo, and M. Uhari. 2004. Ultrastructure of Streptococcus pneumoniae after exposure to xylitol. J. Antimicrob. Chem. 54:225-228. [DOI] [PubMed] [Google Scholar]
- 43.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 44.Trahan, L., M. Bareil, L. Gauthier, and C. Vadeboncoeur. 1985. Transport and phosphorylation of xylitol by a fructose phosphotransferase system in Streptococcus mutans. Caries Res. 19:53-63. [DOI] [PubMed] [Google Scholar]
- 45.Trahan, L. 1995. Xylitol: a review of its actions on mutans streptococci and dental plaque—its clinical significance. Int. Dent. J. 45(Suppl.):77-92. [PubMed] [Google Scholar]
- 46.Tuompo, H., J. H. Meurman, K. Lounatmaa, and J. Linkola. 1983. Effect of xylitol and other carbon sources on the cell wall of Streptococcus mutans. Scand. J. Dent. Res. 91:17-25. [DOI] [PubMed] [Google Scholar]
- 47.Uhari, M., T. Tapiainen, and T. Kontiokari. 2000. Xylitol in preventing acute otitis media. Vaccine 19(S1):S144-S147. [DOI] [PubMed]
- 48.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate: sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187-207. [DOI] [PubMed] [Google Scholar]
- 49.Waldor, M. K., D. I. Friedman, and S. L. Adhya (ed.). 2005. Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 50.Williams, A. C., and H. G. M. Edwards. 1994. Fourier transform Raman spectroscopy of bacterial cell walls. J. Raman Spectrosc. 25:673-677. [Google Scholar]
- 51.Wood, W. A. 1961. Ribitol and d-arabitol utilization by Aerobacter aerogenes. J. Biol. Chem. 236:2190-2195. [PubMed] [Google Scholar]
- 52.Yokota, S., A. Harimaya, and K. Sato. 2007. Colonization and turnover of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in otitis-prone children. Microbiol. Immunol. 51:223-230. [DOI] [PubMed] [Google Scholar]