Abstract
Cyanobacteria of the genera Synechococcus and Prochlorococcus are the most abundant photosynthetic organisms on earth, occupying a key position at the base of marine food webs. The cynS gene that encodes cyanase was identified among bacterial, fungal, and plant sequences in public databases, and the gene was particularly prevalent among cyanobacteria, including numerous Prochlorococcus and Synechococcus strains. Phylogenetic analysis of cynS sequences retrieved from the Global Ocean Survey database identified >60% as belonging to unicellular marine cyanobacteria, suggesting an important role for cyanase in their nitrogen metabolism. We demonstrate here that marine cyanobacteria have a functionally active cyanase, the transcriptional regulation of which varies among strains and reflects the genomic context of cynS. In Prochlorococcus sp. strain MED4, cynS was presumably transcribed as part of the cynABDS operon, implying cyanase involvement in cyanate utilization. In Synechococcus sp. strain WH8102, expression was not related to nitrogen stress responses and here cyanase presumably serves in the detoxification of cyanate resulting from intracellular urea and/or carbamoyl phosphate decomposition. Lastly, we report on a cyanase activity encoded by cynH, a novel gene found in marine cyanobacteria only. The presence of dual cyanase genes in the genomes of seven marine Synechococcus strains and their respective roles in nitrogen metabolism remain to be clarified.
Cyanase (EC 4.2.1.104) converts cyanate to carbon dioxide and ammonia in a bicarbonate-dependent reaction: NCO− + HCO3− + 2H+ → 2CO2 + NH3 (16). The enzyme is encoded by cynS and is found in a wide range of organisms: Cyanobacteria, Proteobacteria (including enterobacteria), some Gram-positive bacteria, fungi, and plants. Transcriptional regulation and enzymatic activity were initially studied in Escherichia coli strain B/1 (1). In response to cyanate addition, transcription was induced as a polycistronic message of cynS, together with a cyanate transporter gene (1). Twenty years later, the protein structure and subunit organization of E. coli CynS were determined at 1.65-Å resolution (38). The cyanase monomer was found to be composed of two domains: an N-terminal domain with similarity to the DNA-binding α-helix bundle motif and an “open-fold” C-terminal domain with no structural homology to other proteins. The dimer structure of the cyanase subunit revealed intertwined C-terminal domains with five dimers forming a decameric cyanase holoenzyme. The proposed active site contains three conserved residues—Arg-96, Glu-99, and Ser-122—so that five catalytic sites found in the active decamer form an inner ring around a hollow core (38).
Cyanase activity in cyanobacteria was first described for the freshwater Synechococcus sp. strain PCC6301 (UTEX 625) and cyanate decomposition did not require preexposure of cells to cyanate (24). Instead, the decomposition of exogenous cyanate by Synechocystis sp. strain PCC6803 and Synechococcus elongatus PCC7942 was found to be light dependent (10). Based on the sequence similarity, cynS was identified in Synechocystis sp. strain PCC6803, Synechococcus elongatus PCC7942 (14), and freshwater Synechococcus sp. strain PCC6301 (10). In the Synechococcus strains, cynS was transcribed as a part of an operon, together with cynABD, encoding a ABC-type cyanate transporter, while in Synechocystis it was cotranscribed with four molybdenum-cofactor biosynthesis genes (14). Transcription of the operon was negatively regulated by ammonium and required the presence of NtcA, a global nitrogen (N) regulator of cyanobacteria (14). Comparative genomics of marine cyanobacteria revealed cynS in the majority of Synechococcus (26, 31) and in some Prochlorococcus (11, 33). The physiological and ecological roles of cyanase in marine cyanobacteria have not yet been elucidated. In the presence of a specific transporter, cyanase may play a role in cyanate assimilation. Marine cyanobacteria strains that possess the cynABD genes, encoding an ABC-type cyanate transporter, grew at near-maximal growth rates with cyanate as the sole N source (17). The CynABD complex was recently shown to also contribute to nitrite uptake in Synechococcus elongatus PCC7942 (22). Conversely, transport systems for CO2, HCO3−, NO3−, NO2−, Cl−, PO42−, and SO42− do not contribute to cyanate acquisition (10). The great majority of cyanobacteria that contain cynS in fact lack the genes for cyanate acquisition, suggesting a role for CynS in the detoxification of internally generated cyanate, which accumulates as a by-product of the urea cycle or via the degradation of carbamoyl phosphate (33). Here, we characterize transcriptional regulation of cynS and the resulting cyanase activity in marine cyanobacteria. Furthermore, we report on a novel source of cyanase activity associated with a conserved hypothetical gene in seven marine Synechococcus strains. Based on this activity, we have named it cynH (for cyanate hydratase), and we refer to this gene as such throughout this report.
MATERIALS AND METHODS
Strains and media.
Prochlorococcus sp. strain MED4 was grown in the seawater-based PRO99 medium (25), while Synechococcus sp. strains WH8102 and WH7803 were grown in artificial seawater medium (41), supplemented to a final concentration of 0.8 mmol of ammonium chloride (NH4Cl; J. T. Baker, Deventer, MO) liter−1, up to mid-log phase. They were maintained at 25 ± 1°C with gentle agitation at 80 to 90 rpm on a model G2 gyratory shaker (New Brunswick Scientific Co., New Brunswick, NJ) with continuous illumination provided by “warm-white” fluorescence tubes at 20 to 25 μmol of photons m−2 s−1. For N nutrition experiments, NH4Cl was replaced with 0.8 mmol of nitrate liter−1, 0.8 mmol of freshly prepared sodium cyanate (NaOCN; Aldrich) liter−1, 0.4 mmol of freshly prepared urea (Amresco) liter−1, or 0.8 mmol of sodium chloride liter−1 to produce an N-free medium. Cultures supplemented with fresh ammonium were used as a control.
For protein expression, we used HMS174 (Novagen/EMD Biosciences, Inc.), Rosetta pLysS (Novagen/EMD Biosciences, Inc.), and BL21-CodonPluS-RIL (Stratagene/Agilent Technologies) E. coli strains. Cloning and propagation of recombinant plasmids was performed according to the protocols of the manufacturers. For a negative control of specificity of the cyanate activity, maltose-binding protein (MBP)-fused NtcA and MBP itself were overexpressed in E. coli strains Rosetta pLysS and BL21-CodonPluS-RIL, respectively.
Sequence analysis.
Protein sequence data of the cynS and cynH genes and their genomic context were obtained from the genome of Synechococcus sp. strain WH7803 available from GenBank, using the DNA sequence viewer and annotation tool Artemis (32). Further genomic data for comparative genomic context study, alignments, and phylogenetic analyses were obtained from the nr public database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Blast.cgi) and the All Metagenomic ORF Peptides subject database of the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA; http://camera.calit2.net) by BLASTN and BLASTP searches. Multiple protein sequence alignments were performed by using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) program (8). Phylogenetic relationships were analyzed with MrBayes 3.1 (15), with nucleotide frequencies and parameters for the GTR+gamma invariant model estimated from the data. Two independent runs of four chains were run for 2 million generations and sampled every 100 generations; comparison of parameter estimates from the two runs indicated convergence (13). The secondary structures of CynS and CynH were determined by using the Jpred Prediction Server (www.compbio.dundee.ac.uk/www-jpred).
Transcriptional regulation.
Cyanobacterial cells (200 ml) grown with different N sources were harvested 3, 6, 9, 12, and 24 h after medium replacement and centrifuged at 10,500 × g for 6 min at 4°C. Cell pellets were resuspended in 0.5 ml of TRI Reagent (Ambion) and immediately frozen at −20°C. Further RNA isolation was performed according to standard procedure recommended by the manufacturer. Prior to the analysis, RNA samples were treated with the DNA-free kit (Ambion) to eliminate DNA contamination, and the RNA purity was confirmed by PCR using the primer pairs listed in Table 1. Final nucleic acid concentrations were determined photometrically (NanoDrop). An ImProm-II reverse transcription system (Promega) kit was used for two-step reverse transcription-PCR (RT-PCR) analysis. For the first step, a standard reaction was applied with the gene-specific reverse primers listed in Table 1. Subsequent PCRs with specific primer sets were performed in a final volume of 50 μl containing 1 to 2 μl of cDNA, 0.5 μmol of each primer liter−1, 0.25 mmol of each deoxyribonucleotide triphosphate liter−1, 1.25 U of Taq DNA polymerase (Peqlab), and 10× PCR buffer containing 15 mmol of MgCl2 liter−1. Using a PTC200 Thermo Cycler (MJ Research, Inc.), the reaction mix was preincubated at 94°C for 5 min, followed by cycles of denaturation at 94°C for 45 s, primer annealing for 30 s (see Table 1 for primer-specific temperatures), and elongation at 72°C for 45 s. Samples of 6 μl were promptly collected after 20, 25, and 30 cycles. Equal volumes of PCR products from the three sets (20, 25, and 30 cycles) were run on 1.5% agarose gels and visualized with ethidium bromide (Sigma). Quantification of gene expression level was performed with ImageJ analysis software (rsbweb.nih.gov/ij/). The set chosen for quantification was always the one before saturation of the PCR amplification was reached (Fig. 1). In the rare case of early saturation, cDNA was PCR amplified for 16 to 18 cycles instead. The reference genes in the RT-PCR analyses were PMM0615 of Prochlorococcus sp. MED4 (cell wall hydrolase/autolysin [COG0388]) and orf0250 of Synechococcus sp. WH7803 (Ycf48-like photosystem II stability/assembly factor [COG4447]). These genes were chosen since they are part of the cyanobacterial core genomes and do not alter their transcript level, as assessed by microarray (H. Zer, A. Singer, and A. F. Post, unpublished data). For Synechococcus sp. WH8102, the reference was the 16S rRNA gene. The transcript levels of the genes of interest determined were normalized to those of the reference genes. The regression lines describing gene expression over time were compared while testing the hypotheses of coincidence, parallelism, and equality of intercepts (as described in Master of Applied Statistics by Pia Veldt Larsen [http://statmaster.sdu.dk/courses/st111/module09/]). The P value was calculated from the F value (www.graphpad.com/quickcalcs/) obtained with the following equation:
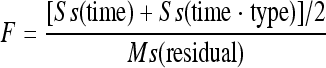 |
(where Ss is the sum of squares and Ms is the mean square) from the multivariate analysis of variance (MANOVA)-derived variables.
TABLE 1.
Sequences of primers used in RT-PCR amplification, their annealing temperatures, and expected PCR product sizes
| Target gene | Primera | Sequence (5′-3′) | Ta (°C)b | Size (bp) |
|---|---|---|---|---|
| Prochlorococcus sp. MED4 | ||||
| cynA | RTcA-Med4 F | GGAGGTAGCTAAGGCTATTT | 52.5 | 198 |
| RTcA-Med4 R | CCTCCTAGATCCCATCTTAT | |||
| cynS | RTcS-Med4 F | CCTACGGATCCTCTTATCTA | 52 | 163 |
| RTcS-Med4 R | CTAGAACCCTATCTCCCTTT | |||
| ntcA | RTnA-Med4 F | AGAGGAGCAGTAAGGTTATC | 51 | 116 |
| RTnA-Med4 R | TCAGACCTATGTCCTGTTAG | |||
| PMM0615 | RTorf1206 F | CCCTGAACTTTATAGACACC | 52 | 202 |
| RTorf1206 R | GACTTTGTCTTCTCCCATAG | |||
| Synechococcus sp. WH8102 | ||||
| cynA | cA-8102 F | GCCTCTATTCACTCTAGTTCCCC | 53 | 828 |
| cA-8102 R | GCGAATTATGCAACAAATCCTACT | |||
| cynS | cS-8102 F | AGGTTTGGGTTGCATCTTTG | 52.5 | 235 |
| cS-8102 R | TCTCCGAAATGCTCCTGAAT | |||
| 16S rRNA | 16S-8102 F | CATCATGCCCCTTACATCCT | 56 | 103 |
| 16S-8102 R | AACTGAGCCACGGTTTATGG | |||
| Synechococcus sp. WH7803 | ||||
| cynS | RTcS-2 F | GGCCACAGCATCAGCGGAGG | 63 | 283 |
| RTcS-2 R | GGTGATCTTCACGCGATCGCC | |||
| orf0252 | RTorf0252 F | GTGCCCGGTATGTCTTCCTT | 59 | 150 |
| RTorf0252 R | ATTGGCGTCTGTGTTGAGGT |
F, forward; R, reverse.
Ta, annealing temperature.
FIG. 1.
Typical example of RT-PCR amplification of cynS cDNA for Synechococcus sp. WH7803. Samples (50 μl) were run for 30 cycles, and 6-μl subsamples were collected at five-cycle intervals. PCR products from each cycle set were run on a 1.5% agarose gel and visualized by staining with ethidium bromide. The band density was determined for the set in which subsamples had clearly not reached saturation phase (arrow), assuming that they most closely resembled the phase of exponential amplification.
Cloning of cyanase genes.
Genomic DNA of Synechococcus sp. WH7803 was extracted by using phenol-chloroform as described previously (28). The complete coding sequences of Synechococcus sp. WH7803 cynS (424 bp) and cynH (201-bp) were PCR amplified with the primer combinations (i) cSEcoRI2-F (5′-AGAAAGGGGAATTCATGAGTTTCGCCGATC-3′) and cSPstI-R (5′-CACACGATTCAAGCTGCAGTTACCATTTTTTGTAAGGAAGG-3′) and (ii) scSEcoRI-F (5′-AGTTCGTGGAATTCCATGAGTGCTCTTTTCCGTTCC-3′) and scSPstI-R (5′-GCCCGAGGGCTGCAGTTACGGGGAGTCGAGATAGG-3′), respectively. The forward/reverse primers contain EcoRI and PstI restriction sites, respectively, to facilitate synthesis of the MBP fusion construct. PCRs (50 μl) were performed with Phusion high-fidelity DNA polymerase (Finnzymes) and 1.2 ng of DNA template μl−1. PCRs were run over 30 cycles of denaturation (98°C, 30 s), annealing (58°C for cynS and 66°C for cynH, 20 s), and elongation (72°C, 20 s), followed by a final 5 min of elongation. PCR products were purified on 1.2% Tris-acetate-EDTA-buffered agarose gels and eluted with the Wizard SV Gel and PCR Clean-Up System. Both the amplicons and pMBP1 vector (kindly provided by P. Sheffield, University of Virginia) were digested with EcoRI and PstI. Purified amplicons (150 ng) were ligated downstream of malE on the pMBP1 vector in a 1:3 molar ratio. In-frame assembly of fusion constructs was verified from DNA sequence analysis after transformation into a suitable E. coli host strain.
Overexpression of cyanase genes.
After transformation into E. coli, the expression of the CynS-MBP and CynH-MBP fusion proteins was tested in several strains suitable for protein overexpression. In order to determine optimal conditions for protein expression, we tested different IPTG (isopropyl-β-d-thiogalactopyranoside) concentrations, temperatures, and incubation times. Accumulation of recombinant protein was then confirmed by SDS-PAGE analysis using whole-cell lysate of IPTG-induced cells. The cells were centrifuged at 20,000 × g for 1 min at 4°C, and the cell pellet was kept overnight at −20°C to ease further lysis. The next day, the cell pellet was resuspended in column buffer (20 mmol Tris-HCl [pH 7.5], 200 mmol of NaCl, and 1 mmol of EDTA liter−1) containing 100 μg of lysozyme ml−1, 1 mmol of phenylmethylsulfonyl fluoride liter−1, 0.1 mg of DNase ml−1, and 0.1 mmol of MgSO4 liter−1; incubated on ice for 30 min; and then sonicated. Membrane debris was removed by centrifugation at 20,000 × g for 15 min at 4°C. Both pellet and soluble fractions were analyzed for recombinant protein accumulation by using SDS-PAGE. The experimental conditions for the expression and accumulation of recombinant proteins in the soluble fraction of specific E. coli strains were as follows: MBP-CynS was expressed in E. coli strain HMS174 incubated for 4 h at 37°C after the addition of 0.4 mmol IPTG liter−1; MBP-CynH was expressed in E. coli strain Rosetta pLysS incubated for 4 h at 22°C after the addition of 0.1 mmol IPTG liter−1; MBP-NtcA was expressed in E. coli strain Rosetta pLysS incubated for overnight at 17°C after the addition of 0.2 mmol IPTG liter−1; and MBP was expressed in E. coli strain BL21-CodonPluS-RIL incubated for 3 h at 37°C after the addition of 0.3 mmol IPTG liter−1. All E. coli strains were supplemented with 100 μg of ampicillin ml−1, and the Rosetta pLysS strain was supplemented with 34 μg of chloramphenicol ml−1.
SDS-PAGE and Western blotting.
The identity of the induced protein was confirmed by Western blotting, where the cell lysate from a noninduced culture, alongside pellet and eluted fractions from an induced culture, were separated on 14.5% acrylamide gels by SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane at 4°C using transfer buffer containing 54 mmol of Tris liter−1, 384 mmol of glycine liter−1, and 20% (vol/vol) methanol. After blocking the membrane with 5% (wt/vol) skimmed milk powder (Difco) dissolved in TBS (10 mmol of Tris-HCl [pH 7.5] liter−1, 250 mmol of NaCl liter−1) with 0.06% (vol/vol) Tween 20, it was washed three times with TBS buffer containing 0.01% Tween 20 (TTBS). Primary antibody (MBP antibody [R29.6]; ab65; Abcam) diluted 1:1,000 was added, followed by incubation overnight at 4°C, and then washed with TTBS buffer and exposed to a blocking buffer containing peroxidase-conjugated anti-mouse IgG secondary antibody (Jackson Immunoresearch Laboratories, Inc.) diluted 1:5,000. Chemiluminescence detection was carried out with an EZ-ECL Enhanced Chemiluminescence Detection Kit for HRP (Biological Industries, Ltd.) using a LAS-3,000 Image Analyzer (Fujifilm).
Purification of recombinant protein.
Purification of CynS-MBP and CynH-MBP fusion proteins and MBP was performed using amylose resins (E8021) according to New England Biolabs protocols. In brief, cell pellets were resuspended in 1/10 culture volume of phosphate buffered saline (PBS, 0.05 mmol liter−1, pH 7.6) complemented with 0.2 mg of lysozyme ml−1, 0.05 mg of DNase ml−1, 10 mmol of MgCl2 liter−1, 1 mmol of dimethyl sulfoxide liter−1, and 1:200 protease cocktail inhibitor, and the cells were disrupted by sonication. After centrifugation at 10,000 × g for 15 min, the supernatant was mixed with the amylose resin, and after binding for 1 h at 4°C, the column was washed gently with PBS buffer. Recombinant protein was eluted after the addition of 10 mmol of free maltose liter−1 in PBS buffer.
Cyanase activity assay.
The in vitro cyanase activity of the Synechococcus sp. WH7803 associated with the CynS and CynH fusion proteins was measured as described previously (1) with small modifications. In brief, 20-μg recombinant protein aliquots were used for activity measurements using two controls. The first control was supplemented with all reaction mixture ingredients except cyanate and tested for background ammonia levels occasionally introduced with the recombinant protein mixture. The second control consisted of the reaction mixture without a protein aliquot and tested the spontaneous decomposition of cyanate. The reactions were initiated by the addition of sodium cyanate (NaOCN; Aldrich) and terminated by the addition of 225 μl of Nessler reagent (K2HgI4; Aldrich) diluted 1:3 with double-distilled water. The temperature of the reaction solution was adjusted to the desired value prior to cyanate addition. Reactions were performed in 48-well plates (Nunc) and analyzed on a Microplate Reader Synergy2 (BioTek Instruments, Inc.) within 5 min after termination of the reaction. The specificity of the CynS and CynH activity was demonstrated in parallel control experiments containing MBP by itself or MBP-NtcA, proteins that lack enzymatic activity. Cyanase inhibition reactions were performed after 200-μmol liter−1 Na-azide additions. All control reactions were performed at 26°C. One unit of cyanase activity was defined as the amount of enzyme required to catalyze the formation of 1 μmol of ammonia per min. Temperature optima for the fusion proteins were estimated by performing enzymatic reactions at five different temperatures in the range of 4 to 50°C. Protein concentrations were determined with a Bradford assay (3).
RESULTS
Phylogenetic analysis.
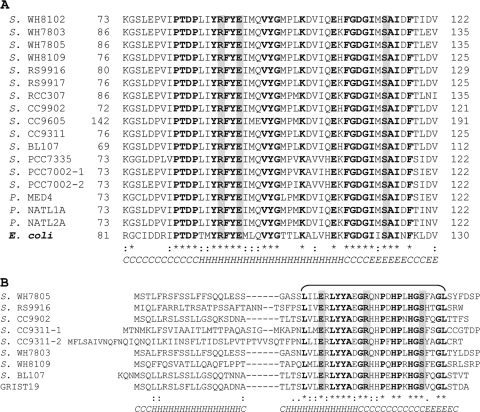
Cyanase, encoded by cynS, is a well-characterized enzyme in E. coli. Orthologs to cynS are commonly found in a wide range of microorganisms, including marine unicellular cyanobacteria. In an attempt to better define cyanase evolution in cyanobacteria, we performed alignments and phylogenetic analyses of both genomic and environmental CynS sequences. Among 12 marine Synechococcus genomes, 11 were found to possess one or more cynS orthologs. The only Synechococcus lacking cynS was WH5701, a strain representative of halotolerant (40), estuarine Synechococcus, ancestral to marine Synechococcus. Among 12 Prochlorococcus genomes only three carried a cynS ortholog (17). Figure 2 shows an alignment of translated cynS sequences of Synechococcus and Prochlorococcus, along with cyanase of E. coli. With an overall 37 to 44% identity, the alignment revealed a higher degree of sequence conservation for the C-terminal region compared to the N-terminal region. Moreover, amino acid residues that are proposed to contribute to the catalytic activity of the E. coli protein (38) were fully conserved in all Synechococcus and Prochlorococcus CynS.
FIG. 2.
Alignment of the amino acid sequences for 17 putative cyanase genes in marine Prochlorococcus (P.) sp. and Synechococcus (S.) sp. strains, along with that of the characterized E. coli cyanase. Fully conserved residues are labeled with “*”, conserved replacements are labeled with “:”, and functional similarity are labeled with “.”. The proposed active-site residues Arg-96, Glu-99, and Ser-122 are shown in boldface against a gray background. The GenBank accession numbers for cyanase sequences in alignment are as follows: Synechococcus sp. strains WH8102 (NP_898579), WH7803 (YP_001226218), WH7805 (ZP_01124911), WH8109 (ZP_05789360), RS9916 (ZP_01471501), RS9917 (ZP_01079240), RCC307 (YP_001228741), CC9902 (YP_378288), CC9605 (YP_382939), CC9311 (YP_732088), BL107 (ZP_01469110), PCC7335 (ZP_05037924), PCC7001 (ZP_05043889, and YP_001733904); Prochlorococcus sp. strains MED4 (NP_892492), NATL1A (YP_001013899), and NATL2A (YP_292581); and E. coli strain K-12 (NP_414874) (4).
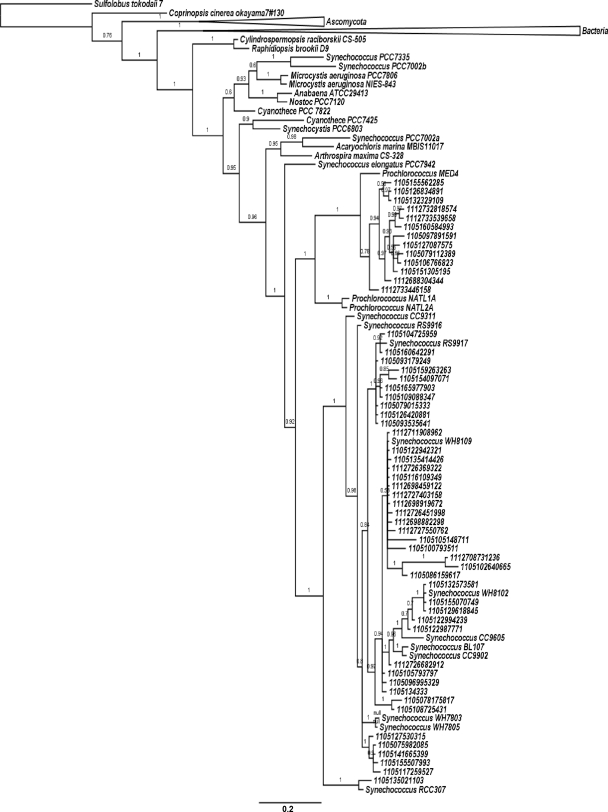
Using CynS from Synechococcus sp. WH7803 and Roseovarius sp. strain 217 as queries in the BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi), we retrieved 107 full-length CynS sequences from genomes of bacterial isolates and strains, 9 fungal CynS sequences, and a single archaeal sequence. In addition, 147 translated open reading frames (ORFs) derived from the Global Ocean Survey (GOS) (27) were retrieved by using the CAMERA BLAST Wizard Tool. After a preliminary alignment, we discarded truncated and incoherent sequences. Subsequently, 90 translated environmental sequences >120 amino acids in length, from surface waters of the Atlantic, Pacific, Indian, and Southern Oceans, were aligned with 29 CynS proteins of known origin. The Archaeal CynS was chosen as an outgroup for the construction of a phylogenetic tree. Tree topology suggests that cyanobacterial cynS evolved from a common ancestor near the base of the bacterial radiation. Of a total of 90 GOS-derived cyanase sequences, 56 clustered with cyanobacterial CynS, and they were affiliated with known Synechococcus and Prochlorococcus CynS (Fig. 3). Of these, 77% clustered with Synechococcus, and the remaining 23% clustered with Prochlorococcus. Branching patterns for cynS closely mirror those of 16S and ntcA topologies (28, 33), suggesting that cyanobacterial cynS distributions resulted from vertical evolution with limited (if at all) contribution of early lateral gene transfer events. Interestingly, cynS has thus far not been identified among any of the marine diazotrophic cyanobacteria.
FIG. 3.
Tree topology resulting from Bayesian phylogenetic analysis of translated environmental cynS sequences (135 amino acids) derived from the Global Ocean Survey database, along with CynS sequences retrieved from GenBank. Detailed relationships among cyanobacterial clades are shown, while bacterial and fungal branches are collapsed for the purpose of presentation. The scale bar provides a distance measure for two substitutions per 100 nucleotides between sequences. Posterior probabilities are given at the nodes using a scale from 0 to 1. The denomination “JCVI PEP” has been omitted from all environmental sequences in order to improve the presentation.
Transcriptional regulation of cynS.
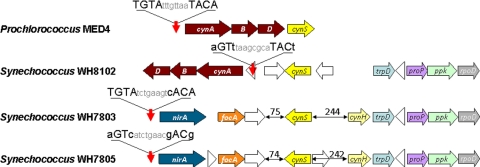
Consistent with the fact that cyanate may serve as an N source, cyanate acquisition genes form an integral part of the regulon controlled by NtcA, a global N-stress regulator in marine cyanobacteria (33). However, NtcA control over cynS transcription has not been clearly established. Whole-genome microarray analyses for Prochlorococcus sp. MED4 (37) showed elevated transcript levels for genes encoding urea (urtA) and cyanate (cynA) transporters in N-deprived cells. However, despite the cynABD and cynS organization as an NtcA-controlled operon, the authors of that study reported that cynS was not differentially expressed (37). In Synechococcus sp. WH8102, cynABD and cynS are separated by 1,932 bp with two putative ORFs (SYNW2488 and SYNW2489) between them. A putative NtcA binding site was found upstream of cynA but not of cynS (Fig. 4), suggesting that cynS transcription may be uncoupled from N stress responses in strain WH8102. Likewise, the promoter region of cynS in Synechococcus sp. WH7803 lacked an NtcA binding motif. Here, we aimed at confirmation of the microarray result for ntcA, cynA, and cynS in N-starved Prochlorococcus sp. MED4 by quantitative RT-PCR. We further expanded transcription studies of these genes to cells supplemented with different N sources (Fig. 5). We further monitored cynA and cynS responses in two Synechococcus strains in an effort to tease apart their contribution to N-scavenging (uptake and conversion of cyanate to ammonium and CO2) and detoxification (cyanate conversion only) pathways.
FIG. 4.
Schematic representation of the genome regions that contain the cyanate utilization genes in Prochlorococcus sp. strain MED4 and Synechococcus spp. strains WH8102, WH7803, and WH7805. Vertical red arrows indicate location of putative NtcA binding sites shown above the arrows, and numbers indicate the nucleotide distance between two adjacent genes. Horizontal arrows indicate cynABD genes encoding for the cyanate transporter (brown), nirA (COG0155, dark blue); focA (COG2116, orange), cynS (bright yellow), cynH (pale yellow), trpD (COG0547E, blue), proP (COG2814G, purple), ppk (COG0855P, green), and rpoD (COG0568K, gray) and ORFs encoding unidentified proteins (white).
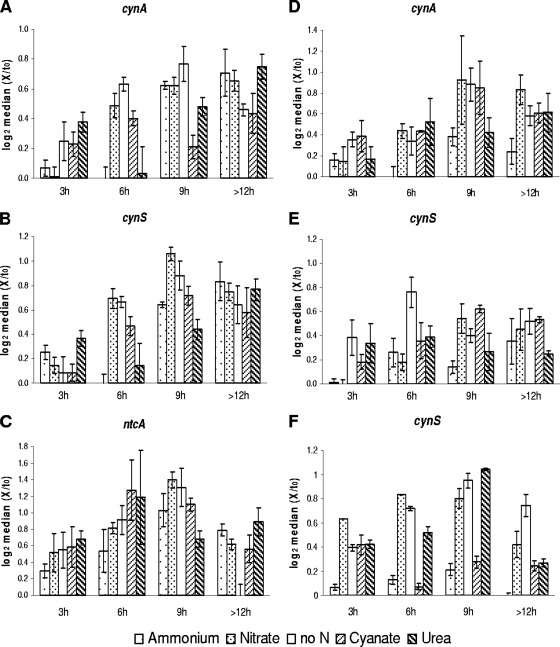
FIG. 5.
Transcript accumulation of cynA (A and D), ntcA (C) and cynS (B, E, and F), in Prochlorococcus sp. MED4 (A to C), Synechococcus sp. WH8102 (D and E), and Synechococcus sp. WH7803 (F) cells grown on ammonium or on alternative N sources or deprived of combined N for 3, 6, 9, and >12 h after resuspension in fresh medium. The data are log2 of median values of three replicates, normalized to their initial transcription level, with 25th to 75th percentiles.
In order to monitor the N status of Prochlorococcus sp. MED4 during the experiment, ntcA transcript levels were determined alongside those of cynA and cynS. ntcA transcript levels, of cells maintained under different N regimes, increased with time before reaching a maximum after 6 h (cyanate, urea) or 9 h (nitrate, no N) of incubation, compared to basal levels in the presence of ammonium. We found as a general trend that during the first 9 h the transcription patterns of cynA and cynS followed that of ntcA in Prochlorococcus sp. MED4, indicating that their transcription occurred in response to changes in N-source and/or its availability (Fig. 5A to C). After 9 h, the transcript had reached steady-state levels or decreased in all treatments. In Synechococcus sp. WH8102, the transcription pattern of cynA (Fig. 5D) was similar to that in Prochlorococcus sp. MED4 (Fig. 5A). However, the increase of cynS transcript was minor and no clear pattern could be discerned in any of the treatments (Fig. 5E). In Synechococcus sp. WH7803 lacking cyanate and urea acquisition capacity, basal transcription of cynS was determined in cells grown with ammonium. After 3 h, cynS transcript accumulated in cells grown with nitrate or urea or in N-free medium. However, in the cyanate-grown culture, transcript levels decreased after 6 h and returned to the levels seen in ammonium-grown cells. After 12 h, cynS remained strongly transcribed in N-starved cells only (Fig. 5F). Figure 6 illustrates the parallel pattern of cynA and cynS transcript accumulation observed for Prochlorococcus sp. MED4 (panel A). In contrast, a significantly different pattern (P < 0.05) was observed in Synechococcus sp. WH8102, suggesting that the cynA and cynS respond to different N signals and controls in this strain.
FIG. 6.
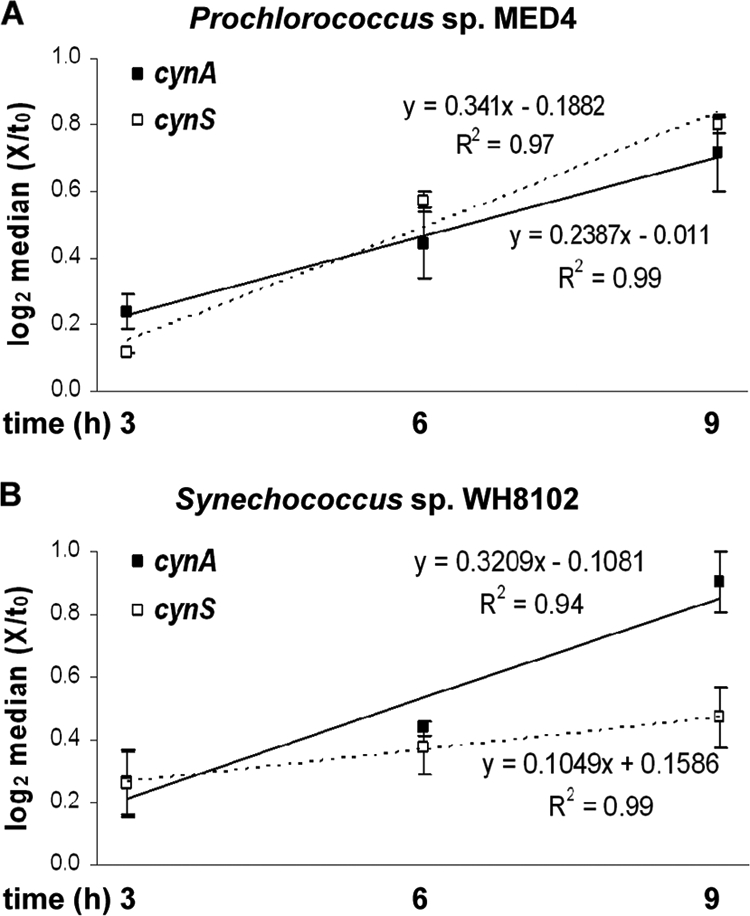
Median log-normalized values of cynA and cynS transcription for Prochlorococcus sp. MED4 (A) and Synechococcus sp. WH8102 (B) cultures grown on alternative N sources or with no N for 3, 6, and 9 h after medium replacement. The error bars represent 25th to 75th percentiles. Linear regression values and R2 values for cynA (continuous) and cynS (dashed) accumulation are presented above and below the respective trend line.
Genomic context of cynS.
The genomic context of cynS was different in different marine cyanobacterial strains. In Prochlorococcus sp. strains NATL1A and NATL2A, cynS was positioned among conserved hypothetical genes. In Prochlorococcus sp. MED4, cynS was positioned immediately downstream of cynABD (Fig. 4), and it was probably transcribed as part of a polycistronic message (Fig. 6), as in E. coli. In marine Synechococcus genomes cynS is confined to a 60-kb region that contains the major N-acquisition genes (33). It is typically found downstream of nirA (assimilatory nitrite reductase) and focA (formate/nitrite transporter) genes and flanked by four genes with a fully conserved localization on these genomes: trpD, proP, ppk, and rpoD (respectively encoding for glycosyl transferase family protein, an unidentified permease, polyphosphate kinase, and an alternative RNA polymerase sigma factor; Fig. 4). Interestingly, Synechococcus sp. WH7805 carries an ORF near cynS that was identified as a cyanate hydratase (i.e., cyanase) in the automated annotation pipeline (Fig. 4). The predicted amino acid sequence appeared unique and orthologs were found in seven marine Synechococcus genomes, as well as on a clone GRIST19 from a metagenomic library obtained from the Atlantic Ocean (GenBank accession no. EU795157). Synechococcus sp. strain CC9311 was found to possess two copies of cynH. Sequence comparison revealed no significant similarity between known CynS sequences and the short protein encoded by cynH. Moreover, CynH could not be assigned to a functional protein family, since no known structural domains were identified in its amino acid sequence by Superfamily 1.73 (42) and Phyre Server 0.2 (20). In the sections below we describe experiments pertaining to cyanase activity associated with overexpressed fusion constructs of WH7803 cynS and cynH.
CynS and CynH overexpression.
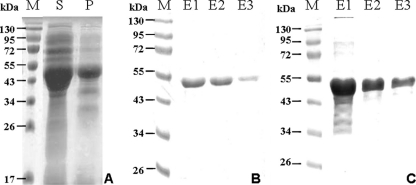
Overexpression of the CynS-MBP and CynH-MBP fusion proteins in different E. coli hosts after IPTG induction was found to be optimal in the E. coli strains HMS174 and Rosetta pLysS, respectively. The recombinant proteins were purified on amylose resin, and the protein presence in the elution fraction was confirmed by SDS-PAGE (Fig. 7). After elution, purified fusion proteins appeared as an ∼50-kDa peptide for CynS-MBP, while the apparent molecular mass of CynH-MBP was slightly smaller (∼48 kDa). Both were in close approximation to the estimated molecular masses for Synechococcus sp. WH7803 CynS (14.3 kDa) and CynH (6.2 kDa) fused to MBP (42.1 kDa). Purified fusion proteins appeared as a single band following the second elution off the maltose resin (Fig. 7B and C). We confirmed the identity of the purified protein by immunoblotting with monoclonal antibodies against MBP (not shown). The levels of fusion protein were below detection for both constructs in crude lysate of noninduced cells but were readily identified in lysate of IPTG-induced cells. A distinct cross-reactivity with the α-MBP antibody was obtained in both supernatant and pellet fractions, implying that the recombinant protein was in part directed to inclusion bodies. In the elution fraction for CynH-MBP, a single band indicated the presence of the fusion protein in a stable configuration. For CynS, however, two bands were detected, presumably representing CynS-MBP and a product resulting from spontaneous cleavage of the fusion protein.
FIG. 7.
(A) IPTG-induced expression of CynS and its accumulation in soluble (lane S) and particulate (lane P) fractions of E. coli cell lysates. (B and C) Purification of CynS (B) and CynH (C) recombinant proteins using amylose resin-based affinity chromatography. M, molecular weight markers; E1 to E3, elution fractions.
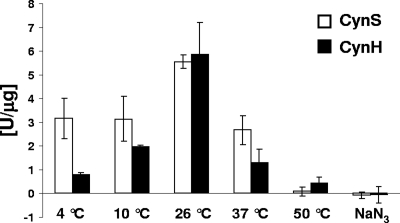
Aliquots of the second eluted fraction for both fusion constructs were subsequently tested for cyanase activity. Measured as ammonium liberated from cyanate, cyanase activity of CynS was maximal (5.56 U mg−1) at 26°C, and rapidly dropped to 50% of this maximum activity at both higher and lower temperatures (Fig. 8). The protein became rapidly inactive at higher temperatures and only residual activity (0.09 U mg−1) was detected at 50°C (Fig. 8). Furthermore, we report here on a distinct cyanase activity associated with the gene product of cynH, identified as a cyanate hydratase in genome annotations, a characterization that thus far lacked experimental evidence. In general, cyanate dependent ammonium accumulation rates were similar to those obtained with CynS: highest cyanase activities (5.87 U mg−1) were measured at 26°C and activities declined at both higher and lower temperatures to 0.80 U mg−1 at 4°C and 0.43 U mg−1 at 55°C. Cyanase activities were derived specifically from either CynS or CynH, as ammonium failed to accumulate when MBP or overexpressed NtcA-MBP (an N regulatory protein) were added to the reaction mix (data not shown). No ammonium accumulation was observed for CynS nor CynH in the presence of the cyanase inhibitor Na-azide (Fig. 8).
FIG. 8.
Characterization of cyanase activity of CynS and CynH fusion constructs at different temperatures and their sensitivity to Na-azide (200 μmol liter−1) addition. Freshly prepared sodium cyanate (2 mmol liter−1) was added to the reaction mix (50-mmol liter−1 PBS buffer [pH 7.6], 3 mmol of sodium bicarbonate liter−1) complemented with aliquots of recombinant protein (20 μg) and incubated at the desired temperature for 10 min. Ammonium accumulation was determined with Nessler reagent. The activities shown are averages from at least nine replicates from three independent experiments.
DISCUSSION
Cyanase serves several functions, the most pronounced being the detoxification of cyanate generated in various metabolic pathways (7, 35). Besides detoxification, microorganisms use cyanase in the assimilation of cyanate from the environment. E. coli transports cyanate into the cell via a cyanate permease encoded by cynX (2, 34). Cyanobacteria utilize cyanate following its acquisition via an ABC-type transport system (10, 17, 24). It has been proposed that cyanate and urea play an important role in the N cycle of marine oligotrophic environments (17). In the present study we characterized the evolution, marine distribution, and transcriptional regulation of cynS (cyanase) and the activity associated with its gene product. We further report on a novel cyanase encoded by cynH in marine Synechococcus.
CynS tree topologies show cyanobacteria as a well-defined branch emerging at the base of the bacterial lineage. Cyanase was found in Synechocystis sp. PCC6803 (19), Synechococcus elongatus PCC7942 (39), the filamentous diazotrophs Anabaena sp. strain ATCC 29413 and Nostoc sp. strain PCC7120 (9), Synechococcus sp. strains PCC7002 and PCC7335 from brackish, estuarine waters (29), toxic bloom-forming Microcystis (18), and members of the unicellular marine Synechococcus and Prochlorococcus (the present study). A total of >60% of the GOS-derived cyanase sequences were identified as cyanobacterial. This includes 10 clones from a hypersaline lagoon that clustered with Synechococcus sp. strain RS9917, a euryhaline ecotype (6, 12).
Cyanobacteria likely acquired cyanase during the very early stages of their evolution. Tree topologies of cyanobacterial CynS matched phylogenies based on 16S rRNA, ITS and ntcA (12, 21, 28, 30, 33), and branching was supported by strong posterior probabilities. Based on these observations, we suggest that cynS was common in ancestral cyanobacteria and cynS was lost from many modern cyanobacteria. Our tree topology suggests that the importance of lateral gene transfer of cynS was minor; however, it might occur among related species. Thus, the estuarine Synechococcus sp. PCC7002 carries two cynS orthologs that share 74% identity at the amino acid level (Synechococcus PCC7002a and PCC7002b in Fig. 3). Synechococcus sp. PCC7002a, encoded by a stand-alone cynS gene, clustered with CynS of the endosymbiont Acaryochloris marina MBIC11017, which is ancestral to Synechococcus sp. PCC7002 (36). The Synechococcus sp. PCC7002b homolog is most closely related to CynS of Synechococcus sp. PCC7335 and partakes in an NtcA-regulated cynABDS operon, which is very similar to our observations for Prochlorococcus sp. strain MED4. Thus, the cyanase gene is involved in lateral gene transfer, suggesting that different CynS may carry out distinct functions in the cyanobacterial cell. We propose that CynS by itself may provide the cell with means to detoxify internally generated cyanate, whereas the cynABDS operon encodes the utilization of external cyanate. The presence of two cyanase homologs on a single genome suggests that both functions play distinct roles in the N metabolism and N assimilation of cyanobacteria.
In an attempt to set apart the cyanate detoxification and utilization functions of cyanase, we studied transcriptional regulation of cynS and cynA alongside that of the N-regulatory gene ntcA. In Prochlorococcus sp. MED4, in contrast to Synechococcus sp. WH8102, cynABDS genes showed coordinated expression in response to N deprivation (Fig. 6). This is in agreement with the gene arrangement in the strains examined (Fig. 4). In Synechococcus sp. WH7803, which lacks the transporter genes, N-independent regulation of cynS was observed. This implies that cynS in Prochlorococcus sp. MED4 takes part in utilization of external cyanate, whereas the presence of cynS in Synechococcus genomes indicates a requirement to detoxify an intracellular cyanate. There are several possible sources for the intracellular cyanate in cyanobacteria. A substrate of the urea cycle, carbamoyl phosphate is known to decompose spontaneously to cyanate and phosphate (2). Urea undergoes spontaneous transformation to cyanate by an isomeric change (5, 23). The origin of urea in Synechococcus and Prochlorococcus cells is unclear, since they lack the arg gene product that facilitates urea hydrolysis (33). However, marine cyanobacteria may convert excess arginine to spermidine by sequential action of arginine decarboxylase (EC 4.1.1.19), agmatine ureaohydrolase (EC 3.5.3.11), and spermidine synthase (EC 2.5.1.16). Hence, despite a lack of arginase (EC 3.5.3.1), the toxic cyanate can transform from urea produced by agmatine ureaohydrolase.
In an attempt to confirm cyanase activity by CynS from unicellular marine cyanobacteria, the cynS was cloned as a fusion construct with MBP (CynS-MBP), overexpressed in an E. coli background, and subsequently purified on amylose resin. Using enzyme assays, we clearly identified CynS as a functionally active cyanase. Similarly to cyanases described in other studies (2, 9, 38), the CynS-MBP construct showed bicarbonate-dependent cyanate degrading activity that was inhibited by Na-azide, the latter preventing binding of substrate to the holoenzyme (38). Cyanase activity was also confirmed for a short peptide, a product of an ORF that was tentatively annotated as cyanate hydratase, and we propose to rename this ORF cynH. The identification of an additional cyanase raises questions about its origin and physiological importance. The cynH gene was found in seven marine Synechococcus genomes, as well as on the metagenomic clone GRIST19. Although located on the same genomic region, cynS is separated from cynH by 210 to 244 bp and is transcribed in the opposite orientation (Fig. 4). It is unlikely that cynH resulted from (partial) gene duplication since its amino acid sequence does not align with any part of cynS. The two genes are thus paralogs. Nine cynH sequences shared a high degree of identity in the C-terminal half (Fig. 9 B) similar to CynS, suggesting that the catalytic domain of CynH is confined to this region. The catalytic site of CynS contains Arg (R), Glu (E), and Ser (S) residues (38) in a configuration conserved across bacteria, fungi, and plants (9). Interestingly, we identified three fully conserved amino acid residues in CynH, identical to those of CynS, and their configuration is reminiscent of the active site of CynS. Moreover, secondary structure predictions indicated the presence of an α-helix followed by a short β-sheet in the C-terminal domain of CynH in agreement with the secondary structure of the C-terminal domain of CynS (38) (Fig. 9). No reliable prediction for tertiary structure of CynH is available due to the low similarity to any known protein and a lack of defined motifs. The dual role the cynS and cynH genes in marine Synechococcus remains to be clarified.
FIG. 9.
(A and B) Amino acid alignments for catalytic site regions of cyanases for 17 CynS sequences (A) and 9 full-length CynH sequences (B) found in marine Synechococcus (S.) and in a single metagenomic clone GRIST19. Identical residues are presented in boldface and labeled “*”, conserved residue replacements are labeled “:”, and functionally similar residues are labeled “.”. The proposed active-site residues Arg (R), Glu (E), and Ser (S) are shown in boldface against a gray background. The bottom line presents the consensus secondary structure predicted by the Jpred prediction server that identifies randomly coiled region (C), α-helix (H), and β-sheet (E) motifs. In panel B, the aligned sequences are cyanate hydratase (ZP_01124909) in Synechococcus sp. WH7805, RS9916_37357 (ZP_01471502) in Synechococcus sp. RS9916, Syncc9902_2288 (YP_378289) in Synechococcus sp. CC9902, sync_2840 and sync_2903 (YP_732028 and YP_732090) in Synechococcus sp. CC9311, SynWH7803_2496 (YP_001226219) in Synechococcus sp. WH7803, SH8109_0550 (ZP_05789530) in Synechococcus sp. WH8109, non-annotated (reverse strand 1870903-1871064) in Synechococcus sp. BL107 (NZ_AATZ00000000), and metagenomic clone GRIST19 (EU795157).
Acknowledgments
Plasmids with recombinant MBP-NtcA constructs were kindly provided by H. Zer and B. Rihtman.
The Niedersachsen State Fund at the Hebrew University, the Israel Science Foundation (grant 135/05), and the NATO Science for Peace program (grant SfP 98216) all provided financial support.
Footnotes
Published ahead of print on 5 November 2010.
REFERENCES
- 1.Anderson, P. M. 1980. Purification and properties of the inducible enzyme cyanase. Biochemistry 19:2882-2888. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P. M., Y. C. Sung, and J. A. Fuchs. 1990. The cyanase operon and cyanate metabolism. FEMS Microbiol. Rev. 87:247-252. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Chin, C. C., P. M. Anderson, and F. Wold. 1983. The amino acid sequence of Escherichia coli cyanase. J. Biol. Chem. 258:276-282. [PubMed] [Google Scholar]
- 5.Dirnhuber, P., and F. Schütz. 1948. The isomeric transformation of urea into ammonium cyanate in aqueous solutions. Biochem. J. 42:628-632. [PubMed] [Google Scholar]
- 6.Dufresne, A., M. Ostrowski, D. J. Scanlan, L. Garczarek, S. Mazard, B. P. Palenik, I. T. Paulsen, N. T. de Marsac, P. Wincker, C. Dossat, S. Ferriera, J. Johnson, A. F. Post, W. R. Hess, and F. Partensky. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbs, S. 2004. Biological degradation of cyanide compounds. Curr. Opin. Biotechnol. 15:231-236. [DOI] [PubMed] [Google Scholar]
- 8.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elleuche, S., and S. Pöggeler. 2008. A cyanase is transcriptionally regulated by arginine and involved in cyanate decomposition in Sordaria macrospora. Fungal Genet. Biol. 45:1458-1469. [DOI] [PubMed] [Google Scholar]
- 10.Espie, G. S., F. Jalali, T. Tong, N. J. Zacal, and A. K.-C. So. 2007. Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria. J. Bacteriol. 189:1013-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman, J. 2003. Genome sequences from the sea. Nature 424:1001-1002. [DOI] [PubMed] [Google Scholar]
- 12.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelman, A., and D. Rubin. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7:457-511. [Google Scholar]
- 14.Harano, Y., I. Suzuki, S.-i. Maeda, T. Kaneko, S. Tabata, and T. Omata. 1997. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5744-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, W. V., and P. M. Anderson. 1987. Bicarbonate is a recycling substrate for cyanase. J. Biol. Chem. 262:9021-9025. [PubMed] [Google Scholar]
- 17.Kamennaya, N. A., M. Chernihovsky, and A. F. Post. 2008. The cyanate utilization capacity of marine unicellular cyanobacteria. Limnol. Oceanogr. 53:2485-2494. [Google Scholar]
- 18.Kaneko, T., N. Nakajima, S. Okamoto, I. Suzuki, Y. Tanabe, M. Tamaoki, Y. Nakamura, F. Kasai, A. Watanabe, K. Kawashima, Y. Kishida, A. Ono, Y. Shimizu, C. Takahashi, C. Minami, T. Fujishiro, M. Kohara, M. Katoh, N. Nakazaki, S. Nakayama, M. Yamada, S. Tabata, and M. M. Watanabe. 2007. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., and S. Tabata. 1997. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 38:1171-1176. [DOI] [PubMed] [Google Scholar]
- 20.Kelley, L. A., and M. J. E. Sternberg. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 21.Lavin, P., P. Gómez, B. González, and O. Ulloa. 2008. Diversity of the marine picocyanobacteria Prochlorococcus and Synechococcus assessed by terminal restriction fragment length polymorphisms of 16S-23S rRNA internal transcribed spacer sequences. Rev. Chil. Hist. Nat. 81:515-531. [Google Scholar]
- 22.Maeda, S.-I., and T. Omata. 2009. Nitrite transport activity of the ABC-type cyanate transporter of the cyanobacterium Synechococcus elongatus. J. Bacteriol. 191:3265-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marier, J. R., and D. Rose. 1964. Determination of cyanate, and a study of its accumulation in aqueous solutions of urea. Anal. Biochem. 7:304-314. [DOI] [PubMed] [Google Scholar]
- 24.Miller, A. G., and G. S. Espie. 1994. Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch. Microbiol. 162:151-157. [Google Scholar]
- 25.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 26.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 27.Parthasarathy, H., E. Hill, and C. MacCallum. 2007. Global ocean sampling collection. PLoS Biol. 5:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penno, S., D. Lindell, and A. F. Post. 2006. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ. Microbiol. 8:1200-1211. [DOI] [PubMed] [Google Scholar]
- 29.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 30.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 32.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 33.Scanlan, D. J., M. Ostrowski, S. Mazard, A. Dufresne, L. Garczarek, W. R. Hess, A. F. Post, M. Hagemann, I. Paulsen, and F. Partensky. 2009. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73:249-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung, Y. C., and J. A. Fuchs. 1988. Characterization of the cyn operon in Escherichia coli K-12. J. Biol. Chem. 263:14769-14775. [PubMed] [Google Scholar]
- 35.Sung, Y. C., and J. A. Fuchs. 1989. Identification and characterization of a cyanate permease in Escherichia coli K-12. J. Bacteriol. 171:4674-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swingley, W. D., M. Chen, P. C. Cheung, A. L. Conrad, L. C. Dejesa, J. Hao, B. M. Honchak, L. E. Karbach, A. Kurdoglu, S. Lahiri, S. D. Mastrian, H. Miyashita, L. Page, P. Ramakrishna, S. Satoh, W. M. Sattley, Y. Shimada, H. L. Taylor, T. Tomo, T. Tsuchiya, Z. T. Wang, J. Raymond, M. Mimuro, R. E. Blankenship, and J. W. Touchman. 2008. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. U. S. A. 105:2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolonen, A. C., J. Aach, D. Lindell, Z. I. Johnson, T. Rector, R. Steen, G. M. Church, and S. W. Chisholm. 2006. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol. Syst. Biol. 2:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, M. A., Z. Otwinowski, A. Perrakis, P. M. Anderson, and A. Joachimiak. 2000. Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure 8:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterbury, J. B., and R. Rippka. 1989. Subsection 1: order Croococcales. Wettsten 1924, emend. Rippka et al., 1979, p. 1728-1746. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 40.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus, p. 71-120. In T. Platt and W. Li (ed.), Photosynthetic picoplankton, vol. 214. Department of Fisheries and Oceans, Ottawa, Ontario, Canada. [Google Scholar]
- 41.Waterbury, J. B., and J. M. Willey. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100-105. [Google Scholar]
- 42.Wilson, D., R. Pethica, Y. Zhou, C. Talbot, C. Vogel, M. Madera, C. Chothia, and J. Gough. 2009. SUPERFAMILY: sophisticated comparative genomics, data mining, visualization, and phylogeny. Nucleic Acids Res. 37:380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]