Abstract
Diffusion of entities inside biofilm triggers most mechanisms involved in biofilm-specific phenotypes. Using genetically engineered hydrophilic and hydrophobic cells of Lactococcus lactis yielding similar biofilm architectures, we demonstrated by fluorescence correlation spectroscopy that bacterial surface properties affect diffusion of nanoparticles through the biofilm matrix.
Bacteria are able to grow adhered to almost any surface, forming architecturally complex communities termed biofilms (10). It is now recognized that genetic and physiological heterogeneities participate in specific biofilm functions, such as dramatic resistance to antimicrobials. The primary mechanism generated by such heterogeneities is diffusion limitation through the three-dimensional organization of the biofilm framework (2, 9, 11, 12). Although extracellular polymeric substances (EPS) are described to possibly influence such diffusion processes, there are no reported studies on the impact of bacterial cell wall properties on such phenomenon. To explore the influence of the bacterial surface properties in intrabiofilm entity mobility, we used genetically engineered hydrophilic and hydrophobic isogenic mutants of Lactococcus lactis described not to affect biofilm architecture (5, 6). Fluorescent nanoparticle diffusion inside biofilms formed by these mutants was analyzed noninvasively by fluorescence correlation spectroscopy (FCS) under two-photon excitation.
Expression of the cell wall-anchored proteinase PrtP increases Lactococcus lactis hydrophobicity but does not affect biofilm architecture.
Lactococcus lactis is a model lactic acid bacterium, generally recognized as safe but phylogenetically closely related to pathogens of the genera Streptococcus and Enterococcus. It has been previously shown that the expression of the major cell wall-anchored proteinase PrtP was responsible for altering L. lactis surface physicochemical properties, shifting the cell envelope from a hydrophilic surface (affinity to hexadecane in a partitioning test, <10%) to an extremely hydrophobic one (affinity to hexadecane, >90%) (4, 5, 7).
We used derivatives of L. lactis subsp. cremoris strain MG1363 expressing or lacking PrtP: PRTP+ (PrtP anchored and active), PRTP* (PrtP anchored and inactive, to separate the influence of the protein itself from its enzymatic activity), and PRTP− (MG1363 carrying a vector plasmid without the prtP gene). In addition, it was recently shown that PrtP expression in L. lactis did not induce architectural modifications during biofilm formation (6). Indeed, no statistical differences were detected between PRTP−, PRTP+, and PRTP* biofilm structural parameters extracted from the confocal laser scanning microscopy image series (P value of >0.05 for biovolume; roughness and thickness extracted by the PHLIP Matlab routine [http://sourceforge.net/projects/phlip]).
FCS measurements of nanoparticle diffusion in L. lactis biofilms.
Biofilms of L. lactis PRTP−, PRTP+, and PRTP* were cultivated at 25°C in single-channel flow cells (BST FC 81) as described previously (6). In brief, flow cells were inoculated with 1 ml of exponential-phase culture adjusted to an optical density at 600 nm (OD600) of 0.1 and left in static conditions for 1 h to allow initial bacterial attachment. After this period, the flow was resumed at 2 ml/h, and surface-associated bacteria were left to grow at 25°C for 24 h. The diffusion of anionic carboxylate-modified fluorescent polystyrene beads of 50-nm radius (FluoSpheres F8801, Invitrogen) inside these biofilms was examined on 30 to 46 measurements performed on at least three independent samples for each strain. The FCS principle is based on monitoring the emission intensity fluctuations due to a small number of molecules passing through the confocal excitation volume. These fluctuations can be quantified in their amplitude and duration by temporally autocorrelating the recorded intensity signal. The FCS two-photon excitation experimental setup was described previously. In short, biofilms covered with a 10 nM fluorescent nanoparticle suspension were mounted under a 63×/1.4 numerical aperture objective and then biphotonically excited at 800 nm by a Ti:Sa pulsed laser (120 femtoseconds, 76 MHz). The fluorescence was then collected through the same objective and separated from the excitation beam by a dichroic mirror before being focused on a photomultiplier (3). Fluorescence intensity fluctuations were analyzed using a commercial correlator and treated with homemade programs (1, 8).
Bacterial cell wall hydrophobicity affects nanoparticle diffusion inside the biofilm matrix.
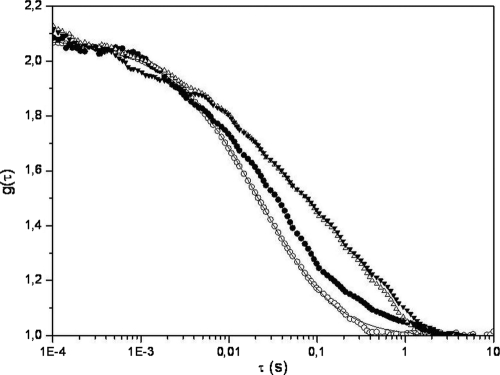
Representative autocorrelation curves [g(τ)] obtained for fluorescent nanospheres within PRTP−, PRTP+, and PRTP* biofilms are presented in Fig. 1. Adjustments of these experimental curves were performed using the Brownian diffusion model with two components (Table 1):
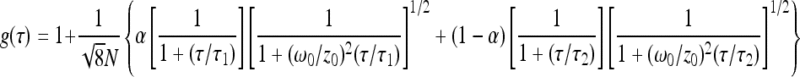 |
where α and 1 − α are the fractions of the molar concentrations of the two diffusive species and τ1 and τ2 are their respective translational diffusion times, N is the number of fluorescent species inside the excitation volume, and ω0/z0 is the lateral/axial radius ratio of the laser beam (ω0/z0 = 0.30 ± 0.03, determined elsewhere [1]).
FIG. 1.
Normalized fluorescence autocorrelation curves [g(τ)] for 0.6 nmol/liter anionic 50-nm-radius nanospheres diffusing in water (○) and in PRTP− (•), PRTP+ (▵), and PRTP* (▾) biofilms. Each curve corresponds to the mean of at least 30 measurements performed on three independent samples.
TABLE 1.
Translational diffusion time (τ and α) parameters obtained from fitting with the equation from the fluorescent correlation curves for anionic latex beads within water and three different L. lactic biofilmsa
| Water or biofilm | τ1 (ms) | α1(%) | τ2(ms) | α2(%) |
|---|---|---|---|---|
| Water | 8.9 ± 0.6 | 92 ± 3 | >100 | 8 ± 3 |
| PRTP− | 8.7 ± 0.7 | 52 ± 9 | 177 ± 43 | 48 ± 9 |
| PRTP+ | 7.5 ± 0.7 | 38 ± 6 | 204 ± 46 | 62 ± 6 |
| PRTP* | 7.8 ± 0.7 | 36 ± 6 | 239 ± 42 | 64 ± 6 |
α1 and α2 are the fractions of the molar concentrations of the two diffusive species, and τ1 and τ2 are their respective translational diffusion times. Values are represented as the means ± standard errors of the means from 30 to 46 measurements from at least three independent experiments for each strain.
An initial diffusion time (τ1) of ∼8 ms was obtained in water and in all biofilms and hence could reasonably be assigned to the free diffusion of monomeric nanospheres. Nonetheless, the proportion of probes (α1) freely diffusing is statistically different (P < 0.05) depending on the surface physicochemical properties of the strains used to grow the biofilms: a lower fraction of fluorescent particles freely diffuses in PRTP+ and PRTP* biofilms composed of hydrophobic cells than in PRTP− biofilms composed of hydrophilic cells, as presented in Table 1. The remaining populations (α2) correspond to probes diffusing with a longer diffusion time (τ2) that could be assigned to species whose diffusion inside L. lactis biofilms is hindered. Although the observed obstruction of diffusion could be interpreted as being caused by particle aggregation, the obtained τ2 value measured in biofilms would unlikely correspond to the diffusion of aggregates larger than 1 μm in diameter (which has never been measured with these nanospheres in such proportions). This τ2 diffusion time may more likely be ascribed to particles colliding with the bacterial envelope. Furthermore, α2 and τ2 values are higher in the case of PRTP+ and PRTP* than in the case of PRTP− (P < 0.05), suggesting softer impacts of nanoparticles with hydrophobic cell walls than with hydrophilic ones. Moreover, similar results were obtained in both PRTP+ and PRTP* biofilms (P > 0.05), suggesting that the presence of the anchored proteinase rather than its enzymatic activity is responsive in hindering particle diffusion inside L. lactis biofilms. It cannot be excluded that modifications to the cell surface may exert an indirect effect on EPS properties that could affect diffusion properties in the biofilm matrix.
To summarize, the presented data demonstrated that bacterial cell wall properties can affect diffusion inside the biofilm matrix. Indeed, in addition to the exopolymeric substances, interfacial bacterial components, such as peptidoglycan, capsules, proteins, S-layer, and pili, among others, could also condition molecule and particle mobility inside biofilms and hence participate in specific biofilm phenotypes.
Acknowledgments
O. Habimana was the recipient of a fellowship from LabHealth under Marie Curie contract MEST-CT-2004-514428.
Confocal laser scanning microscopy was performed at the MIMA2 microscopy platform (http://voxel.jouy.inra.fr/mima2).
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Briandet, R., P. Lacroix-Gueu, M. Renault, S. Lecart, T. Meylheuc, E. Bidnenko, K. Steenkeste, M. N. Bellon-Fontaine, and M. P. Fontaine-Aupart. 2008. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl. Environ. Microbiol. 74:2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryers, J. D., and F. Drummond. 1998. Local macromolecule diffusion coefficients in structurally non-uniform bacterial biofilms using fluorescence recovery after photobleaching (FRAP). Biotechnol. Bioeng. 60:462-473. [PubMed] [Google Scholar]
- 3.Gulot, E., P. Georges, A. Brun, M. P. Fontaine-Aupart, M. N. Bellon-Fontaine, and R. Briandet. 2002. Heterogeneity of diffusion inside microbial biofilms determined by fluorescence correlation spectroscopy under two-photon excitation. Photochem. Photobiol. 75:570-578. [DOI] [PubMed] [Google Scholar]
- 4.Haandrikman, A. J., R. Meesters, H. Laan, W. N. Konings, J. Kok, and G. Venema. 1991. Processing of the lactococcal extracellular serine proteinase. Appl. Environ. Microbiol. 57:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habimana, O., C. Le Goff, V. Juillard, M. N. Bellon-Fontaine, G. Buist, S. Kulakauskas, and R. Briandet. 2007. Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habimana, O., M. Meyrand, T. Meylheuc, S. Kulakauskas, and R. Briandet. 2009. Genetic features of resident biofilms determine attachment of Listeria monocytogenes. Appl. Environ. Microbiol. 75:7814-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kok, J., and G. Buist. 2003. Genetics of proteolysis in Lactococcus lactis, p. 189-223. In B. J. B. Wood and P. J. Warner (ed.), Genetics of lactic acid bacteria, vol. 7. Kluwer Academic/Plenum, New York, NY.
- 8.Lacroix-Gueu, P., R. Briandet, S. Leveque-Fort, M. N. Bellon-Fontaine, and M. P. Fontaine-Aupart. 2005. In situ measurements of viral particles diffusion inside mucoid biofilms. C. R. Biol. 328:1065-1072. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence, J. R., G. M. Wolfaardt, and D. R. Korber. 1994. Determination of diffusion coefficients in biofilms by confocal laser microscopy. Appl. Environ. Microbiol. 60:1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez, D., H. Vlamakis, and R. Kolter. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart, P. S., and M. J. Franklin. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199-210. [DOI] [PubMed] [Google Scholar]