Abstract
This study aimed to isolate and characterize treponemes present in the bovine gastrointestinal (GI) tract and compare them with bovine digital dermatitis (BDD) treponemes. Seven spirochete isolates were obtained from the bovine GI tract, which, on the basis of 16S rRNA gene comparisons, clustered within the genus Treponema as four novel phylotypes. One phylotype was isolated from several different GI tract regions, including the omasum, colon, rumen, and rectum. These four phylotypes could be divided into two phylotype pairs that clustered closest with each other and then with different, previously reported rumen treponemes. The treponemes displayed great genotypic and phenotypic diversity between phylotypes and differed considerably from named treponeme species and those recently reported by metagenomic studies of the bovine GI tract. Phylogenetic inference, based on comparisons of 16S rRNA sequences from only bovine treponemes, suggested a marked divergence between two important groups. The dendrogram formed two major clusters, with one cluster containing GI tract treponemes and the other containing BDD treponemes. This division among the bovine treponemes is likely the result of adaptation to different niches. To further differentiate the bovine GI and BDD strains, we designed a degenerate PCR for a gene encoding a putative virulence factor, tlyC, which gave a positive reaction only for treponemes from the BDD cluster.
Treponema species are typically anaerobic, fastidious, highly motile, spiral microorganisms and may be found in the oral cavity, digestive tract, and genital areas of humans, animals, and insects (20, 32). Several treponeme taxa are associated with disease, such as the human syphilis infectious agent Treponema pallidum (30), various Treponema species associated with human periodontal infections (10), and several treponeme phylotypes involved in bovine digital dermatitis (BDD) (4). In contrast, several treponemes have been reported to be commensal, living as symbionts in the gastrointestinal (GI) tracts of animals and insects. The spirochetes Treponema bryantii and Treponema saccharophilum have been isolated from the rumen of cows (26, 36), and another three novel treponeme taxa have been isolated from GI material of pigs (6, 23). In insects, Treponema azotonutricium and Treponema primitia were isolated from the guts of termites (15).
The relative paucity of data regarding treponemes and their locations in animal tissues is primarily the result of difficulties associated with their isolation, cultivation, and purification. More frequently, molecular techniques such as 16S rRNA gene clone libraries have been used to identify treponemes within animal samples but without subsequent culture. While no targeted studies of bovine GI tract treponeme 16S rRNA genes have been reported, several global bacterial studies showed that there are several phylotypes of treponemes in the bovine GI tract (11, 29, 38, 44). Although only two bovine GI tract treponemes have been proposed as novel taxa (26, 36), considerable work has suggested phenotypic and physiological diversity among the bovine rumen treponemes (25, 35, 45, 46). However, it has been more than 20 years since the majority of the bovine rumen treponeme work was reported, and as a result, important data such as 16S rRNA gene sequences have not been described for many of these bacteria. Furthermore, since those initial studies, an infectious lameness attributed largely to treponemes, known as digital dermatitis, has appeared in cows and more recently has also been identified in sheep (3a, 21). We have been able to isolate treponemes from BDD lesions and, along with others, have identified high associations of treponemes with this infection (13, 14, 17, 22). As there have been no recent isolation and characterization studies of bovine GI tract treponemes, we considered it important to attempt the isolation of treponemes from the bovine GI tract. Such isolations might identify treponemes similar to BDD treponemes or yield useful commensal control microorganisms for comparison studies. To this end, we have isolated four phylotypes of treponemes from the bovine GI tracts of cattle and subjected them to comparisons with BDD treponemes and previously reported bovine GI tract treponemes.
MATERIALS AND METHODS
Isolation and cultivation.
Samples were taken from different parts of the GI tracts of Holstein-Friesian dairy cattle immediately after slaughter. Approximately 30-ml samples of GI contents were taken from each location, as shown in Table 1. These GI tract samples were stored on ice for transport to the laboratory. The samples were transferred into an anaerobic cabinet (85% N2, 10% H2, and 5% CO2 at 36°C), and 3 loopfuls from the center of the sample was inoculated into oral treponeme enrichment broth (OTEB; Anaerobe Systems, Morgan Hill, CA) containing antibiotics and supplemented with 10% fetal calf serum (FCS) or 10% rabbit serum (RS) and incubated for 1 to 3 days before subculture. Antibiotics for broth and plate subculture were initially rifampin (5 μg/ml) and enrofloxacin (5 μg/ml) as described previously for the isolation of BDD treponemes from the United Kingdom (13), and the amount of rifampin was later increased to 25 μg/ml, resulting in final concentrations of antibiotics used for the isolation of BDD treponemes in the United States (42). Bacteria from broth cultures were then subcultured onto fastidious anaerobe agar (FAA) plates (LabM, Bury, United Kingdom) supplemented with 5% defibrinated sheep blood, 10% FCS, and antibiotics, as described above, for ∼2 weeks. Single colonies from FAA plates were inoculated into growth medium without antibiotics, and subculturing was repeated if cultures were deemed impure by phase-contrast microscopy and 16S rRNA gene sequencing. Isolates were stored at −80°C in growth medium containing 10% glycerol.
TABLE 1.
Spirochetes isolated from bovine gastrointestinal tracts in this study
| Sample | Biopsy date (day/mo/yr) | Farm and cowa | Location of isolation | BDD statusb | Selection antibioticsc | Treponeme(s) isolatedd |
|---|---|---|---|---|---|---|
| 1 | 6/5/05 | Farm 1, cow 1 | Rectum | + | 5r5e | IF |
| 2 | 6/5/05 | Farm 1, cow 2 | Rectum | − | 5r5e | IF |
| 3 | 13/9/05 | Farm 2, cow 1 | Rectum | + | 5r5e | IF |
| 4 | 13/9/05 | Farm 2, cow 2 | Rectum | + | 5r5e | IF |
| 5 | 13/9/05 | Farm 2, cow 3 | Rectum | + | 5r5e | IF |
| 6 | 14/9/05 | Farm 3, cow 1 | Rectum | + | 25r5e | IF |
| 7 | 14/9/05 | Farm 3, cow 1 | Rumen | + | 25r5e | IF |
| 8 | 14/9/05 | Farm 3, cow 2 | Rectum | + | 25r5e | IF |
| 9 | 15/12/05 | Farm 4, cow 1 | Rumen | + | 25r5e | IF |
| 10 | 15/12/05 | Farm 4, cow 1 | Rectum | + | 25r5e | IF |
| 11 | 15/12/05 | Farm 4, cow 1 | Cecum | + | 25r5e | IF |
| 12 | 15/12/05 | Farm 4, cow 1 | Abomasum | + | 25r5e | AC3 |
| 13 | 15/12/05 | Farm 4, cow 1 | Omasum | + | 25r5e | OC1 |
| 14 | 15/12/05 | Farm 4, cow 1 | Colon | + | 25r5e | CC2 |
| 15 | 4/01/06 | Farm 5, cow 1 | Rectum | + | 25r5e | IF |
| 16 | 4/01/06 | Farm 5, cow 2 | Rectum | + | 25r5e | CHPA |
| 17 | 15/11/06 | Farm 6, cow 1 | Urine | + | 25r5e | IF |
| 18 | 15/11/06 | Farm 6, cow 1 | Rectum | + | 25r5e | IF |
| 19 | 15/11/06 | Farm 6, cow 2 | Urine | + | 25r5e | IF |
| 20 | 15/11/06 | Farm 6, cow 2 | Rectum | + | 25r5e | IF |
| 21 | 16/6/09 | Farm 7, bull 1 | Rumen | − | 25r5e | RU1, RU2 |
| 22 | 16/6/09 | Farm 7, bull 2 | Rectum | − | 25r5e | RE1 |
| 23 | 16/6/09 | Farm 7, heifer 1 | Rumen | − | 25r5e | IF |
| 24 | 16/6/09 | Farm 7, heifer 1 | Abomasum | − | 25r5e | IF |
| 25 | 16/6/09 | Farm 7, heifer 2 | Rectum | − | 25r5e | IF |
| 26 | 16/6/09 | Farm 7, heifer 2 | Abomasum | − | 25r5e | IF |
| 27 | 7/10/09 | Farm 8, cow 1 | Rectum | + | 25r5e | IF |
All farms listed were in Cheshire, and all animals tested were Holstein-Friesian cows.
Whether the animal had an active BDD lesion (+) or no lesion present (−).
Antibiotic concentration used in medium and on agar plates. Numbers are micrograms/milliliter. r, rifampin; e, enrofloxacin.
IF, isolation failed.
Reference strains.
Reference strains T19, T56, T320A, T354B, T3552B, and T354A are two representatives from each of the three unique treponeme groups previously isolated from BDD lesions (13). Treponema medium ATCC 700293T and Treponema phagedenis biotype Reiter were grown and DNA was extracted as previously described (12, 13).
Gene sequencing and phylogenetic analyses.
Genomic DNA was extracted from spirochete cultures as previously described (13) and stored at −20°C. Spirochete 16S rRNA gene PCRs were carried out as described previously (7). A PCR assay targeting the dinitrogenase reductase gene (nifH) in spirochetes was carried out as described previously (19). A PCR assay targeting the gene encoding a putative hemolysin/extracellular matrix binding protein (TlyC) used degenerate primers (tlyCdegenF [5′-GAGGTKATGATWCCGCGTAT-3′] and tlyCdegenR [5′-CTTATGTSCKTCCATRTC-3′]). These primers were chosen by aligning the tlyC sequences of Treponema pallidum subsp. pallidum (strain Nichols) and Treponema denticola ATCC 35405 using CLUSTALW (41). Stringent PCR conditions were identified by using a Mastercycler gradient thermocycler (Eppendorf, Germany). PCRs used Taq polymerase (Qiagen, United Kingdom) according to the manufacturer's instructions (with the addition of magnesium to a final concentration of 3 mM), with 1 μl of the DNA template per 25-μl reaction volume and incubation at 95°C for 5 min, followed by 35 cycles of 94°C for 1 min, 43.5°C for 3 min, and 72°C for 3 min, with a final extension step of 72°C for 5 min. Amplified PCR products were sequenced commercially, and completed genes were assembled by using PREGAP4 from the Staden package (34). Gene sequences were aligned by using CLUSTALW and trimmed, and phylogenetic trees were calculated with the unweighted-pair group method using average linkages (UPGMA) (bootstrap values based on 10,000 iterations) using nucleotide substitution rates calculated according to the Kimura two-parameter model implemented in MEGA4 (39).
Enzyme activities.
Enzyme profiles for the spirochete isolates were determined by using the Apizym system (bioMeriéux, Lyon, France) according to the manufacturer's instructions, and each test was carried out in triplicate. The validation of Apizym testing used Treponema vincentii ATCC 35580 as a test microorganism, with enzyme activities identified that were identical to those reported previously (31).
Electron microscopy.
The morphologies of the microorganisms were examined by using transmission electron microscopy as described previously (8), except that spirochetes were taken directly from liquid cultures.
Nucleotide sequence accession numbers.
16S rRNA gene sequences for seven strains of bovine GI tract treponemes (OC1, CC2, AC3, Ru1, CHPA, Re1, and Ru2) were submitted to GenBank and given accession numbers GU566695 to GU566701, respectively. GenBank nifH accession numbers for treponeme strains Ru1 and AC3 and tlyC accession numbers for the BDD treponemes T3552B, T354A, T354B, T320A, T19, and T56 and the human treponemes Treponema medium and Treponema phagedenis were GU566702 to GU566711, respectively.
RESULTS
Spirochete isolation and growth characteristics.
Seven spirochete isolates were obtained from bovine gastrointestinal sites (Table 1). Initial isolation attempts using rifampin (5 μg/ml) and enrofloxacin (5 μg/ml) resulted in a heavy contamination of cultures, so the rifampin concentration was increased to 25 μg/ml after ensuring that representatives of all three BDD phylogroups could grow with this increased antibiotic concentration. Most strains required ∼2 repeated passages on plates before noncontaminated, single-spirochete-isolate cultures were obtained. To identify optimal serum for growth in liquid medium, isolated strains were inoculated into OTEB containing either no serum, 10% FCS, or rabbit serum (RS). Of the seven spirochetes isolated, they all exhibited equivalent growth with or without serum present in culture and on plates. The use of blood in FAA plates along with serum did not enhance growth. All spirochetes isolated took 2 to 3 days to reach stationary phase in OTEB cultures and 5 days to reach final colony size/growth on FAA plates (Table 2). Colonies were typically individual, circular, convex, and translucent to a final size of 0.5 to 2 mm, with no local hemolysis observed.
TABLE 2.
Comparisons between bovine gastrointestinal and BDD treponemes
| Strain | Isolation material | Nearest relative (% shared identity) | Growth condition (days of growth)a | Presence of: |
Reference | |||
|---|---|---|---|---|---|---|---|---|
| Hemolysis | Serum dependence | tlyC geneb | nifH geneb | |||||
| OC1 | GI tract | T. bryantii (90.0) | NSSR (2-3) | − | − | − | − | This study |
| AC3 | GI tract | T. zioleckii (89.3) | NSSR (2-3) | − | − | − | + | This study |
| CC2 | GI tract | T. bryantii (90.0) | NSSR (2-3) | − | − | − | − | This study |
| CHPA | GI tract | T. bryantii (87.7) | NSSR (2-3) | − | − | − | − | This study |
| RE1 | GI tract | T. bryantii (90.0) | NSSR (2-3) | − | − | − | − | This study |
| RU1 | GI tract | T. zioleckii (90.2) | NSSR (2-3) | − | − | − | + | This study |
| RU2 | GI tract | T. bryantii (90.0) | NSSR (2-3) | − | − | − | − | This study |
| T19 | BDD lesion | T. medium (99.6) | RS (9) | + | + | + | − | 12 |
| T56 | BDD lesion | T. medium (99.6) | RS (9) | + | + | + | − | 12 |
| T320A | BDD lesion | T. phagedenis (100) | FCS (7) | − | + | + | − | 12 |
| T354A | BDD lesion | T. phagedenis (100) | FCS (7) | − | + | + | − | 12 |
| T3552B | BDD lesion | T. denticola (95.7) | FCS (4) | + | + | + | − | 12 |
| G819CN | BDD lesion | T. denticola (95.7) | FCS (4) | + | + | + | − | 12 |
The preferred OTEB serum supplement for optimal growth is described (determined by total cell numbers). The number of days for optimal growth is shown in parentheses. NSSR, no serum supplement required.
Presence as identified by PCR using genus/eubacterium-specific degenerate primers.
16S rRNA gene analysis.
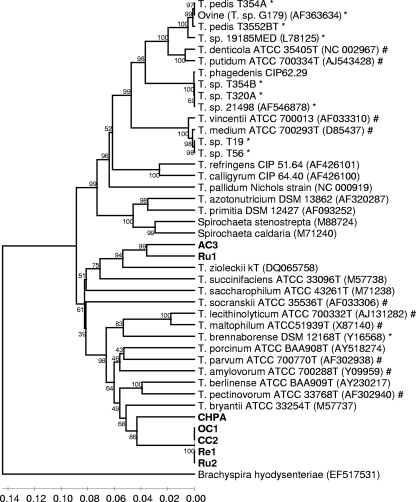
Approximately 1,320 bp of the 16S rRNA gene was sequenced for each of the seven gastrointestinal spirochetes, and sequences were initially aligned against a large number of relevant isolated 16S rRNA gene sequences. After phylogenetic tree construction, the seven isolates were separated into four distinct phylotypes within the genus Treponema. Phylotype 1 consisted of four strains (OC1, CC2, Ru2, and Re1) with identical 16S rRNA gene sequences, suggesting that they are the same species. Phylotype 1 clustered specifically with phylotype 2 (represented by one isolate, CHPA) (Fig. 1), and the two shared 91.8% 16S rRNA gene sequence identity with one another. Of the valid Treponema species, T. bryantii, a bovine rumen spirochete, was most closely related to phylotypes 1 and 2 (Fig. 1), sharing 90.0% and 87.7% 16S rRNA gene sequence identities, respectively. Phylotypes 3 (represented by strain AC3) and 4 (represented by strain Ru1) also clustered together and shared 92.9% 16S rRNA gene sequence identity with one another. These two phylotypes clustered with the sheep rumen spirochete species Treponema zioleckii kT and shared 89.3% and 90.2% 16S rRNA gene sequence identities with it, respectively. Phylogenetic inference from the alignment of treponeme 16S rRNA sequences revealed that the genus Treponema was split into two deep-rooted clusters (Fig. 1). All bovine GI tract-associated spirochetes, including those obtained in this study, lay in the same cluster, which also included several human oral treponemes. The majority of BDD-associated treponemes lay in the cluster that did not contain the bovine GI tract-associated treponemes.
FIG. 1.
Phylogenetic tree of bovine GI tract treponemes based on an alignment of 16S rRNA gene sequences for comparison with other isolated treponemes. Comparisons are over 1,320 aligned bases showing a relationship between strains isolated here (boldface type) and 16S rRNA gene sequences of relevant isolated strains. Bootstrap confidence levels are shown as percentages of nodes, and only values above 40% are shown. GenBank accession numbers are shown in parentheses next to each strain. *, previously reported 16S rRNA gene sequences from BDD lesions; #, previously reported 16S rRNA gene sequences from human oral periodontal infections.
In a second analysis, the 16S rRNA genes of the bovine GI tract spirochetes were compared to treponeme sequences from bovine samples only. This analysis included 16S rRNA clones from three previously reported metagenomic studies of the bovine rumen (11, 38, 44) and a previously reported bovine feces metagenomic study that identified nine clones representative of 5 treponeme phylotypes (29). As with the initial spirochete dendrogram, the treponeme 16S rRNA gene sequences formed two distinct groups/clusters, with one cluster consisting of all BDD treponeme sequences (with just one exception) and one consisting of all the bovine GI tract treponemes (Fig. 2). This distribution contrasted with that of human oral spirochetes, the phylotypes of which were present equally in both clusters.
FIG. 2.
Phylogenetic tree of bovine GI tract treponemes based on an alignment of 16S rRNA gene sequences for comparison with bovine treponemes. The phylogenetic tree was based on 16S rRNA gene sequence comparisons over 1,320 aligned bases showing the relationship between strains isolated here (boldface type) and other relevant bovine treponeme 16S rRNA gene sequences. Bootstrap confidence levels are shown as percentages of nodes, and only values above 40% are shown. GenBank accession numbers are shown in parentheses next to each strain or 16S rRNA gene fragment clone. *, previously reported 16S rRNA gene sequences from BDD lesions.
The phylogenetic cluster containing GI tract-associated treponemes contained many deep branch lengths, and the rumen and rectum (fecal) treponemes seemed to populate the cluster equally, with no apparent GI region subgrouping. In terms of nearest relatives in this bovine treponeme dendrogram, bovine GI tract phylotypes 1 and 2 were located closer to several bovine fecal treponeme clones than Treponema bryantii, sharing 90.1% and 87.8% 16S rRNA gene sequence identities with treponeme clones EMP R20 and EMP F10, respectively. As the ovine rumen spirochete Treponema zioleckii was now omitted, phylotypes 3 and 4 were now located closest to U.S. bovine rumen strain Treponema sp. strain CA, sharing 88.1% and 88.7% 16S rRNA gene sequence identities, respectively.
nifH analysis.
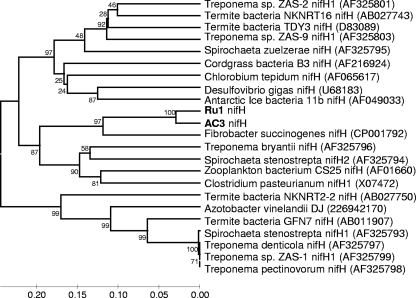
PCR-based amplification of partial nifH sequences (320 bp) was attempted with the seven GI tract treponemes isolated in this study, six BDD treponemes previously isolated in our laboratory (13), and the two Treponema species most closely related to the BDD treponemes, namely, Treponema medium and Treponema phagedenis. PCR products could be obtained only from two of the seven rumen strains (AC3 and Ru1) and none of the BDD treponemes or their nearest relatives. Approximately 320 bp of nifH was sequenced for each of the two positive isolates. A dendrogram (Fig. 3) of the GI treponeme nifH sequences aligned with spirochete nifH sequences, other nifH sequences previously reported to be highly similar to spirochetal nifH (19), and gene sequences with high BLAST scores (1) was constructed. The dendrogram shows that the nifH genes of these two strains clustered together closely with a sequence identity of 94.4% and then clustered secondarily to Fibrobacter succinogenes S85 (79.6%) and then to T. bryantii (∼74%) and Spirochaeta stenostrepta (∼73%).
FIG. 3.
Phylogenetic tree based on alignment of nifH sequences showing the relationship of bovine spirochetes isolated in this study (boldface type) with other relevant sequences. Comparisons are over ∼320 aligned bases. GenBank accession numbers are shown in parentheses next to each strain. Bootstrap confidence levels are shown as percentages of nodes, and only values above 40% are shown.
tlyC analysis.
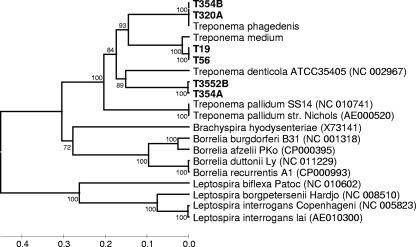
The panel of seven GI tract treponemes, six BDD treponemes, and two of the nearest BDD treponeme relatives were subjected to a PCR designed to detect a putative hemolysin gene, tlyC. PCR products of the predicted size were observed for the six BDD treponemes and for their two nearest relatives but not for any of the GI tract treponemes. Approximately 490 bp of tlyC was sequenced for the six BDD isolates and the close relatives Treponema medium and Treponema phagedenis. The tlyC gene sequences were then aligned to a number of previously reported, relevant treponeme tlyC sequences. After phylogenetic tree construction (Fig. 4), the 6 isolates could be divided into three distinct groups. The T. medium-like BDD treponeme tlyC sequences were identical and were most similar to that of T. medium, sharing 96.6% sequence identity. The tlyC sequences of the two T. phagedenis-like BDD treponemes were identical to that of T. phagedenis. The T. denticola-like BDD treponeme tlyC sequences shared 99.5% gene similarity with one another and were most similar to that of T. denticola albeit at a modest level of similarity (75.3%).
FIG. 4.
Phylogenetic tree based on alignment of tlyC sequences showing the relationship of BDD spirochetes (boldface type) with other previously reported spirochete sequences. Comparisons are over ∼490 aligned bases. GenBank accession numbers are shown in parentheses next to each strain. Bootstrap confidence levels are shown as percentages of nodes, and only values above 40% are shown.
Enzyme activities.
The enzyme activities of the bovine GI tract-associated spirochetes compared with other relevant treponemes are presented in Table 3 . Unfortunately, strains OC1, CC2 (two of the four strains belonging to phylotype 1), and AC3 (the only representative of phylotype 3) could not be revived from frozen stocks and were therefore not included in this study. The enzyme activity profiles of the two remaining representatives of phylotype 1 were indistinguishable from one another. The enzyme activity profile for each of the two remaining phylotypes tested was unique and different from patterns of previously designated Treponema species. Only β-galactosidase enzyme activity was present in all bovine GI treponemes that we tested; otherwise, there was no discernible enzyme pattern shared across these spirochetes.
TABLE 3.
Enzyme activities of bovine gastrointestinal spirochetes compared with those of other relevant treponemes
| Species, group, or phylotypea | Strain | Presence of enzyme activityb |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkaline phosphatase | C4 esterase | C8 esterase lipase | C14 lipase | Leucine arylamidase | Valine arylamidase | Cystine arylamidase | Trypsin | Chymotrypsin | Acid phosphatase | Naphtholphospho-hydrolase | α-Galactosidase | β-Galactosidase | β-Glucuronidase | α-Glucosidase | β-Glucosidase | N-Acetyl-β-glucosaminidase | α-Mannosidase | α-Fucosidase | ||
| T. brennaborensec | DSM 12168T | + | + | + | − | − | − | − | − | − | + | + | − | + | − | + | − | + | − | − |
| T. mediumd | ATCC 700293T | + | + | + | − | + | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| T. vincentiic | ATCC 35580 | − | − | − | − | + | − | − | − | − | + | + | − | + | − | − | − | + | − | − |
| T. phagedenise | Reiter | − | − | − | − | − | − | − | − | − | + | − | − | + | + | − | − | + | − | − |
| T. putidumf | ATCC 700334T | + | + | + | − | + | − | − | + | + | + | + | + | + | − | + | + | − | − | − |
| T. denticolaf | ATCC 35405T | − | + | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − |
| BDD group 1d | 5 isolates | + | + | + | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| BDD group 2d | 14 isolates | + | + | + | − | − | − | − | − | − | + | + | − | + | + | − | − | + | − | + |
| BDD group 3d | 4 isolates | − | + | + | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − |
| GI phylotype 1 | Re1 | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | − | − | − |
| GI phylotype 1 | Ru2 | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | − | − | − |
| GI phylotype 2 | CHPA | − | + | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − |
| GI phylotype 4 | Ru1 | − | − | + | − | + | − | − | − | − | − | − | − | + | − | − | + | − | − | − |
| T. berlinenseg | 7CPL208T | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − |
| T. porcinumg | 14V28T | − | + | − | − | − | − | − | − | − | + | + | − | − | − | + | − | − | − | − |
| T. socranskii subsp. socranskiic | ATCC 35536T | + | + | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − |
| T. pectinovorumc | ATCC 33768T | − | + | + | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − |
| T. amylovorumc | (HA2PT) | + | + | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | + |
| T. maltophilumc | BRT | + | + | + | − | − | − | − | − | − | + | + | + | − | − | + | − | − | − | + |
Nearest species designated relatives (according to 16S rRNA gene sequence identity) are shown above the bovine treponemes, and previously reported bovine and ovine isolates are shown below.
Determined by the Apizym system.
Apizym results were previously reported (31).
Apizym results were previously reported (13).
Apizym results were previously reported (9).
Apizym results were previously reported (43).
Apizym results were previously reported (24).
Spirochete morphology.
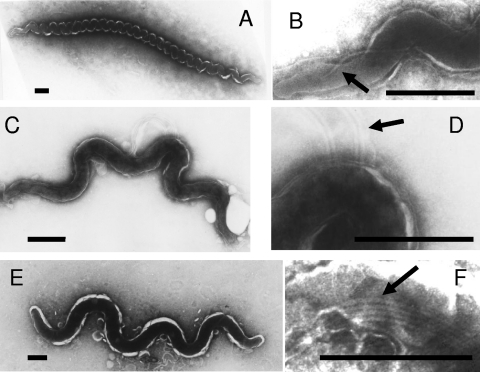
Transmission electron microscopy showed that the different bovine GI tract-associated spirochetes were different on the basis of cell morphology, although they all shared common treponemal morphological characteristics. Phylotype 1 (Ru2 and Re1) spirochetes were 7 to 10 μm long and 0.2 to 0.30 μm wide, with 16 to 28 even windings tightly coiled and one periplasmic flagellum (Fig. 5 A and B). Phylotype 2 (CHPA) spirochetes were 1 to 5 μm long and 0.15 to 0.25 μm wide, with 2 to 5 uneven coils and 4 periplasmic flagella (Fig. 5C and D). Phylotype 4 (Ru1) spirochetes had cell dimensions of 5 to 9 μm in length and 0.40 to 0.50 μm in width, with 3 to 5 even coils and 4 periplasmic flagella (Fig. 5E and F).
FIG. 5.
Electron micrographs of negatively stained cells of novel GI tract treponemes. (A and B) Re1 (phylotype 1) (A) exhibiting a single flagellum (B). (C and D) CHPA (phylotype 2) (C) exhibiting four flagella (D). (E and F) Ru1 (phylotype 4) (E) exhibiting four flagella (F). Flagella are indicated by arrows. Bars, 0.5 μm.
DISCUSSION
In this study, a total of seven spirochetes were isolated from the bovine GI tract. This is a significant advance, as there have been few previous reports of bovine GI tract spirochete isolations and none since BDD has emerged as a severe disease in cattle. As these microorganisms are very difficult to isolate and maintain in vitro, their recent identification in the bovine GI tract resulted solely from 16S rRNA gene-centered metagenomic studies of bovine rumen and feces (11, 29, 38, 44). The bovine GI tract spirochetes isolated here have 16S rRNA gene sequences that identify them as all belonging to the genus Treponema, and furthermore, they can be characterized into four phylotypes. All phylotypes have less than 97% sequence identity with the nearest treponemes, suggesting that they are representatives of new species (33), but further phenotypic characterizations are required to clarify this. The different phylotypes had different, specific Apizym profiles and were very different morphologically, which, along with their diverse 16S rRNA gene sequences (<93% similarity to each other or to the nearest treponeme sequences), is in agreement with the great physiological diversity previously reported for bovine rumen treponemes (25). All phylotypes were novel in morphology, exhibiting little similarity to previously reported bovine GI tract spirochetes, including those for which no 16S rRNA gene sequences were reported (25, 26, 35, 36, 45, 46). The spirochetes were isolated from different parts of the bovine GI tract; one phylotype was isolated from several different GI tract regions, including the omasum, colon, rumen, and rectum. Isolation from these different regions along with metagenomic data identifying treponemes as being present in cow feces (29) suggest that treponemes are not limited to the rumen of the bovine GI tract.
One reason for obtaining bovine GI tract treponemes was to compare them with the only other reported bovine treponemes: those associated with BDD lesions. BDD lesions most commonly occur on the hind feet of cattle, so it is feasible that they are introduced here from contaminated fecal material. However, we determined that BDD-associated treponemes were different from those present in the bovine GI tract. In terms of phenotype, the GI tract treponemes grew considerably faster than the BDD treponemes that we previously isolated (Table 2). In contrast to the BDD treponemes, the addition of serum or blood did not enhance the growth of the GI tract treponemes, identifying them as being serum independent, and no hemolysis was observed. These functional characteristics are differences that might be expected when host-adapted pathogens and commensals are compared.
Phylogenetic inference based on a comparison of 16S rRNA sequences from bovine GI tract and BDD-associated treponemes suggested a marked divergence between the two groups. The dendrogram formed two major clusters, which could be assigned as one cluster belonging to BDD treponemes and the other belonging to the GI tract treponemes. The division of Treponema into two deep-rooted clusters was described previously by Paster et al. (28) and has been a common feature of many previously reported treponeme dendrograms (10, 14, 24, 27, 43). Bovine GI tract-associated phylotypes were found to belong to the cluster that, with one exception, did not contain the BDD-associated phylotypes. However, although this observation suggests that divergence between clusters may underlie adaptation to different niches, it is important that treponemes associated with the human oral cavity populate both clusters of the tree (Fig. 1). The exception mentioned above, and, hence, the only BDD treponeme that lies within the cluster containing GI tract-associated species, is Treponema brennaborense (31). Although this species was isolated from a BDD lesion, several recent studies identified it as being absent or having a very low association with BDD, and it was suggested that it may be fecal contaminant and/or a secondary invader in BDD lesions (17, 22). This is in sharp contrast to the majority of treponemes present in the BDD dendrogram cluster, with many of these having been reported to have high-level associations with BDD (14, 17, 22).
Given that bovine treponemes appear to fall into two large subgroups, we attempted to try and identify markers for commensalism or virulence to differentiate these two groups. Free-living and termite gut spirochetes exhibit nitrogen fixation and have a nitrogenase gene (nifH) (19). As a nifH homologue has also been found in the GI tract-associated spirochete Treponema bryantii (19), we set out to identify the distribution of this gene in our treponeme collection. The nifH gene was identified in only two of the seven GI tract treponemes and none of the BDD treponemes. The potential absence of the nifH gene from the majority of GI treponemes makes it a poor marker for treponemes from the GI tract, while the PCR assay failing to identify nifH in the BDD treponemes coincides with previous reports that it is absent from many spirochetal pathogens (19).
In an attempt to further delineate these two large groups of bovine treponemes, we developed a degenerate PCR for tlyC, a gene originally defined as encoding a hemolysin in the spirochete Brachyspira hyodysenteriae (40). We found that tlyC was present in all three of the BDD treponeme groups and their nearest relatives, but we were unable to identify the gene in GI tract treponemes. In terms of bovine samples, this makes tlyC PCR a useful marker for BDD treponemes. As to the potential function of this gene, we have observed hemolysis for only two of the three BDD groups that we can isolate (13), yet the PCR was positive for all three of these novel groups. Interestingly, the hemolytic properties of TlyC have recently been questioned, and for Leptospira interrogans (another pathogenic spirochete), it was shown that the predicted TlyC cell surface protein has extracellular matrix (ECM) binding properties (3). Tissue attachment is an import virulence trait in spirochetes (2, 3, 5, 37), and TlyC may represent part of tissue attachment machinery which the BDD treponemes may have but which bovine GI tract treponemes do not.
On the basis of Apizym enzyme detection, we could not identify any specific trends that differentiated between the bovine GI tract and BDD treponemes. In terms of reported functional classifications, members of the BDD treponeme cluster such as human T. denticola and T. vincentii were previously reported to be asaccharolytic (18). Interestingly, T. denticola can degrade glucose using the Embden-Meyerhof pathway (maybe making it not truly “asaccharolytic”); however, it was previously reported to use this only as a minor energy source, preferentially fermenting amino acids (16). In contrast, many of the oral and rumen treponemes from the lower cluster were reported to be saccharolytic, with many being named after their sugar-fermenting activities (Fig. 1). The difference in distribution between human oral, bovine GI tract, and BDD treponemes across the phylogenetic tree may therefore be representative of host niches; e.g., the bovine GI tract presents an abundance of carbohydrates, and the bovine foot may present mostly protein, while in the human mouth, there may be both carbohydrates and proteins available. Another potential common factor for the treponemes belonging to the BDD treponeme group is serum dependence (Table 2). Using OTEB and FAA plates, the BDD treponemes that we isolated appeared to be serum dependent, whereas the GI tract treponemes did not. Interestingly, human T. denticola, T. refringens, and T. phagedenis were originally considered serum dependent (32), and while some complex media have now been produced for some of these treponemes, which do not contain serum (including T. denticola), it remains that serum enhances the growth of these microbes but does not enhance or, in fact, inhibits many of the saccharolytic treponemes in the bovine GI tract treponeme cluster (24).
Here, we have isolated and characterized several treponemes from the bovine GI tract and made comparisons with BDD treponemes. When previous GI tract treponeme identification studies are included, analyses of GI treponemes suggest that they are genotypically far more diverse than the BDD treponemes. Using phylogenetics we have identified that GI tract and BDD treponemes form their own genetic clusters and reported that saccharolytic ability and serum dependence may represent important characteristics for each of these clusters, respectively. Hence, in terms of bovine treponemes at least, serum dependence may underpin the ability to cause disease. These observations of bovine treponeme phylogeny and relationship with ecology and etiology may allow us to better understand the pathogenesis of BDD. Furthermore, this study has identified a putative surface-exposed ECM binding protein in BDD treponemes, which may be responsible for tissue attachment (3) and could be studied further as a potential vaccine candidate. The bovine GI treponemes described here are now a valuable resource for further study by those interested in commensal versus pathogenic treponemes and may provide a valuable insight into a variety of disease processes.
Acknowledgments
This work was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) CEDFAS research grant (BBE0189201).
Footnotes
Published ahead of print on 5 November 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Cameron, C. E., E. L. Brown, J. M. Kuroiwa, L. M. Schnapp, and N. L. Brouwer. 2004. Treponema pallidum fibronectin-binding proteins. J. Bacteriol. 186:7019-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, E., A. S. Barbosa, R. M. Gomez, A. M. Cianciarullo, P. Hauk, P. A. Abreu, L. C. Fiorini, M. L. Oliveira, E. C. Romero, A. P. Goncales, Z. M. Morais, S. A. Vasconcellos, and P. L. Ho. 2009. Leptospiral TlyC is an extracellular matrix-binding protein and does not present hemolysin activity. FEBS Lett. 583:1381-1385. [DOI] [PubMed] [Google Scholar]
- 3a.Cheli, R., and C. M. Mortellaro. 1974. Digital dermatitis in cattle, p. 208-213. In P. Gallarati (ed.), Proceedings of the 8th International Conference on Diseases of Cattle, Milan, Italy.
- 4.Choi, B. K., H. Nattermann, S. Grund, W. Haider, and U. B. Gobel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47:175-181. [DOI] [PubMed] [Google Scholar]
- 5.Cullen, P. A., D. A. Haake, and B. Adler. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28:291-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cwyk, W. M., and E. Canale-Parola. 1979. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch. Microbiol. 122:231-239. [DOI] [PubMed] [Google Scholar]
- 7.Demirkan, I., S. D. Carter, C. Winstanley, K. D. Bruce, N. M. McNair, M. Woodside, and C. A. Hart. 2001. Isolation and characterisation of a novel spirochaete from severe virulent ovine foot rot. J. Med. Microbiol. 50:1061-1068. [DOI] [PubMed] [Google Scholar]
- 8.Demirkan, I., H. F. Williams, A. Dhawi, S. D. Carter, C. Winstanley, K. D. Bruce, and C. A. Hart. 2006. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 101:948-955. [DOI] [PubMed] [Google Scholar]
- 9.Dettori, G., R. Grillo, P. Cattani, A. Calderaro, C. Chezzi, J. Milner, K. Truelove, and R. Sellwood. 1995. Comparative study of the enzyme activities of Borrelia burgdorferi and other non-intestinal and intestinal spirochaetes. New Microbiol. 18:13-26. [PubMed] [Google Scholar]
- 10.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, J. E., N. R. McEwan, A. J. Travis, and R. J. Wallace. 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86:263-281. [DOI] [PubMed] [Google Scholar]
- 12.Evans, N. J., J. M. Brown, I. Demirkan, R. Birtles, C. A. Hart, and S. D. Carter. 2009. In vitro susceptibility of bovine digital dermatitis associated spirochaetes to antimicrobial agents. Vet. Microbiol. 136:115-120. [DOI] [PubMed] [Google Scholar]
- 13.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, W. D. Vink, R. W. Blowey, C. A. Hart, and S. D. Carter. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet. Microbiol. 130:141-150. [DOI] [PubMed] [Google Scholar]
- 14.Evans, N. J., J. M. Brown, I. Demirkan, P. Singh, B. Getty, D. Timofte, W. D. Vink, R. D. Murray, R. W. Blowey, R. J. Birtles, C. A. Hart, and S. D. Carter. 2009. The association of unique, isolated treponemes with bovine digital dermatitis lesions. J. Clin. Microbiol. 47:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graber, J. R., J. R. Leadbetter, and J. A. Breznak. 2004. Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the first spirochetes isolated from termite guts. Appl. Environ. Microbiol. 70:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hespell, R. B., and E. Canale-Parola. 1971. Amino acid and glucose fermentation by Treponema denticola. Arch. Mikrobiol. 78:234-251. [DOI] [PubMed] [Google Scholar]
- 17.Klitgaard, K., M. Boye, N. Capion, and T. K. Jensen. 2008. Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rRNA gene analysis and fluorescence in situ hybridization. J. Clin. Microbiol. 46:3012-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koseki, T., I. Ishikawa, M. Umeda, and Y. Benno. 1995. Characterization of oral treponemes isolated from human periodontal pockets. Oral Microbiol. Immunol. 10:271-277. [DOI] [PubMed] [Google Scholar]
- 19.Lilburn, T. G., K. S. Kim, N. E. Ostrom, K. R. Byzek, J. R. Leadbetter, and J. A. Breznak. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495-2498. [DOI] [PubMed] [Google Scholar]
- 20.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 21.Naylor, R. D., P. K. Martin, J. R. Jones, and M. C. Burnell. 1998. Isolation of spirochaetes from an incident of severe virulent ovine footrot. Vet. Rec. 143:690-691. [PubMed] [Google Scholar]
- 22.Nordhoff, M., A. Moter, K. Schrank, and L. H. Wieler. 2008. High prevalence of treponemes in bovine digital dermatitis—a molecular epidemiology. Vet. Microbiol. 131:293-300. [DOI] [PubMed] [Google Scholar]
- 23.Nordhoff, M., D. Taras, M. Macha, K. Tedin, H. J. Busse, and L. H. Wieler. 2005. Treponema berlinense sp. nov. and Treponema porcinum sp. nov., novel spirochaetes isolated from porcine faeces. Int. J. Syst. Evol. Microbiol. 55:1675-1680. [DOI] [PubMed] [Google Scholar]
- 24.Norris, S. J., B. J. Paster, A. Moter, and U. B. Göbel. 2006. The genus Treponema, p. 211-234. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 6. Springer Science, New York, NY. [Google Scholar]
- 25.Paster, B. J., and E. Canale-Parola. 1982. Physiological diversity of rumen spirochetes. Appl. Environ. Microbiol. 43:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paster, B. J., and E. Canale-Parola. 1985. Treponema saccharophilum sp. nov., a large pectinolytic spirochete from the bovine rumen. Appl. Environ. Microbiol. 50:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paster, B. J., and F. E. Dewhirst. 2006. The phylogenetic diversity of the genus Treponema, p. 10-18. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
- 28.Paster, B. J., F. E. Dewhirst, W. G. Weisburg, L. A. Tordoff, G. J. Fraser, R. B. Hespell, T. B. Stanton, L. Zablen, L. Mandelco, and C. R. Woese. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton, T. G., A. J. Scupham, S. M. Bearson, and S. A. Carlson. 2009. Characterization of fecal microbiota from a Salmonella endemic cattle herd as determined by oligonucleotide fingerprinting of rDNA genes. Vet. Microbiol. 136:285-292. [DOI] [PubMed] [Google Scholar]
- 30.Radolf, J. D., K. R. O. Hazlett, and S. A. Lukehart. 2006. Pathogenesis of syphilis, p. 197-236. In J. D. Radolf and S. A. Lukehart (ed.), Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
- 31.Schrank, K., B. K. Choi, S. Grund, A. Moter, K. Heuner, H. Nattermann, and U. B. Gobel. 1999. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 49(Pt. 1):43-50. [DOI] [PubMed] [Google Scholar]
- 32.Smirbert, R. M. 1984. Genus III Treponema, p. 49-57. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 33.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 35.Stanton, T. B., and E. Canale-Parola. 1979. Enumeration and selective isolation of rumen spirochetes. Appl. Environ. Microbiol. 38:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanton, T. B., and E. Canale-Parola. 1980. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 127:145-156. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2:e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tajima, K., R. I. Aminov, T. Nagamine, K. Ogata, M. Nakamura, H. Matsui, and Y. Benno. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29:159-169. [Google Scholar]
- 39.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 40.ter Huurne, A. A., S. Muir, M. van Houten, B. A. van der Zeijst, W. Gaastra, and J. G. Kusters. 1994. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb. Pathog. 16:269-282. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trott, D. J., M. R. Moeller, R. L. Zuerner, J. P. Goff, W. R. Waters, D. P. Alt, R. L. Walker, and M. J. Wannemuehler. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J. Clin. Microbiol. 41:2522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyss, C., A. Moter, B. K. Choi, F. E. Dewhirst, Y. Xue, P. Schupbach, U. B. Gobel, B. J. Paster, and B. Guggenheim. 2004. Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int. J. Syst. Evol. Microbiol. 54:1117-1122. [DOI] [PubMed] [Google Scholar]
- 44.Yang, S., S. Ma, J. Chen, H. Mao, Y. He, D. Xi, L. Yang, T. He, and W. Deng. 2010. Bacterial diversity in the rumen of Gayals (Bos frontalis), Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences. Mol. Biol. Rep. 37:1309-1317. [DOI] [PubMed] [Google Scholar]
- 45.Ziolecki, A. 1979. Isolation and characterization of large treponemes from the bovine rumen. Appl. Environ. Microbiol. 37:131-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziolecki, A., and M. Wojciechowicz. 1980. Small pectinolytic spirochetes from the rumen. Appl. Environ. Microbiol. 39:919-922. [DOI] [PMC free article] [PubMed] [Google Scholar]