Abstract
The bovine pathogen Streptococcus uberis was assessed for biofilm growth. The transition from planktonic to biofilm growth in strain 0140J correlated with an upregulation of several gene products that have been shown to be important for pathogenesis, including a glutamine ABC transporter (SUB1152) and a lactoferrin binding protein (gene lbp; protein SUB0145).
Approximately 33% of bovine mastitis cases within the United Kingdom are attributable to Streptococcus uberis, and infections can be both persistent and resistant to antimicrobial treatment (10, 17). In this study, we assess the ability of S. uberis to form biofilms in vitro. We also analyze the S. uberis clinical isolate 0140J under planktonic and biofilm growth conditions in vitro by using gel electrophoresis liquid chromatography (GeLC)-tandem mass spectrometry (MS/MS) and demonstrate that the changes that 0140J undergoes in the transition from planktonic to biofilm growth include factors strongly implicated in pathogenesis.
Biofilm formation by S. uberis isolates.
To establish biofilm formation among different strains of S. uberis, we analyzed S. uberis isolates 0140J (a strain of high virulence for the lactating bovine mammary gland), EF20, Y38, C197C, C221, and C198 (a strain obtained from a healthy bovine mammary gland and not associated with pathogenesis) (see Table S1 in the supplemental material) in TSBM medium {tryptic soy broth without dextrose [Difco Labs], supplemented with 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.6, and 20% [vol/vol] sterile skim milk}, using a microtiter plate biofilm formation assay (4). All isolates formed biofilms in vitro, with C198 forming the smallest biomass (Fig. 1).
FIG. 1.
Biofilm formation by S. uberis clinical isolates. Quantitative biofilm formation of S. uberis isolates after 24 h in TSBM. Strains 0140J, EF20, Y38, C197C, and C221 were isolated from diseased cows and form significantly more biofilm biomass than strain C198, isolated from a healthy cow. The asterisk denotes the statistical significance of biofilm formation between 0140J and C198 (P ≤ 0.05).
A previously elucidated difference between strain C198 and all other strains tested is that C198 lacks the hasAB gene cluster and is, as a consequence, acapsular (J. Leigh, personal communication). In support of capsule production being involved in 0140J biofilm growth in vitro, S. uberis capsule size has been shown to increase in the presence of bovine milk-related constituents (12) and the addition of 20% (vol/vol) bovine milk to medium was found to promote 0140J biofilm formation in vitro (data not shown).
Differential proteomic analysis of Streptococcus uberis 0140J biofilm development.
To determine factors involved in S. uberis biofilm growth and development, we compared the proteomes of 0140J planktonic cells (8 h) with those of biofilm cells (8 h), as well as comparing 8-h biofilm cells with 36-h biofilm cells.
Biofilms were grown in tube reactors (1) that were initially inoculated with S. uberis strain 0140J in TSBM for 1 h under no-flow conditions. Both biofilm and planktonic cells were grown at 37°C in TSB supplemented with 10% (vol/vol) lactose (46 g/liter). Protein extraction was carried out essentially as previously described (6). Protein samples (30 ng) were loaded onto 10% acrylamide gels (Protean III system; Bio-Rad) and stained (GelCode blue; Pierce) according to manufacturer's instructions. Gel lanes were cut into 13 to 15 equal-size gel slices, and each slice was processed as previously described (19). Capsular supernatant fractions were directly processed (13). Peptides were resuspended and separated (3, 18). Eluted peptides were analyzed by tandem mass spectrometry (LTQ ion trap mass/LTQ Orbitrap FT-MS; ThermoScientific), and spectra (m/z range, 350 to 1,800) were searched against the annotated S. uberis 0140J genome (23) (SEQUEST, BioWorks v3.3; Thermo Scientific).
The transition from 8-h planktonic growth to 8-h biofilm growth in 0140J correlated with an increase in proteins predicted to be involved in adhesion (SUB0837 and SUB1152), glutamine transport (SUB1152), internalization (SUB1212), and sugar metabolism (SUB0135, SUB0235, SUB0161, and SUB0750) (see Tables S2 and S3 in the supplemental material). This is of interest with regard to pathogenesis of infection as such processes have been implicated in virulence and/or biofilm growth in other streptococcal species (7, 8, 16). For example, a glutamine transport glnP mutant of Streptococcus pneumoniae showed decreased adherence to human pharyngeal epithelial cells (9).
The development of 0140J biofilms correlated with the downregulation of a relatively large number of proteins (i.e., 34) involved in metabolism, cell wall synthesis, and cell division, suggestive of a decrease in growth rate/metabolic activity as a biofilm ages (from 8 to 36 h) (see Tables S4 and S5 in the supplemental material).
Although GeLC-MS/MS alone is not a quantitative technique, it was noticeable that some proteins, although expressed in planktonic and biofilm cells, showed marked differences in peptide coverage between these growth types, which may indicate a quantitative difference in abundance of the protein. Quantitative reverse transcription (qRT-PCR) was carried out on these candidates to further assess and quantify putative differential expression at the transcriptional level.
Quantitative verification of proteomic analyses during 0140J biofilm growth.
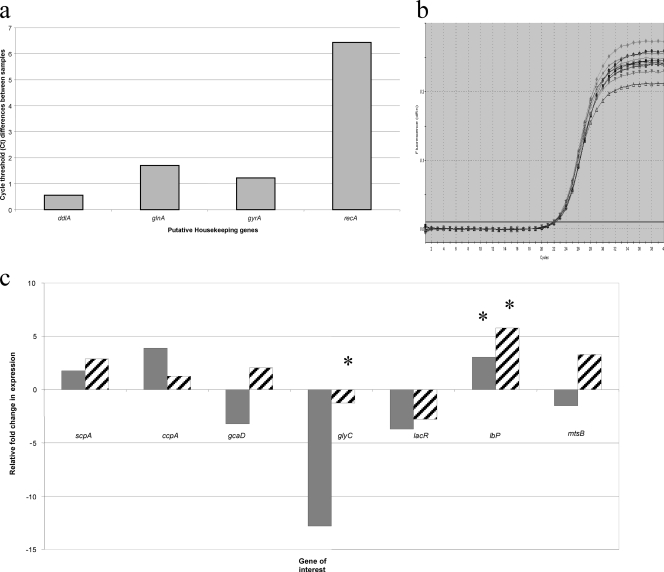
RNA isolation and cDNA synthesis were carried out as previously described (21) (see Table S1 in the supplemental material). qRT-PCR was normalized to ddlA (gene 7391862; SUB1257) due to ddlA showing the least variation in expression across all samples (Fig. 2 a and b). Changes in transcriptional expression between 8-h planktonic cells and 8-h biofilm cells as well as 8-h biofilm and 36-h biofilm cells were calculated by the comparative threshold cycle (ΔΔCT) method (15, 21) and plotted as the fold change in mRNA expression levels.
FIG. 2.
Transcriptional analysis in 0140J planktonic and biofilm cells. (a) Difference in CT values across all planktonic and biofilm cDNAs from reference gene candidates ddlA, glnA, gyrA, and recA. (b) Example of raw qRT-PCR data showing amplification curves of ddlA in 8-h planktonic, 8-h biofilm, and 36-h biofilm cDNA. (c) Relative fold change in expression of scpA, ccpA, gcaD, glyC, lacR, lbp, and mtsB in 8-h biofilm cells. The gray bars represent expression in early biofilm cells, in comparison to planktonic cells. The striped bars represent expression in early biofilm cells, in comparison to late biofilm cells. glyC and lbp show significant fold change between 8 h of planktonic growth and 8 h of biofilm growth (>9.06- and 2.72-fold change, respectively). lbp shows the only significant fold change in expression between 8-h and 36-h biofilm growth (>2.96-fold change). Asterisks denote statistical significance in difference in expression, as normalized by the reference gene ddlA (P ≤ 0.05).
The transition from 8-h planktonic to 8-h biofilm growth was accompanied by a significant increase in lbp (SUB0145) expression (>2.7-fold) (Fig. 2c) and decrease in glyC (>9-fold). Mutation of SUB0145 has recently been shown to impair virulence for the bovine lactating gland (11). In addition to iron, lactoferrin (Lf) is also capable of binding other divalent metal ions (such as manganese, which has been shown to be involved in S. uberis virulence via the mtu operon [20]). Intriguingly, the bovine Lf concentration is greatest during the dry period, which is also the period in which the highest recorded rates of S. uberis infections occur (22). However, lbp expression significantly decreased (2.9-fold) in 36-h biofilms in comparison to 8-h biofilms (Fig. 2c). As glyC has been shown to be an osmoprotectant (14), our data suggest that 8-h biofilm cells are not under greater osmotic stress than their planktonic counterparts. However, it is interesting that a >9-fold decrease in mRNA levels of glyC in 8-h biofilms, in comparison to 8-h planktonic cells, still resulted in GlyC detection by GeLC-MS/MS.
In conclusion, we present evidence of S. uberis biofilm formation in which clinical isolates had greater biofilm biomass than a strain obtained from a healthy cow. Differential expression analysis at the translational level during 0140J biofilm growth suggest that glutamine transport, adherence interactions, and sugar metabolism are important processes in early biofilm growth, while at the transcriptional level, an increase in mRNA levels of lbp was apparent.
Supplementary Material
Acknowledgments
We thank Terence Field (IAH, United Kingdom) for the aseptic collection of sterile bovine milk from uninfected cows, Peter Splatt (University of Exeter, United Kingdom) for valuable assistance with biofilm imaging, and Deborah Taylor (University of Sydney, Australia) for advice with regard to qRT-PCR.
This work was funded by BBSRC grants BBS/S/P/2003/10459 and 85/S12893.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Blondeau, F., B. Ritter, P. D. Allaire, S. Wasiak, M. Girard, N. K. Hussain, A. Angers, V. Legendre-Guillemin, L. Roy, D. Boismenu, R. E. Kearney, A. W. Bell, J. J. Bergeron, and P. S. McPherson. 2004. Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U. S. A. 101:3833-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Hill, A. W., J. M. Finch, T. R. Field, and J. A. Leigh. 1994. Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow. FEMS Immunol. Med. Microbiol. 8:109-117. [DOI] [PubMed] [Google Scholar]
- 7.Kiliç, A. O., L. Tao, Y. Zhang, Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J. Bacteriol. 186:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes sigmaB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 10.Kossaibati, M. A., M. Hovi, and R. J. Esslemont. 1998. Incidence of clinical mastitis in dairy herds in England. Vet. Rec. 143:649-653. [DOI] [PubMed] [Google Scholar]
- 11.Leigh, J. A., S. A. Egan, P. N. Ward, T. R. Field, and T. J. Coffey. 2010. Sortase anchored proteins of Streptococcus uberis play major roles in the pathogenesis of bovine mastitis in dairy cattle. Vet. Res. 9-10:41-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews, K. R., B. M. Jayarao, A. J. Guidry, E. F. Erbe, W. P. Wergin, and S. P. Oliver. 1994. Encapsulation of Streptococcus uberis: influence of storage and cultural conditions. Vet. Microbiol. 39:361-367. [DOI] [PubMed] [Google Scholar]
- 13.Meza, J. E., C. A. Miller, and S. M. Fischer. 2004. Improved tryptic digestion of proteins using 2,2,2-trifluoroethanol (TFE), poster ABRF (2004). Excellence Microfluidics. Association of Biomolecular Resource Facilities, Agilent Technologies, Santa Clara, CA.
- 14.Pichereau, V., S. Bourot, S. Flahaut, C. Blanco, Y. Auffray, and T. Bernard. 1999. The osmoprotectant glycine betaine inhibits salt-induced cross-tolerance towards lethal treatment in Enterococcus faecalis. Microbiology 145:427-435. [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pullinger, G. D., T. J. Coffey, M. C. Maiden, and J. A. Leigh. 2007. Multilocus-sequence typing analysis reveals similar populations of Streptococcus uberis are responsible for bovine intramammary infections of short and long duration. Vet. Microbiol. 119:194-204. [DOI] [PubMed] [Google Scholar]
- 18.Qian, W. J., T. Liu, M. E. Monroe, E. F. Strittmatter, J. M. Jacobs, L. J. Kangas, K. Petritis, D. G. Camp II, and R. D. Smith. 2005. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4:53-62. [DOI] [PubMed] [Google Scholar]
- 19.Schirle, M., M. A. Heurtier, and B. Kuster. 2003. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 2:1297-1305. [DOI] [PubMed] [Google Scholar]
- 20.Smith, A. J., P. N. Ward, T. R. Field, C. L. Jones, R. A. Lincoln, and J. A. Leigh. 2003. MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 71:4842-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor, D. L., P. N. Ward, C. D. Rapier, J. A. Leigh, and L. D. Bowler. 2003. Identification of a differentially expressed oligopeptide binding protein (OppA2) in Streptococcus uberis by representational difference analysis of cDNA. J. Bacteriol. 185:5210-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vangroenweghe, F., I. Lamote, and C. Burvenich. 2005. Physiology of the periparturient period and its relation to severity of clinical mastitis. Domest. Anim. Endocrinol. 29:283-293. [DOI] [PubMed] [Google Scholar]
- 23.Ward, P. N., M. T. Holden, J. A. Leigh, N. Lennard, A. Bignell, A. Barron, L. Clark, M. A. Quail, J. Woodward, B. G. Barrell, S. A. Egan, T. R. Field, D. Maskell, M. Kehoe, C. G. Dowson, N. Chanter, A. M. Whatmore, S. D. Bentley, and J. Parkhill. 2009. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Microbiol. 10:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.