Abstract
Xanthomonas arboricola pv. pruni, the causal agent of bacterial spot disease of stone fruit, is considered a quarantine organism by the European Union and the European and Mediterranean Plant Protection Organization (EPPO). The bacterium can undergo an epiphytic phase and/or be latent and can be transmitted by plant material, but currently, only visual inspections are used to certify plants as being X. arboricola pv. pruni free. A novel and highly sensitive real-time TaqMan PCR detection protocol was designed based on a sequence of a gene for a putative protein related to an ABC transporter ATP-binding system in X. arboricola pv. pruni. Pathogen detection can be completed within a few hours with a sensitivity of 102 CFU ml−1, thus surpassing the sensitivity of the existing conventional PCR. Specificity was assessed for X. arboricola pv. pruni strains from different origins as well as for closely related Xanthomonas species, non-Xanthomonas species, saprophytic bacteria, and healthy Prunus samples. The efficiency of the developed protocol was evaluated with field samples of 14 Prunus species and rootstocks. For symptomatic leaf samples, the protocol was very efficient even when washed tissues of the leaves were directly amplified without any previous DNA extraction. For samples of 117 asymptomatic leaves and 285 buds, the protocol was more efficient after a simple DNA extraction, and X. arboricola pv. pruni was detected in 9.4% and 9.1% of the 402 samples analyzed, respectively, demonstrating its frequent epiphytic or endophytic phase. This newly developed real-time PCR protocol can be used as a quantitative assay, offers a reliable and sensitive test for X. arboricola pv. pruni, and is suitable as a screening test for symptomatic as well as asymptomatic plant material.
Xanthomonas arboricola pv. pruni (31) (synonym, Xanthomonas campestris pv. pruni [Smith]) is a Gram-negative plant-pathogenic bacterium that causes bacterial spot disease of stone fruits. X. arboricola pv. pruni has been reported to affect a wide range of Prunus species, such as plum, nectarine, peach, apricot, cherry, almond, and ornamental species (19, 26, 32). The disease was first described for Japanese plum in North America in 1903 (28), and since then, it has been reported to occur in many of the major stone-fruit-producing areas of the world (3, 4). Symptoms occur on leaves, fruits, and twigs, ranging from necrotic angular lesions on leaves and sunken lesions on fruits to cankers on twigs. X. arboricola pv. pruni can be very damaging when severe infections occur on highly susceptible cultivars (27).
International trade has led to the dissemination of X. arboricola pv. pruni through contaminated material used for propagation (11). Moreover, the bacterium overwinters in buds and leaf scars, which act as efficient sources of primary inocula for spring infections (34). Because of its negative economic impact, X. arboricola pv. pruni is considered a quarantine organism by European Union phytosanitary legislation (see reference 1 and amendments therein) and by the European and Mediterranean Plant Protection Organization (EPPO) (2).
As no effective chemical control is available, the introduction and dissemination of X. arboricola pv. pruni should be avoided by eliminating contaminated plant material from nurseries and plantations. Effective quarantine measures require rapid and highly sensitive methods to detect X. arboricola pv. pruni in propagative material or new reservoirs. Moreover, the information provided by such methods could reveal new potential sources of X. arboricola pv. pruni inocula.
Currently, only visual inspections looking for symptoms are performed to certify plants as being X. arboricola pv. pruni free in stone fruit nurseries. In order to diagnose bacterial spot disease, laborious and time-consuming methods are advised, based on bacterial isolation followed by identification through biochemical tests, protein profiling (SDS-PAGE), fatty acid methyl-ester (FAME) profiling, immunofluorescence (IF), repetitive-sequence-based PCR (REP-PCR) analysis, and pathogenicity confirmation testing (3). An important improvement was the development of a conventional PCR protocol for the specific detection of a 943-bp DNA fragment of a gene sequence for a putative protein related to an ABC transporter ATP-binding system in X. arboricola pv. pruni (18). However, although this protocol offers a specific approach to diagnose the pathogen in symptomatic plants, it is not sensitive enough to detect X. arboricola pv. pruni in asymptomatic plants. In this study, one such previously reported sequence (18) was targeted to develop a specific and sensitive real-time PCR method to detect X. arboricola pv. pruni in naturally infected symptomatic or asymptomatic samples.
MATERIALS AND METHODS
Bacterial strains.
Bacteria utilized in this study are listed in Table 1. Strains of X. arboricola pv. pruni were grown on YPGA medium (25) (5 g of yeast extract [Difco], 5 g of bacteriological peptone [Difco], 10 g of glucose, 20 g of agar, and distilled water to 1 liter [pH 7.0 to 7.2]) for 3 to 4 days at 25°C. Other bacteria were grown on King's B medium (12) at 25°C.
TABLE 1.
Bacterial strains tested
| Strain(s)a | Host | Country of isolation | Sourceb |
|---|---|---|---|
| Agrobacterium tumefaciens C58 | Prunus avium | United States | CFBP |
| Agrobacterium vitis IVIA 339-26 | Vitis vinifera | Spain | IVIA |
| Brenneria quercina IVIA 2389-1 | Quercus sp. | Spain | IVIA |
| Clavibacter michiganensis subsp. michiganensis IVIA 2869 | Lycopersicon esculentum | Spain | IVIA |
| Clavibacter michiganensis subsp. sepedonicus NCPPB 2140 | Solanum tuberosum | NCPPB | |
| Erwinia amylovora | |||
| CFBP 1232T | Pyrus communis | United Kingdom | CFBP |
| CFBP 1430 | Crataegus oxyacantha | France | CFBP |
| CFBP 2150 | Rubus sp. | United States | CFBP |
| CFBP 2301 | Pyracantha sp. | France | CFBP |
| CFBP 2585 | Sorbus sp. | Ireland | CFBP |
| Erwinia billingiae | |||
| NCPPB 661T | Pyrus communis | United Kingdom | NCPPB |
| NCPPB 1261 | Malus sylvestris | United Kingdom | NCPPB |
| Erwinia piriflorinigrans CFBP 5881, CFBP 5882, CFBP 5883, CFBP 5884, CFBP 5885, CFBP 5586, CFBP 5587, CFBP 5888T | Pyrus communis | Spain | CFBP |
| Erwinia pyrifoliae CFBP 4171, CFBP 4172T, CFBP 4173, CFBP 4174 | Pyrus pyrifolia | South Korea | CFBP |
| Erwinia sp. strains CFBP 4243 and CFBP 4244 | Pyrus pyrifolia | South Korea | CFBP |
| Erwinia tasmaniensis | |||
| NCPPB 4357T | Malus domestica | Australia | NCPPB |
| Et4/99 | Malus domestica | Australia | |
| NCPPB 4358 | Pyrus communis | Australia | NCPPB |
| Pseudomonas corrugata IVIA 1765 | Lycopersicon esculentum | Spain | IVIA |
| Pseudomonas savastanoi pv. savastanoi ITM 317 | Olea europaea | Italy | ITM |
| Pseudomonas syringae pv. mori IVIA 2488-1 | Morus sp. | Spain | IVIA |
| Pseudomonas syringae pv. syringae IVIA 2716 | Prunus persica | Spain | IVIA |
| Pseudomonas syringae pv. tomato IVIA 1733-3 | Lycopersicon esculentum | Spain | IVIA |
| Xanthomonas arboricola pv. corylina | |||
| CFBP 1846 | Corylus avellana | France | CFBP |
| RIPF X08, RIPF X10, RIPF X18, RIPF X23 | Corylus avellana | Poland | RIPF |
| Xanthomonas arboricola pv. fragariae CFBP 6771* | Fragaria x ananassa | Italy | CFBP |
| Xanthomonas arboricola pv. juglandis | |||
| IVIA 1317-1a | Juglans regia | Spain | |
| RIPF X04, RIPF X05, RIPF X06 | Juglans sp. | Poland | RIPF |
| Xanthomonas arboricola pv. populi CFBP 3123* | Populus x canadensis | Netherlands | CFBP |
| Xanthomonas arboricola pv. pruni | |||
| CFBP 2535* | Prunus salicina | New Zealand | CFBP |
| CFBP 411 | Unknown | United States | CFBP |
| CFBP 1311 | Unknown | Canada | CFBP |
| CFBP 3917 | Prunus persica | Italy | CFBP |
| CFBP 5722 | Prunus persica | Brazil | CFBP |
| CFBP 5231 | Prunus sp. | Argentina | CFBP |
| ISPaVe B4, ISPaVe B6 | Prunus salicina | Italy | M. Scortichini |
| Xcp 2, Xcp 4 | Prunus salicina | South Africa | L. Mansvelt |
| IVIAc | Prunus spp. | Spain | IVIA (this study) |
| CITAc | Prunus spp. | Spain | CITA (this study) |
| Xanthomonas citri subsp. citri 306 | Citrus sinensis | Brazil | IAPAR |
| Pantoea agglomerans IVIA FSO55 | IVIA | ||
| Pseudomonas fluorescens IVIA 2521-1 | Spain | IVIA |
T, type strains; *, pathotype strains.
CFBP, Collection Française de Bactéries Phytopathogènes, Angers, France; IVIA, Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain; NCPPB, National Collection of Plant Pathogenic Bacteria, Sand Hutton, York, United Kingdom; ITM, Istituto Tossine e Micotossine da Parassiti Vegetali, CNR, Bari, Italy; RIPF, Research Institute of Pomology and Floriculture, Skierniewice, Poland; CITA, Centro de Investigación y Tecnología Agroalimentaria de Aragón, Zaragoza, Spain; IAPAR, Instituto Agronômico do Paraná, Londrina, Brazil.
One hundred forty-nine other Spanish X. arboricola pv. pruni strains from Prunus amygdalus, P. armeniaca, P. persica, and P. salicina cultivars and Santa Lucia SL-64 (P. mahaleb) and Barrier (P. persica × P. davidiana) rootstocks were tested for specificity evaluation.
DNA extraction.
DNA from pure bacterial cultures was obtained by heat treatment (96°C for 10 min) or by a simple extraction method previously described (13). DNA concentration and purity were determined by using an ND-1000 spectrophotometer (NanoDrop; Thermo Fisher Scientific), and the preparation was used for PCR or stored at −20°C.
DNA from plant-bacterium mixtures was obtained through different procedures: (i) heat treatment (96°C for 10 min), (ii) a simple extraction method (13), (iii) cetyltrimethylammonium bromide (CTAB) extraction (8), and (iv) a DNeasy Plant minikit (Qiagen), according to the manufacturer's instructions.
Primers and probe design for real-time PCR.
Primers and probes were designed by using ABI PRISM Primer Express software (version 2; Applied Biosystems). Three sets of primers were initially designed from three different regions of a gene encoding a putative protein in X. arboricola pv. pruni with similarity to an ABC transporter ATP-binding system previously reported (18) (see Fig. S1 in the supplemental material). The new primers and the probe, as well as PCR product sequences, were compared to sequences available in databases by BLAST (http://www.ncbi.nlm.nih.gov/BLAST) in order to evaluate the theoretical X. arboricola pv. pruni specificity.
Conventional and real-time PCR conditions.
Conventional PCR was performed as previously described (18). Briefly, DNA templates (5-μl samples) were included in a 25-μl reaction mixture volume containing 0.2 mM (each) deoxynucleotide triphosphates (dNTPs) (Boehringer-Mannheim), 1.2 U of Tth polymerase (Biotools), 1 μM primers, 2 mM MgCl2, 4% dimethyl sulfoxide, 5% glycerol in 10 mM Tris-HCl (pH 8.0), and 50 mM KCl. Amplifications were performed with a thermal cycler (Perkin-Elmer 9600) programmed for one cycle of 4 min at 95°C, followed by 30 cycles of 1 min at 92°C, 1 min at 53°C, and 2 min at 72°C and a final extension period of 5 min at 72°C. After PCR amplification, DNA samples were electrophoresed in 1% agarose gels in 1× TBE buffer (8.9 mM Tris, 0.25 mM Na2EDTA, 0.89 mM boric acid [pH 8.3]), stained with ethidium bromide, and photographed.
Three sets of primers designed for real-time PCR were initially assayed by using Sybr green master mix (QuantiMix Easy SYG kit; Biotools). In order to optimize reaction conditions, different concentrations of primers (0.2 μM, 0.4 μM, and 1 μM) were tested. For TaqMan PCRs, Sybr green was replaced by master mix (QuantiMix Easy Probes kit; Biotools), and different concentrations (150, 200, and 250 nM) of probe (Applied Biosystems) were tested. In both cases, amplifications were performed in a 25-μl volume containing 12.5 μl master mix and 2.5 μl of sample. Real-time PCR amplifications were conducted with a Smartcycler (Cepheid Inc.) and consisted of an initial denaturation step at 95°C for 5 min followed by 45 cycles, each one consisting of 1 min at 95°C and 1 min at 59°C.
Specificity of PCR assays.
The specificity of the real-time PCR protocol was assessed with heat-treated cultures (described above) of 159 strains of X. arboricola pv. pruni from different geographical origins, 12 bacterial strains representative of other closely related Xanthomonas species, 34 non-Xanthomonas phytopathogenic strains, and two strains of saprophytic bacteria (Table 1). Purified DNA from Prunus sp. samples was also tested to discard false-positive results from healthy material. All PCRs included X. arboricola pv. pruni strain ISPaVe B4 as a positive control and master mix only as a negative control. Each bacterial strain was assayed twice.
Sensitivity and efficiency of the detection method.
Real-time PCRs were performed by using (i) pure cultures of X. arboricola pv. pruni after heat treatment or simple DNA extraction (13) and (ii) heat-treated samples or DNA extracts from washed or comminuted Prunus sp. leaves spiked with X. arboricola pv. pruni suspensions at different concentrations.
To evaluate sensitivity in pure cultures, a 10-fold dilution series was prepared from strain ISPaVe B4 grown for 72 h at a concentration range from 107 to 10 CFU ml−1. The bacterial concentration was confirmed by spectrophotometry (Thermo Spectronic) and plate counting on YPGA medium. To evaluate sensitivity and efficiency in plant material, 1 g [fresh weight] of leaf tissue was taken from peach [Prunus persica (L.) Batsch cv. Catherine], almond [Prunus amygdalus Batsch, syn. P. dulcis (Miller) D. A. Webb cv. Guara], peach × almond hybrid GF-677 (P. persica × P. amygdalus) rootstock, or Japanese plum (P. salicina Lindley cv. Golden Japan); either washed or slightly comminuted with a pestle in 15 ml of sterile distilled water; and incubated at room temperature for 15 or 5 min, respectively. One hundred ten microliters of bacterial suspension from 107 to 10 CFU ml−1 was added to 990-μl aliquots of each washed or comminuted leaf preparation. Thus, each spiked sample contained decreasing amounts of bacteria, ranging from 106 to 1 CFU ml−1 (final concentrations). Real-time PCR was performed on all 10-fold dilution series of pure cultures and spiked samples to determine the sensitivity and efficiency of the method. In all assays, appropriate negative controls containing no template DNA were subjected to the same procedure; they included heat-treated samples and DNA extracts from nonspiked washed or comminuted leaf samples as well as master-mix-only samples. Strain ISPaVe B4 was included as a positive control for all reactions. Duplicates of controls and samples were subjected simultaneously to PCR analysis. Three independent repetitions of all sensitivity experiments were carried out, and in all of them, duplicates of each sample were run. For the purpose of this study, the limit of detection in each matrix was defined as the lowest target amount giving positive results in at least three of the six total reactions performed in the three independent repetitions. Serial dilutions of the washed and comminuted tissues of those spiked samples containing 106, 103, and 102 CFU ml−1 (final concentrations) of X. arboricola pv. pruni were plated onto YPGA medium with 250 mg liter−1 cycloheximide (Sigma-Aldrich) to confirm the presence of the bacterium.
Linear regression curves, plotting the threshold cycles (CTs) of each reaction against the logarithmic values of X. arboricola pv. pruni DNA concentrations, were constructed. The determination coefficient of each curve (R2) was calculated for each of the DNA concentrations extracted from each bacterial dilution series obtained as described above. The slope of the curves (k) was used to determine the average amplification efficiency (E) with the equation E = 10[−1/k], where E = 2 corresponds to 100% efficiency (23). Each regression curve was constructed from at least two independent serial dilutions and two PCRs for each bacterial concentration per series. Regression lines were statistically compared to determine significant differences among slopes and intercepts to infer efficiency and sensitivity differences in PCRs from different materials and the two protocols described above. In addition, the average efficiency for each extraction/plant material combination was calculated and subjected to analysis of variance, and the means were separated by a least significant difference (LSD) test for each comparison (P < 0.05). All statistical analyses were performed by using Statgraphics Plus for Windows 4.1 (Statistical Graphics).
Real-time PCR analyses of leaf and bud samples from nurseries and orchards.
The real-time PCR assay was evaluated for X. arboricola pv. pruni detection on symptomatic and asymptomatic leaves and dormant buds of Prunus spp. collected from nurseries and orchards. Five peach leaf samples of Prunus persica cv. Baby Gold 5 (M1 to M5) showing bacterial spot disease symptoms were collected from orchards. Each sample (approximately 1 g [fresh weight] of leaf tissue) was either washed or comminuted in 15 ml of sterile distilled water and incubated as indicated above. Serial dilutions of comminuted tissue were also assayed. Amplifications were repeated twice. Aliquots of these samples were plated onto YPGA medium with 250 mg liter−1 cycloheximide and incubated at 25°C for 3 to 4 days. Alternatively, DNA extraction from comminuted tissue was performed by a simple DNA extraction method (13). A conventional PCR protocol (18) was also evaluated.
Leaves from asymptomatic plants of different cultivars of almond, peach, nectarine (Prunus persica var. nectarina), flat peach (Prunus persica var. platycarpa), European plum (Prunus domestica L.), cherry [Prunus avium (L.) L. and P. cerasus L.], and Japanese plum as well as the rootstocks Santa Lucía SL-64 (Prunus mahaleb L.), Barrier (P. persica × P. davidiana), Myrobalan (Prunus cerasifera), Garnem (GxN-15) (P. amygdalus × P. persica), Adesoto (Prunus insititia), and GF-677 and GxN (P. amygdalus × P. persica) were collected in nurseries and orchards from June to October. A total of 280 leaf samples were processed by heat treatment of washed tissue samples in a first set of analyses. In a second set of analyses, a further 117 asymptomatic leaf samples were collected in nurseries and orchards and processed by both heat treatment of washed tissue samples and DNA extraction of comminuted tissue as indicated above. Most samples came from trees from areas suspected of X. arboricola pv. pruni infestation.
A total of 285 dormant buds of different cultivars of almond, peach, apricot [Prunus armeniaca (L.) Batsch], European plum, and Japanese plum and Myrobalan, GxN, and GF-677 rootstocks were collected from the middle of January to the end of March in nurseries and orchards. Ten milliliters of sterile distilled water was added to each sample (20 buds, approximately 0.7 g), and they were processed by DNA extraction from comminuted tissues as indicated above. Real-time PCR analyses were performed on all samples, and appropriate negative controls were included in all real-time PCR assays.
Selected samples that tested positive or negative were subjected to bacterial isolation to confirm the accuracy of the real-time PCR detection. Aliquots from these extracts and their dilutions (up to 1:1,000) were plated as described above. Those colonies morphologically resembling X. arboricola pv. pruni were purified and further identified by biochemical tests, conventional PCR (18), and real-time PCR, and representative isolates were inoculated in a detached-leaf bioassay (24).
RESULTS
Specificity of the real-time PCR assay.
Primer set Xap-2F (forward) (5′-TGG CTT CCT GAC TGT TTG CA-3′) and Xap-2R (reverse) (5′-TCG TGG GTT CGC TTG ATG A-3′), designed to obtain a PCR product of 72 bp, was finally selected from among those designed from the ABC transporter gene, to be used in combination with the TaqMan probe Xap-2P (5′-6-carboxyfluorescein [FAM]-TCA ATA TCT GTG CGT TGC TGT TCT CAC GA-6-carboxytetramethylrhodamine [TAMRA]-3′) (see Fig. S1 in the supplemental material). Further amplification reactions were performed at optimized concentrations: 0.4 μM each primer and 150 nM TaqMan probe.
A BLAST analysis showed similarity between the sequence corresponding to the ABC transporter used for the conventional (18) and real-time PCR primers and two sequences available in the databases. A high level of similarity (81%) was found between the sequence corresponding to the 72-bp PCR product obtained with real-time PCR primers Xap-2F and Xap-2R and a sequence (GenBank accession number FP565176.1) from a Xanthomonas albilineans strain (GPE PC 73). Moderate similarity (51%) was found with the sequence of a homologous gene (accession number FN392235.1) from an Erwinia pyrifoliae strain (DSM 12163 = CFBP 4172T). No other significant matches were found.
In addition to this in silico evaluation, the specificity of real-time PCR was tested with DNA from the bacterial strains listed in Table 1. All X. arboricola pv. pruni strains gave consistent positive results. Nondesired specific PCR products were obtained only from Xanthomonas arboricola pv. corylina and Xanthomonas citri subsp. citri strains. In the latter organism, a considerably higher CT than that obtained with the same bacterial concentration of X. arboricola pv. pruni was observed. No other bacterial species or Prunus samples yielded CT values.
Sensitivity and efficiency of the detection method.
A different sensitivity of the real-time PCR detection method was achieved from pure cultures of heated bacteria or DNA extracted (Table 2): positive PCR results were obtained from 103 CFU ml−1 or 102 CFU ml−1 after heat treatment or DNA extraction, respectively.
TABLE 2.
Limit of detection of real-time PCR analyses of pure culture cells of Xanthomonas arboricola pv. pruni strain ISPaVe B4 in cell suspensions and in spiked samples
| Sample | Heat treatment |
DNA extraction |
||||
|---|---|---|---|---|---|---|
| CFU ml−1 | Mean E ± SDa | Avg CT ± SDb | CFU ml−1 | Mean E ± SDa | Avg CT ± SDb | |
| Bacteria | 103 | 2.11 ± 0.11 | 33.95 ± 3.01 | 102 | 1.86 ± 0.05 | 39.28 ± 2.28 |
| Spiked washed leaves | ||||||
| Peach | 102 | 1.92 ± 0.02 | 40.62 ± 3.16 | 102 | 1.92 ± 0.02 | 36.06 ± 0.26 |
| Almond | 102 | 1.95 ± 0.17 | 37.33 ± 0.86 | 102 | 1.88 ± 0.04 | 36.6 ± 0.55 |
| GF-677 | 102 | 2.03 ± 0.02 | 38.84 ± 2.10 | 102 | 1.98 ± 0.08 | 36.42 ± 1.61 |
| Japanese plum | 105 | 1.62 ± 0.02 | 33.41 ± 0.82 | 102 | 1.96 ± 0.05 | 37.36 ± 0.11 |
| Spiked comminuted leaves | ||||||
| Peach | 104 | 1.82 ± 0.24 | 27.80 ± 1.93 | 102 | 1.90 ± 0.05 | 34.54 ± 1.05 |
| Almond | 103 | 1.97 ± 0.08 | 33.37 ± 0.81 | 102 | 1.98 ± 0.02 | 34.25 ± 1.13 |
| GF-677 | 103 | 2.02 ± 0.12 | 33.49 ± 0.87 | 102 | 1.99 ± 0.09 | 34.09 ± 1.20 |
| Japanese plum | —c | 1.62 ± 0.02 | —c | 102 | 2.11 ± 0.05 | 39.78 ± 2.35 |
E is the average efficiency of amplification, calculated using the formula E = 10[−1/k], where k is the regression line slope. Means and standard deviations were calculated from at least two regression lines calculated from at least two PCRs per bacterial concentration and two independent assays.
Average CT values were calculated for at least three PCRs from the lowest bacterial concentration detected.
—, no fluorescence above the threshold was recorded.
When spiked plant samples were analyzed without prior DNA extraction, the detection level achieved from spiked washed tissue samples of peach, almond, and GF-677 rootstock samples was very high, about 102 CFU ml−1. However, only 105 CFU ml−1 could be detected when spiked Japanese plum wash tissue samples were analyzed. In general, a lower sensitivity was observed when comminuted tissue without DNA extraction was analyzed. Again, the case of Japanese plum was especially significant, because amplification was completely inhibited when no DNA extraction was performed (Table 2). The sensitivity level of the reactions performed using purified DNA from spiked samples was satisfactory for both washed and comminuted tissues (102 CFU ml−1), reaching a sensitivity equal to that obtained after DNA purification from pure bacterial cultures. No apparent sensitivity differences were found among the different plant materials analyzed (Table 2). Although the simple DNA extraction method (13) and the CTAB method (8) showed similar sensitivities in preliminary assays at different primer concentrations and both gave a PCR efficiency rate close to 2 (data not shown), the simple extraction (13) was retained for further assays to avoid the use of toxic compounds such as phenol and chloroform. The kit from Qiagen did not provide as satisfactory results as the other DNA extraction methods.
In at least one of the three independent repetitions of the sensitivity experiments, X. arboricola pv. pruni colonies were recovered on culture plates from washed or comminuted spiked samples, containing 106, 103, or 102 CFU ml−1 (final concentrations) of the bacterium.
Dilutions from some of the heat-treated spiked comminuted samples were performed before the real-time PCR was performed in order to try to reduce the inhibitor's concentrations and to improve sensitivity. No sensitivity improvement generally resulted in most of the cases after the dilution of peach, almond, or GF-677 extract samples (with the exception of 103 CFU ml−1 spiked peach samples). Moreover, amplification efficiencies from DNA dilutions of those samples were always close to 2, as described above. However, in Japanese plum samples, the dilution of the heat-treated samples improved the PCR sensitivity for the spiked samples from 106 to 104 CFU ml−1. While undiluted samples and a 1:10 dilution resulted in negative results, a 1:100 dilution gave positive results, with PCR efficiencies close to 2 (about 3 cycles between each dilution) when serial dilutions were analyzed (see Table S1 in the supplemental material).
Calibration curves obtained by using primers and a TaqMan probe based on the ABC transporter gene to detect and quantify X. arboricola pv. pruni bacteria were performed for each plant material/DNA extraction combination analyzed. Plotting the cycle number versus the log concentration of the bacteria gave straight linear regression lines in all cases, with correlation coefficients (r) of −0.87 in the worst case, with Japanese plum material, and an r value of up to −0.99 in most other cases. Data from two independent bacterial dilution series were used to calculate all the calibration curves. Amplification efficiencies (10[−1/slope]) for each extraction method/plant material combination ranged from 1.6 for washed Japanese plum material to around 2 in most other cases (Table 2).
When average PCR efficiencies were analyzed and compared, no significant differences were found between washed and comminuted material for peach, almond, and GF-677. No significant PCR efficiency differences were shown after either DNA extraction or heat treatment. However, for Japanese plum, the highest efficiency was always obtained when the DNA was extracted by the protocol described above (Table 2). Evaluation of regression lines gave results similar to those described above for efficiency average comparisons. No significant differences (P > 0.1) were shown among the slopes of the regression lines obtained from the PCRs performed on almond, peach, and GF-677 samples after either heat treatment or DNA extraction (13). Moreover, no significant differences (P > 0.1) were shown between the regression lines from comminuted or washed material for those samples. For Japanese plum, a significantly lower slope (P < 0.01) on the regression line was obtained from the washed sample series after heat treatment, in comparison to the other washed or comminuted plant materials. No amplification resulted from comminuted samples without DNA extraction, revealing total PCR inhibition.
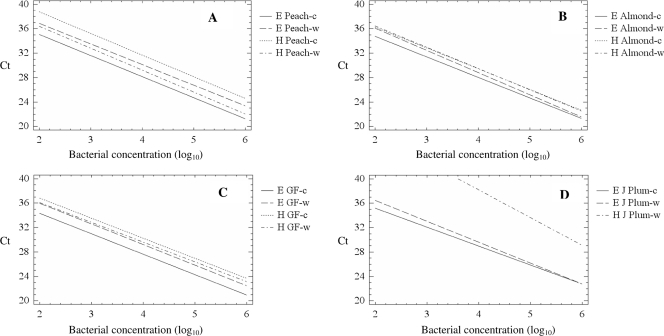
Significant differences (P < 0.01) in the intercepts of the regression lines were shown between heat-treated or DNA-extracted samples (13) for all plant materials analyzed. Lower intercepts, and therefore higher sensitivities, were usually obtained when DNA extraction (13) was performed. Only in the case of washed tissue from peach was the intercept average of the regression lines higher for heat-treated than for DNA-extracted samples (Fig. 1).
FIG. 1.
Calibration curves obtained for dilution series performed for peach (A), almond (B), GF-677 (C), and Japanese plum (J Plum) (D) using primers and a TaqMan probe to detect and quantify X. arboricola pv. pruni bacteria. DNA was amplified after heat treatment (H) or extraction (E) (13) from washed (w) or comminuted (c) material. Data from two independent bacterial dilution series were used to calculate all the calibration curves.
Detection of X. arboricola pv. pruni in nursery and orchard samples.
The five peach leaf samples (M1 to M5) displaying bacterial spot symptoms yielded positive results by real-time PCR when either heat-treated washed tissues or undiluted DNA extracts from comminuted tissue were analyzed. However, undiluted comminuted tissues without DNA extraction released inhibitory compounds and lacked PCR products in one sample (M1). PCR inhibition was solved by performing DNA extraction (13) (see Table S2 in the supplemental material). Samples M1 and M3 displayed the lowest CT values in the analysis of washed tissues, indicating their high bacterial concentrations (Table S2). Positive real-time PCRs were obtained from dilutions of up to 10−5 of these comminuted plants, with the exception of sample M5. Straight-line regression plots were performed with serial dilutions as described above, and the calculated efficiencies were all close to 2. According to the data inferred from the regression lines obtained as described above for peach material, these samples probably harbored bacterial populations of at least 106 CFU ml−1 (Fig. 1A). No amplification was obtained with control samples derived from healthy plant tissues. X. arboricola pv. pruni infection was confirmed for samples M1 to M5 by bacterial isolation on YPGA medium with cycloheximide. However, conventional PCR (18)-positive reactions occurred with the 10−1 dilution only (see Table S2 in the supplemental material).
Asymptomatic leaves from Prunus species and cultivars, as well as rootstocks, from nurseries and orchards were analyzed. When 280 asymptomatic washed leaf samples were analyzed by real-time PCR without prior DNA extraction, 33 of them (11.8%) yielded positive results. CT values ranged from 23.58 to 40.98, indicating different bacterial concentrations theoretically ranging from at least 105 to 102 CFU ml−1 based on regression lines obtained from peach, almond, or GF-677 rootstock samples (Fig. 1A, B, and C). Nineteen representative samples showing CT values ranging from 27.69 to 40.98 were serially diluted (1:10 to 1:1,000) and plated onto YPGA medium with cycloheximide. X. arboricola pv. pruni colonies were recovered from 57.9% of these analyzed samples (Table 3 ). In order to compare heat treatment and DNA extraction on comminuted material, 117 asymptomatic Prunus samples were analyzed by real-time PCR. CT values ranging from 28.55 to 41.50 from 9.4% of the samples were obtained after DNA extraction, whereas no amplification resulted from washed tissues (Table 4 ). According to the regression lines shown in Fig. 1, the bacterial concentration in those samples theoretically ranged from 106 to less than 102 CFU ml−1.
TABLE 3.
Analyses of asymptomatic leaf samples from nurseries and orchards based on real-time PCR of washed leaf tissues without DNA extraction
| Host(s)a | Cultivar or rootstock | No. of washed samples positive by real-time PCR/total no. of washed samples analyzed | CT rangee | No. of positive isolation samples/total no. of isolation samples analyzedb |
|---|---|---|---|---|
| Prunus amygdalus | Unknown | 1/10 | 33.13 | 1/1 |
| Prunus persica | Unknown | 4/13 | 31.34-40.49 | 0/1 |
| Prunus mahaleb | Santa Lucía SL-64 | 16/55 | 31.24-40.98 | 7/13 |
| P. persica × P. davidiana | Barrier | 11/36 | 23.58-39.39 | 3/4 |
| Prunus amygdalus | Variousc | 0/63 | — | ND |
| Prunus avium | Unknown | 0/6 | — | ND |
| Prunus persica, P. persica var. nectarina, P. persica var. platycarpa | Variousd | 1/42 | 33.76 | ND |
| Prunus amygdalus × P. persica | GxN-15 Garnem | 0/16 | — | ND |
| Prunus cerasifera | Myrobalan | 0/1 | — | ND |
| Prunus insititia | Adesoto | 0/23 | — | ND |
| Prunus persica × P. amygdalus | GF-677 | 0/15 | — | ND |
| Total | 33/280 | 23.58-40.98 | 11/19 |
Samples were collected from June to October.
Isolation was achieved from samples testing positive by real-time PCR analysis with CT values ranging from 27.69 to 40.98. ND, not determined.
Prunus amygdalus cv. Guara, Ferragnes, Ferraduel, Largueta, and Marcona and other unknown cultivars.
Prunus persica cv. Badia, Big Top, Nectatop, Sweet Cap, Veruela, VP 58, 0286, E 37, SBN, and ASF and other unknown cultivars.
—, no fluorescence above the threshold was recorded.
TABLE 4.
Comparative real-time PCR analyses of washed tissue and comminuted samples of asymptomatic leaves collected in nurseries and orchards following DNA extraction
| Host(s)a | Cultivar or rootstock | No. of positive samples by real-time PCR/total no. of samples by method |
CT range | |
|---|---|---|---|---|
| Washed tissue | Comminuted and DNA extraction | |||
| Prunus amygdalus | Variousb | 0/8 | 1/8 | 37.53 |
| Prunus armeniaca | Moniquí | 0/2 | 2/2 | 36.04-41.50 |
| Prunus persica | Almameb C12 | 0/10 | 1/10 | 38.74 |
| Prunus mahaleb | Santa Lucía SL-64 | 0/20 | 7/20 | 28.55-39.93 |
| Prunus amygdalus × P. persica | GxN | 0/10 | 0/10 | —d |
| Prunus avium, Prunus cerasus | Variousc | 0/31 | 0/31 | — |
| Prunus domestica | Unknown | 0/4 | 0/4 | — |
| Prunus spp. | Unknown | 0/18 | 0/18 | — |
| Prunus insititia | Adesoto | 0/4 | 0/4 | — |
| Prunus persica × P. amygdalus | GF-677 | 0/10 | 0/10 | — |
| Total | 0/117 | 11/117 | 28.55-41.50 | |
Samples were collected from June to October.
Prunus amygdalus cv. Blanquerma, Ferraduel, and Mardía and other unknown cultivars.
Prunus avium cv. Blanća de Provenza, Castañera, Chinook, Cristobalina, De la Pinta, Ebony, Gilpeck, Larian, Merton Glory, Mollar de Cáceres, Pico colorado, Pico negro, Producta, Ramillete, Spalding, Sparkle, Star, Taleguera brillante, Temprana de Sot C, Temprana de Sot G, Velvet, and Vic and Prunus cerasus cv. Guindo S, Guindo de Toro, Negra de Serra, Reina Hortensia, and other unknown cultivars.
—, no fluorescence above the threshold was recorded.
When DNA extracts from comminuted tissues of 285 dormant Prunus buds from nurseries or orchards with previous episodes of X. arboricola pv. pruni infection were analyzed by real-time PCR, CT values ranging from 21.33 to 41.62 were obtained for 9.1% of the samples. Based on the regression line obtained as described above, those CT values showed bacterial concentrations theoretically ranging from 106 to less than 102 CFU ml−1 (Fig. 1A, B, and D). The isolation of the pathogen was assayed for 48 representative samples (23 of which tested positive and the other 25 of which tested negative by real-time PCR), and X. arboricola pv. pruni recovery was achieved for only 9 of these positive samples (39%), with CT values ranging from 21.33 to 33.70, suggesting bacterial concentrations theoretically ranging from 106 to 102 CFU ml−1. No X. arboricola pv. pruni colonies were found on plates from samples that were negative by real-time PCR (Table 5).
TABLE 5.
Real-time PCR analyses of comminuted buds collected in nurseries and orchards following DNA extraction
| Host(s)a | Cultivar or rootstock | No. of positive samples by real-time PCR/total no. of samples analyzed for comminuted DNA extraction | CT range | No. of positive isolation samples/total no. of samples analyzedb |
|---|---|---|---|---|
| Prunus amygdalus | Variousc | 9/15 | 21.33-38.03 | 5/14 |
| Prunus persica | Variousd | 11/43 | 31.24-39.47 | 2/31 |
| Prunus cerasifera | Myrobalan | 3/31 | 30.86-33.63 | 2/3 |
| Prunus amygdalus | Variousc | 2/99 | 31.54-32.51 | ND |
| Prunus persica | Variousd | 1/33 | 41.62 | ND |
| Prunus armeniaca | Moniquí | 0/3 | —f | ND |
| Prunus amygdalus × P. persica | GxN | 0/5 | — | ND |
| Prunus persica × P. amygdalus | GF-677 | 0/26 | — | ND |
| Prunus salicina, Prunus domestica | Variouse | 0/30 | — | ND |
| Total | 26/285 | 21.33-41.62 | 9/48 |
Samples were collected from January to March.
Isolation was achieved for samples testing positive by real-time PCR with CT values ranging from 21.33 to 33.70. Isolation was not achieved for 25 samples that tested negative by PCR. ND, not determined.
Prunus amygdalus cv. Achaak 506, AL-VT 363, Argentina, Aspirilla 547, Ayles, Barter 189, Biota 530, Bulbuente 549, Castañeda 368, Constantí, Coop Mañan 550, Cosa Trova 320, Ferralise 349, Filipo Ceo 360, Forastero, Kata 514, Largueta, Largueta VT 366, Marcona, Marinada, Merino, Pau 234, Picantilli 106, Rana 259, Rachele 258, Spilo 517, Taitona 242, Tarragonés, Texas, Tree 516, Truoito, Vayro, and Vivot 241 and other unknown cultivars.
Prunus persica cv. Amarillo Octubre, Baby Gold 5, Baladin VT, Calabacero, Calante, Campillo Rocho, Catherine, Evaisa, Gallur, Jeromo, Jerónimo, Jesca, Maluenda 2375, Manolito, Maruja Tejar, Michelini, Miraflores, Montaced, Montamar, Paraguayo Jota, Red Robin, Roig de Aiotona, Rojo-Amarillo Septiembre, Rojo del Rito, Roza, San Jaime, San Lorenzo, Sudanell 2211, Sudanell 2213, Sudanell 2349, Summer Grand, and Zaragozano and other unknown cultivars.
Prunus salicina cv. Golden Japan and other unknown cultivars.
—, no fluorescence above the threshold was recorded.
DISCUSSION
A reliable PCR assay was necessary to implement standard detection procedures for the quarantine bacterium X. arboricola pv. pruni for the analysis of propagation material in nurseries, certification schemes, and import controls and to discern new reservoirs of this pathogen. Here, a real-time PCR approach was developed based on a putative ABC transporter gene previously described (18) and also sequenced in Spanish isolates (data not shown). This ABC transporter seems to be highly conserved in X. arboricola pv. pruni, which is consistent with the genetic homogeneity of this species observed by using other methods (5, 33). As expected, when this new detection protocol was applied to a collection of 159 X. arboricola pv. pruni strains from a variety of geographical areas, all of them yielded positive amplification results. Moreover, no amplification was obtained either from other bacterial pathogens and Prunus microbiota or from most of the selected plant-pathogenic bacteria from other hosts, thus supporting results of previous studies targeting this sequence for conventional PCR (18). However, this study also revealed the existence of homologous genes in different bacteria, including two Xanthomonas strains as well as Erwinia pyrifoliae. A positive PCR resulted when amplifications were performed by using DNA from Xanthomonas citri subsp. citri or Xanthomonas arboricola pv. corylina, although in one of the cases, the CT value was higher than that obtained with X. arboricola pv. pruni strains. Since X. arboricola pv. corylina and X. citri subsp. citri are hazelnut and citrus pathogens, respectively, which have never been reported to occur on Prunus, these results do not limit the reliability of the newly developed real-time PCR method for the detection of X. arboricola pv. pruni. In fact, this result may encourage the development of similar strategies based on this homologous gene sequence to detect these plant pathogens in their respective hosts.
The sensitivity threshold of 102 CFU ml−1 for the X. arboricola pv. pruni real-time PCR assay developed was at least equal to those obtained with similar assays reported previously for other phytopathogenic bacteria (9, 29, 30), and it surpasses the sensitivity levels reported previously for the conventional PCR method (18), which is, as far as we know, the only PCR method described for this pathogen. In addition, the methodology described not only detects the bacteria but also gives information regarding the bacterial population level.
In general, direct PCR amplification of bacterial DNA present in plant tissue has been confirmed to be problematic due to low populations in some cases but mainly because of the presence of inhibitory compounds. Such compounds may reduce the success of PCR by exerting a direct effect on the polymerase or by binding to DNA (6, 14, 15, 16, 20, 21). In our work, the amplification of X. arboricola pv. pruni was possible, even without DNA extraction, for washed tissues from most of the plant materials. On the other hand, a considerable reduction in sensitivity was observed for comminuted plant material suspensions, especially from Japanese plum extracts. However, when a simple DNA extraction method (13) was applied, the sensitivity of the method significantly increased, and a limit of detection of 102 CFU ml−1 was reached for both comminuted and washed material.
For washed material, DNA extraction was required only for the reliable detection of X. arboricola pv. pruni in Japanese plum but not in peach, almond, or GF-677. Bacterial DNA from Japanese plum samples was especially difficult to amplify, probably due to high concentrations of PCR inhibitors, even on the leaf surfaces. Inhibitor release also caused PCR failure and a reduction in sensitivity for all the materials assayed when comminuted samples were analyzed. Similar PCR efficiencies were obtained for almost all the dilution series made from comminuted or washed material, suggesting that the different sensitivities shown among them were due to different DNA recovery success rates rather than to the differential presence of compounds that interfere with polymerase activity. That is not the case for Japanese plum samples, since the PCR efficiency obtained from that material was significantly lower than that from the other Prunus species when no DNA extraction was applied. In such case, the presence of a high PCR-inhibitory concentration could be partially solved by sample dilution that improved PCR detection and minimized the risk of false-negative results.
As shown above, real-time PCR could detect the pathogen from symptomatic or asymptomatic samples with no time-consuming DNA extractions in many cases. This information is of interest and should be considered when a large number of samples must be analyzed, for example, when monitoring the disease in nurseries or orchards, as discussed below. However, such a method may not always ensure reliable detection when low pathogen populations are present or due to the presence of compounds that inhibit the PCR, especially in some plant materials. Inhibitor compounds may include those from the plant itself but also control products applied during crop management, as described previously by some authors (14, 15, 22). The use of a previous DNA extraction step is therefore appropriate for samples which may have a low X. arboricola pv. pruni level, such as asymptomatic samples or those showing special problems for amplification, like Japanese plum.
Our studies of the efficiency and sensitivity of the detection method developed were supported by the results obtained from naturally infected plants collected from nurseries or orchards. We have demonstrated that low populations of X. arboricola pv. pruni could be detected by real-time PCR from simple washed tissues of samples with no DNA extraction or from comminuted tissue following DNA extraction. Each method has its advantages and disadvantages: direct analysis of washed tissues offers high-throughput potential, and therefore, it could be suitable for large-scale screening assays; however, it cannot ensure reliable detection from samples with low pathogen numbers or when a high PCR inhibitor concentration is expected. On the other hand, DNA extraction is time-consuming and limits large-scale applications, but it is desirable for reducing the likelihood of false-negative results. The choice of the methodology for the processing of samples before amplification will depend on the number and type of samples to be analyzed, the expected target populations, as well as their expected amount of inhibitors. In our experience, analysis of washed tissue samples could be advisable for the large-scale screening of plant propagation material when a large number of positive samples is expected, although a further DNA extraction step may be required to confirm true-negative results.
The X. arboricola pv. pruni level in plant samples analyzed here, estimated from the regression lines calculated for each plant host, showed clear differences between symptomatic and asymptomatic samples. Higher CT values generally obtained from nonsymptomatic material were indicative of lower X. arboricola pv. pruni levels and associated with the presence of low colony numbers on isolation plates. However, the relationship between low CTs and high numbers of colonies on isolation plates was not always true, and sometimes, low CT values were obtained from samples where no bacteria were isolated. The difficulty of isolating X. arboricola pv. pruni from plant material might explain this issue, and an underestimation of the X. arboricola pv. pruni population by isolation could be due to the accompanying microbiota that can interfere with the appearance of X. arboricola pv. pruni colonies. In addition, we must also consider the possibility of the existence of a nonculturable condition of this bacterium, as described previously for other Xanthomonas species (7, 10, 17). Viable but nonculturable bacteria would be not considered part of the bacterial population and may therefore be undervalued by culture plating but may be included in the estimation by PCR.
Real-time PCR not only gives an easily interpretable and highly specific result in a quantitative manner, it also provides additional information about the likelihood of a final positive detection. In addition to the efficient analysis of symptomatic leaves, the newly developed real-time PCR enables the detection of the bacterium on asymptomatic leaves during early spring as well as on dormant buds in winter, and it could therefore be suitable for the accurate screening of contaminated nursery propagation material.
Supplementary Material
Acknowledgments
This work was partially supported by an agreement between Subdirección General de Sanidad de la Producción Primaria, MARM, Spain, and IVIA and has been performed in the framework of Cost Action 873. A.P.-B. thanks the Instituto Nacional de Investigaciones Agrarias of Spain and the Centro de Investigación y Tecnología Agroalimentaria of Aragón for a contract within the INIA-CC AA Programme.
We also thank Charles Manceau, Lucienne Mansvelt, Joanna Pulawska, and Marco Scortichini for kindly providing bacterial strains and Mónica Sáenz, Elisa Ferragud, Javier Peñalver, and M. Luisa Palazón for their technical assistance.
Footnotes
Published ahead of print on 29 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anonymous. 2000. Council directive 2000/29/EC of 8 May 2000 on protective measures against the introduction into the community of organisms harmful to plants or plant products and against their spread within the community. Official J. Eur. Communities L169(43):1-112. [Google Scholar]
- 2.Anonymous. 2003. Data sheets on quarantine organisms. Xanthomonas arboricola pv. pruni. European and Mediterranean Plant Protection Organization, Paris, France.
- 3.Anonymous. 2006. EPPO standards PM 7/64. (1) Diagnostics Xanthomonas arboricola pv. pruni. Bull. EPPO 36:129-133. [Google Scholar]
- 4.Anonymous. 2007. Distribution maps of quarantine pests for Europe. European and Mediterranean Plant Protection Organization, Paris, France. http://pqr.eppo.org/datas/XANTPR/XANTPR.pdf.
- 5.Boudon, S., C. Manceau, and J.-P. Nottéghem. 2005. Structure and origin of Xanthomonas arboricola pv. pruni populations causing bacterial spot of stone fruit trees in Western Europe. Phytopathology 95:1081-1088. [DOI] [PubMed] [Google Scholar]
- 6.De Boer, S. H., L. J. Ward, X. Li, and S. Chittaranjan. 1995. Attenuation of PCR inhibition in the presence of plant compounds by addition of BLOTTO. Nucleic Acids Res. 23:2567-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Campo, R., P. Russi, P. Mara, H. Mara, M. Peyrou, I. Ponce de León, and C. Gaggero. 2009. Xanthomonas axonopodis pv. citri enters the VBNC state after copper treatment and retains its virulence. FEMS Microbiol. Lett. 298:143-148. [DOI] [PubMed] [Google Scholar]
- 8.Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12:13-15. [Google Scholar]
- 9.Dreo, T., K. Gruden, C. Manceau, J. D. Janse, and M. Ravnikar. 2007. Development of a real-time PCR-based method for detection of Xylophilus ampelinus. Plant Pathol. 56:9-16. [Google Scholar]
- 10.Ghezzi, J. I., and T. R. Steck. 1999. Induction of the viable but non-culturable condition in Xanthomonas campestris pv. campestris in liquid microcosms and sterile soil. FEMS Microbiol. Ecol. 30:203-208. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, C. A., and M. J. Hattingh. 1986. Transmission of Xanthomonas campestris pv. pruni in plum and apricot nursery trees by budding. HortScience 21:995-996. [Google Scholar]
- 12.King, E. O., M. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocianin and fluorescein. J. Lab. Clin. Med. 44:401-407. [PubMed] [Google Scholar]
- 13.Llop, P., P. Caruso, J. Cubero, C. Morente, and M. M. López. 1999. A simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J. Microbiol. Methods 37:23-31. [DOI] [PubMed] [Google Scholar]
- 14.López, M. M., E. Bertolini, E. Marco-Noales, P. Llop, and M. Cambra. 2006. Update on molecular tools for detection of plant pathogenic bacteria and viruses, p. 1-46. In J. R. Rao, C. C. Fleming, and J. E. Moore (ed.), Molecular diagnostics. Current technologies and applications. Horizon Bioscience, Norfolk, United Kingdom.
- 15.López, M. M., E. Bertolini, A. Olmos, P. Caruso, M. T. Gorris, P. Llop, R. Penyalver, and M. Cambra. 2003. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 6:233-243. [DOI] [PubMed] [Google Scholar]
- 16.López, M. M., P. Llop, A. Olmos, E. Marco-Noales, M. Cambra, and E. Bertolini. 2009. Are molecular tools solving the challenge posed by detection of plant pathogenic bacteria and viruses? Curr. Issues Mol. Biol. 11:13-46. [PubMed] [Google Scholar]
- 17.Ordax, M., E. Marco-Noales, M. M. López, and E. G. Biosca. 2006. Survival strategy of Erwinia amylovora against copper: induction of the viable-but-nonculturable state. Appl. Environ. Microbiol. 72:3482-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagani, M. C. 2004. An ABC transporter protein and molecular diagnoses of Xanthomonas arboricola pv. pruni causing bacterial spot of stone fruits. Ph.D. thesis. University of North Carolina, Raleigh, NC. http://www.lib.ncsu.edu/theses/available/etd-10042004-232356.
- 19.Palacio-Bielsa, A., M. Roselló, M. A. Cambra, and M. M. López. 2010. First report on almond in Europe of bacterial spot disease of stone fruits caused by Xanthomonas arboricola pv. pruni. Plant Dis. 94:786. [DOI] [PubMed] [Google Scholar]
- 20.Palacio-Bielsa, A., M. A. Cambra, and M. M. López. 2009. PCR detection and identification of plant-pathogenic bacteria: updated review of protocols (1989-2007). J. Plant Pathol. 91:249-297. [Google Scholar]
- 21.Pich, U., and Y. Schubert. 1993. Midiprep method for isolation of DNA from plants with a high content of polyphenolics. Nucleic Acids Res. 21:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulawska, J., M. Maes, T. Deckers, and P. Sobiczewski. 1997. The influence of pesticide contamination on detecting epiphytic Erwinia amylovora using PCR, vol. 62, no. 3b, p. 959-962. Mededelingen Faculteit Landbouwkundige en Toegepaste Biologische Wetenschappen Universiteit Gent, Ghent, Belgium. [Google Scholar]
- 23.Ramakers, C., J. M. Ruijter, R. H. L. Deprez, and A. F. M. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 24.Randhawa, P. S., and E. L. Civerolo. 1985. A detached-leaf bioassay for Xanthomonas campestris pv. pruni. Phytopathology 75:1060-1063. [Google Scholar]
- 25.Ridé, M. 1969. Bactéries phytopathogènes et maladies bactériennes des végétaux. In C. V. M. Bourgin (ed.), Les bactérioses et les viroses des arbres fruitiers. Ponsot, Paris, France.
- 26.Ritchie, D. F. 1995. Bacterial spot, p. 50-52. In J. M. Ogawa, E. I. Zehr, G. W. Bird, D. F. Ritchie, K. Uriu, and J. K. Uyemoto (ed.), Compendium of stone fruit diseases. APS Press, St. Paul, MN.
- 27.Ritchie, D. F. 1999. Sprays for control of bacterial spot of peach cultivars having different levels of disease susceptibility, 1998. Fungicide Nematicide Tests 54:63. [Google Scholar]
- 28.Smith, E. 1903. Observations on a hitherto unreported bacterial disease, the cause of which enters the plant through ordinary stomata. Science 17:456-457. [Google Scholar]
- 29.Tambong, J. T., K. N. Mwange, M. Bergeron, T. Ding, F. Mandy, L. M. Reid, and X. Zhu. 2008. Rapid detection and identification of the bacterium Pantoea stewartii in maize by TaqMan real-time PCR assay targeting the cpsD gene. J. Appl. Microbiol. 104:1525-1537. [DOI] [PubMed] [Google Scholar]
- 30.Vandroemme, J., S. Baeyen, J. Van Vaerenbergh, P. De Vos, and M. Maes. 2008. Sensitive real-time PCR detection of Xanthomonas fragariae in strawberry plants. Plant Pathol. 57:438-444. [Google Scholar]
- 31.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 32.Young, J. M. 1977. Xanthomonas pruni in almond in New Zealand. N. Z. J. Agric. Res. 20:105-107. [Google Scholar]
- 33.Zaccardelli, M., P. Ceroni, and U. Mazzucchi. 1999. Amplified fragment length polymorphism fingerprinting of Xanthomonas arboricola pv. pruni. J. Plant Pathol. 81:173-179. [Google Scholar]
- 34.Zaccardelli, M., M. F. Consiglio, and U. Mazzucchi. 1995. Detection of Xanthomonas campestris pv. pruni in symptomless peach trees in winter. Phytopathol. Mediterr. 34:199-203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.