Abstract
TrwC is a bacterial protein involved in conjugative transfer of plasmid R388. It is transferred together with the DNA strand into the recipient bacterial cell, where it can integrate the conjugatively transferred DNA strand into its target sequence present in the recipient cell. Considering that bacterial conjugation can occur between bacteria and eukaryotic cells, this protein has great biotechnological potential as a site-specific integrase. We have searched for possible TrwC target sequences in the human genome. Recombination assays showed that TrwC efficiently catalyzes recombination between its natural target sequence and a discrete number of sequences, located in noncoding sites of the human genome, which resemble this target. We have determined the cellular localization of TrwC and derivatives in human cells by immunofluorescence and also by an indirect yeast-based assay to detect both nuclear import and export signals. The results indicate that the recombinase domain of TrwC (N600) has nuclear localization, but full-length TrwC locates in the cytoplasm, apparently due to the presence of a nuclear export signal in its C-terminal domain. The recombinase domain of TrwC can be transported to recipient cells by conjugation in the presence of the helicase domain of TrwC, but with very low efficiency. We mutagenized the trwC gene and selected for mutants with nuclear localization. We obtained one such mutant with a point A904T mutation and an extra peptide at its C terminus, which maintained its functionality in conjugation and recombination. This TrwC mutant could be useful for future TrwC-mediated site-specific integration assays in mammalian cells.
Bacterial conjugation is a specialized mechanism to transfer plasmid DNA from a donor to a recipient bacterial cell. Under laboratory conditions, conjugative DNA transfer has been described to occur from bacteria to Saccharomyces cerevisiae (19), plants (3), and mammalian cells (45). The molecular process is related to the naturally occurring interkingdom gene transfer process from Agrobacterium tumefaciens to plant cells (47).
Mechanistically, bacterial conjugation has been described as a two-step process involving conjugative DNA processing, followed by active DNA transport (30). DNA processing is driven by a nucleoprotein complex called relaxosome. Within it, a relaxase protein introduces a site- and strand-specific nick into the origin of transfer (oriT), remaining covalently bonded to the 5′ end of the DNA strand to be transferred. For the DNA transport, a set of proteins constitute a type IV secretion system (T4SS) that forms a transmembranal channel. Transfer of the DNA molecule is proposed to occur in two steps: (i) the secretion of the relaxase protein (with the attached DNA strand) through the T4SS and (ii) pumping out of the remaining DNA molecule by a specialized ATPase (30). Thus, the relaxase also functions as a pilot protein guiding the DNA molecule into the recipient cell, where it presumably recircularizes the transferred DNA strand (10).
TrwC is the relaxase of the conjugative system of plasmid R388 (31). Its 966 amino acids comprise several functional domains, as shown in Fig. 1a. The crystal structure of the N-terminal relaxase domain has been determined (17). It shares a central structural domain with the HUH family of proteins, which include the rolling-circle replication initiation proteins (5) and the Rep protein of adeno-associated virus (AAV) (21), which catalyzes the integration of the viral genome into a unique site in the human genome (41). The C-terminal domain shows oligomerization and DNA helicase activities. Both the relaxase and the helicase domains expressed in the same cell reconstitute TrwC function in conjugation to a certain extent (32).
FIG. 1.
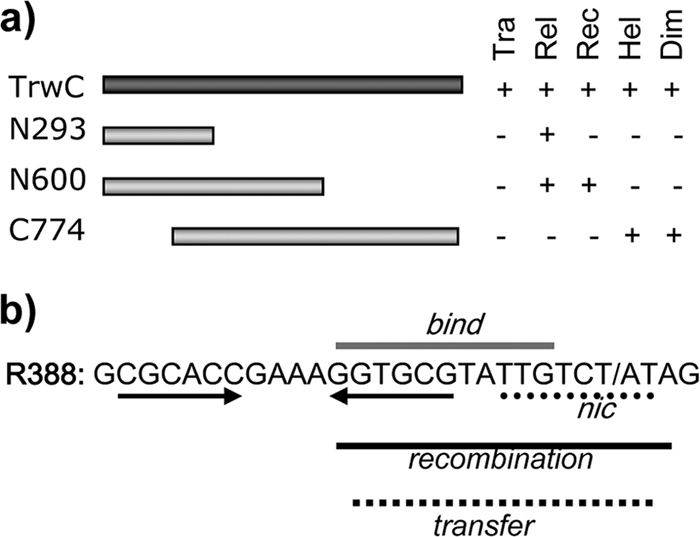
Functional dissection of TrwC and its target oriT sequence. (a) Map of the TrwC protein showing its functional domains. Tra, transfer (functionality in conjugation); Rel, in vitro relaxase activity; Rec, in vivo site-specific recombinase activity; Hel, DNA helicase activity; Dim, dimerization ability. The data were drawn from references 7 and 32. (b) DNA sequence of the central R388 oriT region (coordinates 201 to 173 from reference 27). The inverted repeat IR2 is indicated with arrows. Horizontal bars show the minimal sequence requirements for different TrwC activities. bind, in vitro TrwC binding on oligonucleotides; nic, in vitro TrwC nicking and strand-transfer activity on oligonucleotides; recombination, in vivo TrwC-mediated site-specific recombination; transfer, conjugal mobilization (data were drawn from references 7 and 33).
TrwC is also a site-specific recombinase capable of promoting efficient recombination between two cognate oriTs on a double-stranded DNA (dsDNA) molecule (28). The relaxosomal component protein TrwA is required for high-efficiency recombination (7). The minimal recognition sequences for efficient TrwC-mediated recombination show different requirements at each oriT locus: a core 17-bp sequence is sufficient to host efficient recombination in locus 1, while an additional 183-bp sequence is required at locus 2 (7), with the nic site located 3′ to locus 1. The recombination reaction is strictly dependent on the nicking and strand-transferase abilities of TrwC, since a mutation in the two catalytic tyrosine residues completely abolished recombination (10). However, the relaxase domain alone was not sufficient to catalyze recombination; the recombinase domain has been assigned to the N-terminal 600 residues of TrwC (7).
Figure 1b shows the R388 oriT sequence requirements for different TrwC activities. The common 17-bp core sequence comprises a highly specific sequence termed the nic site, where the scissile phosphate lies, and a 5′ region involving the recognition hairpin formed by an inverted repeat, IR2 (17). This is the essential core sequence required in vivo for both conjugal mobilization (33) and site-specific recombination (7). When the distal half of IR2 is removed, TrwC binding affinity is decreased only 3-fold, and TrwC-mediated in vivo site-specific recombination is slightly affected (7). In contrast, the sequence 6+2 (TTGTCT/AT) is absolutely required for TrwC binding, single-strand nicking, and strand transfer reactions (33). Similar results were reported for the related TraI relaxase of the F plasmid (46), where mutations affecting the nucleotides immediately adjacent to the nic site had a strong detrimental effect on conjugation.
Once TrwC enters the recipient cell it is fully active, as shown by complementation of a trwC-deficient mutant in the recipient (10). Notably, TrwC can also catalyze integration of a conjugatively transferred oriT-containing molecule into a recipient dsDNA oriT target (10). To our knowledge, this ability of TrwC to act as a site-specific integrase is unique among relaxases; even the related F TraI protein cannot catalyze a similar reaction (7). Protein VirD2 of Agrobacterium tumefaciens, functionally related to conjugative relaxases, has been postulated to reach the recipient plant cell, where it directs nuclear import of the transferred DNA and aids T-DNA integration into the plant genome, preserving the integrity of the T-DNA 5′ end (34). However, this integration is not site specific. In addition to VirD2, conjugative relaxases TraI and MobA from plasmids RP4 and RSF1010, respectively, have been localized within the nucleus of human cultured cells (40). However, integration assays in plants showed a specific deficiency of MobA in aiding genomic integration of the transferred DNA (2).
Given the observed capacity for bacterial conjugation to transmit DNA to mammalian cells, this feature of TrwC as a site-specific integrase upon entering the recipient cell suggests a potential biotechnological use of TrwC as an engineering integrase delivered in vivo by conjugation, together with the transferred DNA (29). There are many prokaryotic recombinases used for genetic manipulation in mammals, but for most of them their target cells need to be modified in advance by addition of the recombinase target sequence to their genomes (42). In the present, we show that TrwC catalyzes recombination between its cognate oriT and human sequences closely resembling the nic site. We also address TrwC nuclear targeting, since a recombinase must target the nucleus in order to work efficiently in eukaryotic cells. By random mutagenesis, we obtained a full-length TrwC mutant targeting the nucleus which is fully functional in conjugation and recombination.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli strain DH5α (16) was used as a host for the recombination assays. For mating assays, strains D1210 (38) and DH5α were used as donors and recipients, respectively. The plasmids used in the present study are listed in Tables 1 and 2. Luria-Bertani broth was used for bacterial growth, supplemented with agar for solid culture. Selective media included antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 20 μg/ml; streptomycin, 300 μg/ml; and trimethoprim, 20 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was supplied at a concentration of 60 μg/ml.
TABLE 1.
Published plasmids used in this study
| Plasmid | Descriptionb | Reference |
|---|---|---|
| pCEFL | Eukaryotic expression vector | 43 |
| pCIG1028 | oriT1-oriT2 recombination substrate | 7 |
| pCIG1030 | As pCIG1028 + trwA | 7 |
| pCIG1051 | pET3a::trwC(N450) | 7 |
| pCIG1073 | As pCIG1028 with oriT1(173-190) | 7 |
| pCIG1099 | pET3a::trwC(N600) | 7 |
| pCMS13 | pKK223-3::oriT(63-330)muta | 7 |
| pET:trwA | pET3a::trwA | 7 |
| pET3:trwAC | pET3a:PtrwA-trwA-trwC | 10 |
| pET29:trwAC | pET29c:PtrwA-trwA-trwC | 10 |
| pNEA3b | lexA-SV40 NLS-gal4AD fusion | 36 |
| pNEA:VirE2 | Control − for nuclear export | 36 |
| pNIA3b | lexA-gal4AD fusion | 36 |
| pNIA:VirD2 | Control + for nuclear import | 36 |
| pNIA:VirE2 | Control − for nuclear import | 36 |
| pSU19 | Cloning vector | 1 |
| pSU1445 | R388:Tn5tac1 in trwC | 28 |
| pSU1483 | pKK223-3::trwC | 15 |
| pSU1534 | pHG327::trwC(C774) | 32 |
| pSU1600 | pET3a::trwC(N293) | 7 |
| pSU1621 | pET3a::trwC | 17 |
Mutation affecting the nic site (TCT/A-to-GAG/A change).
SV40 NLS, NLS of simian virus 40.
TABLE 2.
Bacterial plasmids constructed in this study
| Plasmid | Descriptiona | Constructionb |
||
|---|---|---|---|---|
| Vector | Insert | Digestion/oligonucleotides (5′-3′) | ||
| Recombination substrates | ||||
| pCIG1110 | oriT1(63-330mut)† | pCIG1028 | pCMS13 | AACTCTAGAACCCAATGCGCATAGCG |
| AACAAGCTTCCTCTCCCGTAGTGTTAC | ||||
| pCIG1116 | oriT1 HuX (15+3) (−7)‡ | pCIG1028 | Human DNA | CCATCTAGATTAGACACAGGCTCTACTCACACAG |
| TACAAGCTTAAAAATTCAACACAGCCTCTAAGTG | ||||
| pCIG1117 | oriT1 Hu5 (15+3) (−10)‡ | pCIG1028 | Human DNA | CCATCTAGATCTAAGATGCAGTAAGATCCCAGAC |
| TACAAGCTTTCAAGATTAGTGAGCAAGAAATGTG | ||||
| pCIG1122 | oriT1 Hu18 (10+2)‡ | pCIG1028 | Human DNA | CCATCTAGACCTTGAACCTATTCTGCCCATA |
| TACAAGCTTTCAAGGCTCTTGATGTTTGAGA | ||||
| pCIG1126 | oriT1 Hu5b (13+1)‡ | pCIG1028 | Human DNA | CCATCTAGAAGCTATGCACAACAGCATGG |
| TACAAGCTTAATCCCAATATTTGACCACCA | ||||
| pCIG1127 | oriT1 Hu7 (12+3)‡ | pCIG1028 | Human DNA | CCATCTAGACCTGGCGATAGAGCAAGACT |
| TACAAGCTTCTGACCACCTGCTCCAAAAT | ||||
| Plasmids used for intragenic complementation assays | ||||
| pCIG1070 | pSU19::TrwC (N450) | pSU19 | pCIG1051 | XbaI/BamHI |
| pCIG1086 | pSU19::TrwC | pSU19 | pSU1621 | XbaI/BamHI |
| pCIG1103 | pSU19::TrwC (N600) | pSU19 | pCIG1099 | XbaI/BamHI |
| pLA35 | pSU19::TrwC (N293) | pSU19 | pSU1600 | XbaI/BamHI |
| Plasmids used for yeast nuclear import/export assays | ||||
| pCIG1133 | pNIA::TrwC (N600) | pNIA3b | pSU1621 | ACCGGATCCCGATGCTCAGTCACATGGT |
| CCAGAATTCACTCGATGGCCTTGGTTTG | ||||
| pCMS2 | pNIA::TrwC (N293) | pNIA3b | pSU1621 | ACCGGATCCCGATGCTCAGTCACATGGT |
| ACCGAATTCAGCTGAAATCTATGCCGAG | ||||
| pCMS5 | pNIA::TrwC-NLS | pNIA3b | pSU1621 | ACCGGATCCCGATGCTCAGTCACATGGT |
| CCAGAATTCTACCTACCTTTCTTTCCGGCCTCCATGCC | ||||
| pCMS9 | pNIA::TrwC | pNIA3b | pSU1621 | ACCGGATCCCGATGCTCAGTCACATGGT |
| ACCGAATTCACCTTCCGGCCTCCATGCC | ||||
| pCMS10 | pNIA::TrwC (C774) | pNIA3b | pSU1621 | CCAGGATCCTTGGAGCCGTCTATAAC |
| ACCGAATTCACCTTCCGGCCTCCATGCC | ||||
| pCMS15 | pNEA::TrwC | pNEA3b | pCMS9 | ACCGGATCCCGATGCTCAGTCACATGGT |
| CCACTCGAGACCTTCCGGCCTCCATGCC | ||||
| pCMS16 | pNEA::TrwC (C774) | pNEA3b | pCMS10 | CCAGGATCCTTGGAGCCGTCTATAAC |
| CCACTCGAGACCTTCCGGCCTCCATGCC | ||||
| pLA44 | pNIA::TrwC*c | pCMS10 | pCMS9 | Mutagenic PCRd |
| GGCTGGCGGTTGGGGGTTA | ||||
| ACCGAATTCACCTTCCGGCCTCCATGCCGCG | ||||
| pLA66 | pNIA::TrwC (A904T) | pCMS10 | pLA44 | ACCGGATCCCGATGCTCAGTCACATGGT |
| ACCGAATTCACCTTCCGGCCTCCATGCC | ||||
| pMTX719 | pNIA::NLS(x5)-TrwC | pCMS9 | Oligose | GATCCCCAAGAAGAAACGGAAGGT |
| GATCACCTTCCGTTTCTTCTTGGG | ||||
| pMTX720 | pNIA::NLS(x2)-TrwC | pCMS9 | Oligos | GATCCCCAAGAAGAAACGGAAGGT |
| GATCACCTTCCGTTTCTTCTTGGG | ||||
| pMTX726 | pNIA::NLS-TrwC | pMTX719 | None | BamHI and religation |
| Plasmids used for immunofluorescence analyses | ||||
| pLA14 | pCEFL::TrwC | pCEFL | pET29:trwAC | ACCAAAGCTTATGCTCAGTCACATGGTATT |
| ACCAGGATCCTTACCTTCCGGCCTCCA | ||||
| pLA27 | pCEFL::TrwC (N293) | pCEFL | pCMS3 | BamHI/EcoRI |
| pLA28 | pCEFL::TrwC (N600) | pCEFL | pCIG1133 | BamHI/EcoRI |
| pLA29 | pCEFL::TrwC (C774) | pCEFL | pCMS10 | BamHI/EcoRI |
†, mutation affecting the nic site (TCT/A-to-GAG/A change); ‡, human sequences in the indicated human chromosomes (HuX, -5, -18, or -7). The “n+n′” format indicates the extent of the consensus sequence around the nic site; variations from consensus are indicated in parentheses (see the text for details and nomenclature).
The first column (Vector) lists the vector plasmids, the second column (Insert) lists the plasmids from which the inserts were obtained, and the third column (Digestion/oligonucleotides) indicates either the restriction enzymes used for cloning or the oligonucleotides used for PCR amplification of the desired fragment, with the restriction sites underlined.
TrwC*, TrwC mutant obtained by random mutagenesis. It carries missense mutation A904T and the additional peptide KVNSCSHGSSRSTRD after residue 964 of TrwC.
This PCR was performed under mutagenic conditions, as described in Materials and Methods. The PCR fragment was digested with BamHI and EcoRI and ligated into the same sites of the pCMS10 backbone.
Oligos, oligonucleotides. NLS sequences were added to the N terminus of TrwC by oligonucleotide hybridization and insertion at BamHI site of pCMS9.
Plasmid constructions.
Plasmids were constructed by using standard methodological techniques (39). Restriction enzymes, Shrimp Alkaline Phosphatase, T4 DNA ligase and T4 polynucleotide kinase were purchased from Fermentas. Vent polymerase was purchased from New England Biolabs. DNA sequences of all cloned PCR segments were determined. Oligonucleotides used in the amplification of human sequences were designed by using the Primer3:WWW primer tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi [37]), setting an annealing temperature of 60°C and 400-bp product size. Oligonucleotide hybridization was performed as described previously (7).
Table 2 includes the details of how each plasmid was constructed. A brief description of each set of plasmids follows.
(i) All substrate plasmids for recombination assays are derivatives of plasmid pCIG1028 (7), which contains two wild-type oriT copies separated by a kanamycin resistance gene and a lacIq repressor gene in vector pSU19. The 1- to 402-bp wild-type sequence of oriT1 copy was substituted by the indicated mutations or human sequences. The latter were amplified from human genomic DNA obtained from a K562 leukemia cell line (American Type Culture Collection).
(ii) For intragenic relaxase-helicase complementation, different trwC 5′-terminal fragments were cloned in vector pSU19 under the control of the lactose promoter. All constructs carry the start codon at the same position with respect to the transcription/translation signals of the vector.
(iii) Constructs to test nuclear import/export in yeast were made on vectors pNIA3b/pNEA3b (36), which carry appropriate sites to make protein fusions with LexA-Gal4AD and LexA-nuclear localization signal (NLS)-Gal4AD, respectively. pCMS5 carries an NLS at the C terminus of TrwC; the oligonucleotide used to amplify trwC added a sequence coding for the peptide GKKGR just before the stop codon. To construct NLS-TrwC derivatives, oligonucleotides, including the coding sequence for the simian virus 40 NLS, were hybridized and 5′ phosphorylated with T4 polynucleotide kinase. The resulting fragment was cloned into the BamHI site of plasmid pCMS9 (pNIA:trwC). Two constructs were selected: pMTX719 contains five inserts that coded for NLSs in direct orientation and a last one in the opposite orientation, without affecting the trwC reading frame, and pMTX720 carries three copies of the insert, two of them in the sense orientation and the middle one in opposite orientation. To obtain a construct with a single NLS fused to the N terminus of TrwC, pMTX719 was digested with BamHI and religated, yielding pMTX726. Construction of pLA44 is detailed in the random mutagenesis section below. Plasmid pLA66 was constructed from pLA44 as indicated in Table 2 to separate mutations present in pLA44.
(iv) For immunofluorescence analysis, trwC and derivatives were subcloned into the pCEFL expression vector (43) and expressed from the eukaryotic promoter elongation factor 1α (EF1α).
TrwC binding assays.
Binding of TrwC-N293 to oligonucleotides was determined by an electrophoresis mobility shift assay as previously described (17). The oligonucleotides used were either R388 (25+8) or those containing the homologous human sequences shown in Fig. 2a. The 25+8 nomenclature refers to the 25 nucleotides 5′ and 8 nucleotides 3′ to the nic site. The 1 nM 5′-labeled (25+8) oligonucleotides were incubated with 0 to 20 nM TrwC-N293 purified as described previously (17) in the presence of 1 μM competitor oligonucleotide (a mixture of three unlabeled unspecific oligonucleotides).
FIG. 2.
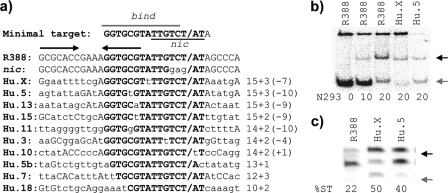
Putative TrwC targets in the human genome. (a) DNA sequences resembling the R388 nic site. The minimal region required for TrwC activity is highlighted in boldface. The nic site is indicated by a slash. Arrows show the inverted repeat recognized by TrwC. Nucleotides identical to the R388 sequence are shown in capital letters. The minimal target corresponds to the 173-190 oriT sequence present in plasmid pCIG1073. The mutation altering the R388 nic site in pCIG1110 is shown below the wild-type R388 sequence. Below, the seven human sequences, including the R388 14+2 sequence with a single mismach, are shown, together with shorter sequences mentioned in the text. Sequences are named indicating the human (Hu) chromosome number and the homologous nucleotides 5′+3′ with respect to the nic site; the mismatch position is indicated in parentheses. (b) TrwC binding assays on the (25+8) oligonucleotides containing the sequences shown in panel a. The amount of TrwC-N293 protein in each assay is indicated at the bottom (in nanograms). Arrows point to free (gray arrow) and retarded, TrwC-bound oligonucleotide (black arrow). (c) TrwC strand-transfer reactions on oligonucleotides, including the indicated R388 and human sequences. Arrows point to the cut product (gray arrow) or strand transfer product (black arrow), which is quantitated at the bottom of the gel (%ST). The assays for panels b and c were performed as described in Materials and Methods.
DNA strand transfer assays.
Oligonucleotides were 5′-labeled and strand-transfer assays were performed as described previously (14). Assays contained a 250 nM concentration of cold “donor” oligonucleotide R388 (12+18) 5′-TGCGTATTGTCT/ATAGCCCAGATTTAAGGA-3′ (the slash indicates the R388 nic site according to Llosa et al. [31]), a 50 nM concentration of the indicated “acceptor” (25+8)-labeled oligonucleotides, and 500 nM TrwC-N293 protein. Gels were scanned on a Molecular Imager FX system, and strand-transfer products were quantified by using Quantity One software (Bio-Rad) and expressed as the percentage of the total label in each lane.
Recombination assays.
Intramolecular recombination was tested as described previously (7). Briefly, the substrate plasmid (pCIG1028 or its derivatives) carries two copies of the R388 oriT. Recombination between the two copies induces expression of the downstream lacZα gene. A plasmid coding for TrwAC under the regulation of the trwA promoter was used as a helper plasmid. A plasmid coding for TrwA only was used as a helper in the negative control. To test the recombination activity of TrwC derivatives in pNIA or pCEFL, plasmid pCIG1030, the same as pCIG1028 but coding also for trwA, was used as a substrate (7). The substrate and helper plasmids were introduced into the lacZΔM15 strain DH5α and plated on selective media with X-Gal. The recombination activity was estimated by the number and size of blue sectors in bacterial colonies.
Mating assays.
Standard mating assays were performed as described previously (14). E. coli D1210 donor cells contained plasmid pSU1445, a TrwC-deficient R388 mutant, which was complemented with plasmids coding for TrwC derivatives in trans. For the relaxase-helicase complementation assays, a combination of compatible plasmids coding for C-terminal and N-terminal domains of TrwC was used to complement the transfer deficiency of plasmid pSU1445. Mating assays were performed in the presence or absence of IPTG (isopropyl-β-d-thiogalactopyranoside), and no significant differences were found (data not shown). A plasmid coding for wild-type TrwC was used as a positive control, and the corresponding empty vector was assayed as a negative control in all cases.
Immunofluorescence assays.
Human embryonic kidney 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. A total of 6 × 105 293T cells were placed in 10 ml of DMEM on glass coverslips and grown for 48 h. Cells were transfected with 7 μg of DNA plus 14 μl of JetPei (Genycell). After 24 h, cells were fixed and permeabilized with methanol for 10 min at −20°C, and immunostaining was performed as previously described (44). Cells were successively incubated with primary anti-TrwC antibody (15) (3 h of incubation at room temperature) and secondary fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Jackson Laboratories), both used at a 1:100 dilution. Coverslips were mounted with antifading mounting medium Vectashield (Vector Laboratories) with DAPI (4′,6′-diamidino-2-phenylindole) to visualize the nucleus. Cell samples were examined by using a Zeiss Imager M1 fluorescence microscope.
Nuclear import/export assays.
Nuclear import/export assays in yeast cells were performed as described previously (36). Saccharomyces cerevisiae strain L40 (22) contains HIS and lacZ genes that are expressed upon binding of the LexA-Gal4AD transcriptional regulators. The pNIA vector codes for a LexA-Gal4AD fusion to which the test protein is fused; if the test protein drives the fusion to the nucleus, then the reporter genes are expressed. L40 cells were transformed by the lithium acetate-polyethylene glycol method (12) with the pNIA/pNEA derivatives. Transformants were grown in tryptophan dropout minimal medium (DO-trp) for plasmid selection. Individual transformants were picked from these plates and grown on liquid DO-trp overnight at 30°C, and 20-μl portions of these cultures were streaked onto DO-trp plates as positive controls and on histidine dropout minimal medium plates (DO-his) to select for HIS expression. The β-galactosidase activity was assayed with X-Gal on permeabilized yeast cells transferred to nitrocellulose filters, as described previously (22).
Random mutagenesis.
Random mutagenesis of full-length TrwC was performed to select for TrwC mutants in pCMS9 entering the yeast nucleus (and thus growing in DO-his medium when introduced in yeast strain L40). Mutations were introduced by mutagenic PCR using a GeneMorph II random mutagenesis kit (Pharmacia) according to the manufacturer's recommendations. We adjusted the reactions to obtain 0 to 3 mutations per kb. Template DNA was pCMS9. Oligonucleotides used were GGCTGGCGGTTGGGGGTTA, annealing in lexA toward the beginning of trwC, and ACCGAATTCACCTTCCGGCCTCCATGCCGCG, annealing at the end of trwC (the sequence complementary to the trwC stop codon is indicated in boldface, and the EcoRI site is underlined). Full-length trwC from the PCR amplified products was obtained by digestion with the enzymes BamHI and EcoRI and cloned into the corresponding sites of pCMS10 (replacing the C774 TrwC fragment, which does not enter the yeast nucleus). In order to maximize the transformation efficiency, ligations containing the pool of mutants were first introduced in E. coli DH5α, and then DNA was extracted from the pool of colonies and introduced into the yeast strain in successive transformation events. About 4,000 colonies were collected from DH5α, from which 34 μg of DNA was extracted and used to transform competent L40 yeast cells, carrying the reporter HIS gene; 1 to 2 μg of DNA was used per L40 aliquot. Transformations were incubated in DO-trp medium for 8 h at 30°C and then 1/10 plated on DO-trp plates (to calculate transformation efficiency), and the rest were plated on DO-his plates. Plasmid DNA was extracted from His+ colonies using a yeast DNA extraction kit (Thermo Scientific) and introduced into DH5α for further analysis.
Western blots.
In order to detect expression of TrwC fusions in yeast, the cells were lysed and processed as follows. A total of ∼108 yeast L40 cells containing the pNIA derivatives were collected by centrifugation and frozen on dry ice. The pellet was then thawed at room temperature and resuspended in 300 μl of 20% trichloroacetic acid (TCA). Next, 300 μl of glass beads (425 to 600 μm; Sigma) was added to the resultant suspension, followed by vortexing for 1 min in order to lyse the cells. The extract was removed to another tube, the glass beads were washed with 300 μl of 5% TCA, and this second extract was added to the first. The mixture of both extracts was centrifuged at 3,000 rpm for 10 min, and the pellet, containing the proteins, was resuspended in 200 μl of 1× “high-pH” Laemmli buffer (ordinary Laemmli sample buffer plus 150 μl of 1 M Tris base and 1 ml of buffer). The samples were boiled for 4 min and centrifuged again at 3,000 rpm for 10 min. Protein samples were transferred to a new tube, discarding the pellets. Then, 5 to 10 μl portions of the samples were subjected to SDS-10% PAGE. The proteins were transferred to nitrocellulose filters, which were blocked and processed as described previously (9). Anti-GAL4-AD antibody (Sigma-Aldrich) for detection of the fusion proteins and secondary antibody (peroxidase-conjugated anti-rabbit IgG from ICN) were used at a 1:10,000 dilution. Detection was performed by using a Supersignal kit (Pierce), and bands were analyzed on a Chemi-Doc apparatus (Bio-Rad). To detect TrwC expression in human cells, 293T cells were grown and transfected as explained for the immunofluorescence analyses, centrifuged, and washed with phosphate-buffered saline, and the pellets were kept at −80°C. About 106 cells were lysed with 30 μl of NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8], 20 mM NaF, 1% NP-40, 1 mM Na3VO4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, and 10 μg of leupeptin/ml). Portions (10 μl) of supernatant were kept for protein quantitation by the Bradford assay. The rest was mixed with an equal volume of 2× Laemmli buffer. Then, 10- to 20-μl samples were boiled, subjected to SDS-PAGE, transferred, and detected as described above, except for the primary antibody anti-TrwC (15), which was used at a 1:10,000 dilution.
Database search.
We searched for sequences homologous to the R388 nic sequence in the human genome by using the “Somewhat Similar Sequences (blastn)” tool against the “human genomic + transcript” NCBI BLAST database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The search was performed using the 16-nucleotide sequence 14+2, corresponding to 14 nucleotides 5′ to the nic site plus 2 nucleotides 3′ to the nic site (see Fig. 2a).
RESULTS
TrwC mediates site-specific recombination between bacterial and human DNA sequences.
The minimal region described as a target for in vivo TrwC-mediated recombination is 14+3 at oriT1 (7). The shortest oligonucleotide that includes determinants for TrwC binding and nicking is 14+2. This segment is also enough to confer basic oriT function (17, 33) (Fig. 1b). We have searched for putative TrwC target sequences in the human genome (Fig. 2a). No perfect 14+2 matches were obtained. The search returned matches corresponding to shorter sequences 12+2 (four times) and 13+1 (once). A short “core” sequence, corresponding to the sequence 10+2, was found >50 times. We repeated the search, allowing for one mismatch within the 14+2 sequence. Seven hits were returned (Fig. 2a). All matches corresponded with either nonannotated or intronic regions of the human genome. The top five human matches shown in Fig. 2a have a mismatch out of the 6+2 region, which is critical for TrwC nicking activity.
It is predicted that TrwC binding will be a prerequisite for targeted integration. We have also shown that TrwC-mediated site-specific recombination reaction requires the nicking and strand-transfer abilities of TrwC, which can be observed in vitro on oligonucleotides containing its target sequence (32). In order to determine whether the sequences found in the human genome could be targets for incoming TrwC-DNA, we performed TrwC binding and strand-transfer reactions with labeled oligonucleotides, including some of the sequences shown in Fig. 2a. We selected two sequences, HuX and Hu5, corresponding to 15+3 sequences carrying a single mismatch in the TrwC binding site. Both putative human targets in chromosomes X and 5 were bound by TrwC roughly with the same affinity as the R388 sequence (Fig. 2b), and both acted efficiently as acceptors of the cut R388 strand donated by TrwC (Fig. 2c).
We tested several of the human sequences as TrwC targets in in vivo site-specific recombination assays. We designed primers amplifying ∼400-bp fragments from the human genome and containing the putative target sequences. We cloned these sequences at the oriT1 locus in the recombination substrate, which carried a wild-type R388 oriT copy at the oriT2 locus. Recombination was assayed as described previously (7). In order to confirm the requirement of an intact nic site for recombination, we tested a substrate carrying a full-length oriT1 but carrying a mutation within the nic site, TCT/A to GAG/A. No recombination was observed (Fig. 3, compare the top and bottom left panels). Constructs containing the human sequences at oriT1 were compared to that containing a minimal oriT1 with the canonical 14+3 R388 sequence. The results are shown in Fig. 3. TrwC is capable of catalyzing recombination between a copy of its cognate oriT and 15+3 sequences containing a single mismatch in the TrwC binding site (top panels 3 and 4), with roughly the same proficiency as with the canonical 14+3 sequence (top of panel 2). None of the constructs containing shorter consensus sequences (12+3, 13+1, or 10+2) behaved as substrates for TrwC-mediated recombination (Fig. 3, bottom panels). Thus, there are a few sequences in the human genome which are targets for TrwC-mediated site-specific recombination, while sequences deviating from the consensus for more than 1 bp or not preserving the nic site are not substrates for recombination.
FIG. 3.
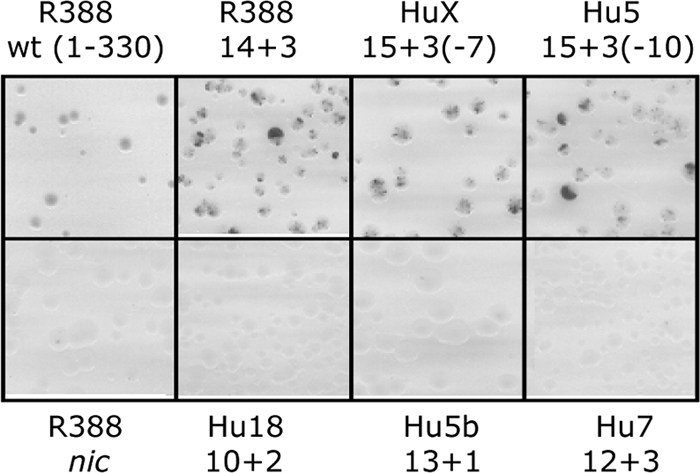
Typical colonies obtained with recombination substrates containing the indicated sequences (as shown in Fig. 2a) cloned at oriT1. oriT2 remains invariable and contains the full-length original R388 oriT. Recombination assays were performed in DH5α strain in the presence of a plasmid coding for both TrwA and TrwC.
Localization of TrwC in human cells.
In order to act as a site-specific integrase in higher organisms, TrwC must reach the nucleus by active transport. Thus, we addressed a study of TrwC localization in eukaryotic cells. Although there is no widely accepted consensus for NLS, two basic clusters of residues form the most accepted bipartite NLS: KRX(10-12)K(K/R)X(K/R) (11). The second cluster on its own can function as a monopartite NLS. TrwC contains two putative NLSs in its sequence: residues 53 to 56 (KRFR) and residues 171 to 190 (KR-X14-KRTR), both within the relaxase domain.
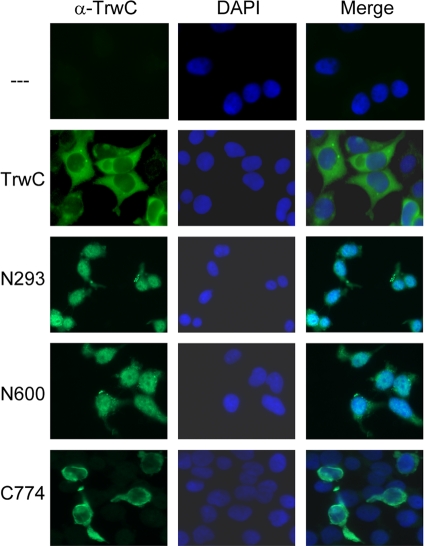
We cloned DNA fragments coding for the relaxase (N293), the recombinase (N600), and the helicase (C774) domains of TrwC, all known to produce stable proteins products (7, 32), and full-length TrwC, into vector pCEFL under the control of the eukaryotic constitutive promoter EF1α. We transfected human 293T cells with these plasmids and confirmed by Western blotting that the proteins were being stably produced in the cell (data not shown). No apparent cytotoxic effect was observed in the cells after 24 h of trwC overexpression. We performed immunofluorescence microscopy to show the cellular distribution of each TrwC domain (Fig. 4). The relaxase and recombinase domains of TrwC (N293 and N600) located preferentially to the nucleus, while the C-terminal helicase domain was cytoplasmic, suggesting that the putative NLSs were functional. However, the localization of full-length TrwC was clearly cytoplasmic.
FIG. 4.
Localization of TrwC and derivatives in human cells. Immunofluorescence images of 293T cells transduced with plasmids coding for TrwC or the indicated TrwC segments. Images are shown at ×40 magnification.
Nuclear import/export assays in yeast.
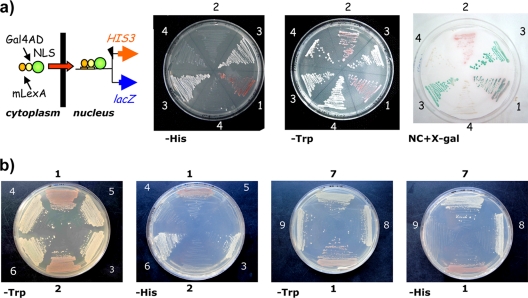
In order to corroborate the above results and to try to understand why full-length TrwC does not localize to the nucleus while shorter N-terminal fragments do, we used a yeast-based genetic assay to look for nuclear import/export signals (36). In the import assay, test proteins are fused to LexA-Gal4AD; if the test protein enables nuclear import, LexA-Gal4AD turns on reporter genes in the yeast strain L40, which are detected by expression of β-galactosidase and/or growth in DO-his plates. We cloned DNA fragments coding for the different TrwC domains into the pNIA vector. To verify that the fusion protein LexA-Gal4AD-TrwC was functional, we confirmed that plasmid pCMS9 (pNIA::trwC) efficiently complemented plasmid pSU1445 (R388 TrwC−) with a similar transfer frequency as a plasmid coding for wild-type TrwC (Table 3). We transformed the constructs into L40 yeast cells and performed Western blots with anti-Gal4AD antibody; the fusion proteins were detected in comparable amounts (data not shown). Controls for the import assay were proteins VirD2, which enters yeast and animal nuclei, and VirE2, which enters only the plant cell nucleus (18, 23). The results (Fig. 5) indicate that the relaxase and recombinase domains of TrwC enter the nucleus, since they drive the fused LexA-Gal4AD to their nuclear targets, leading to expression of HIS and growth on DO-his plates, while growth was not observed with either the fused helicase domain or full-length TrwC, correlating with the results obtained in human cultured cells.
TABLE 3.
Transfer frequencies of R388 trwC mutant pSU1445 when complemented by different TrwC derivativesa
| Complementation type and plasmid | TrwC protein | Transfer frequencyb |
|---|---|---|
| Complementation by TrwC fusion proteins | ||
| pSU1483 | TrwC | 1.0 × 10−1 |
| pCMS9 | LexA-Gal4AD-TrwC | 4.0 × 10−1 |
| pCMS15 | LexA-NLS-Gal4AD-TrwC | 6.0 × 10−1 |
| pCMS5 | LexA-Gal4AD-TrwC-NLS | 4.0 × 10−1 |
| pMTX720 | LexA-Gal4AD-NLSx2-TrwC | 1.1 × 100 |
| pMTX726 | LexA-Gal4AD-NLS-TrwC | 1.3 × 100 |
| pLA44 | LexA-Gal4AD-TrwC mut | 1.3 × 10−1 |
| pNIA3b | None | <10−7 |
| Relaxase-helicase intragenic complementation | ||
| pCIG1086+pSU1534 | TrwC + C774 | 2.1 × 10−1 |
| pLA35+pSU1534 | N293 + C774 | 1.1 × 10−5 |
| pCIG1070+pSU1534 | N450 + C774 | 1.1 × 10−5 |
| pCIG1103+pSU1534 | N600 + C774 | 3.1 × 10−5 |
| pSU1534 | C774 | <10−7 |
Matings were performed as explained in Materials and Methods. Donor strains carried plasmid pSU1445 (R388 TrwC−) and the plasmid(s) indicated in the first column, which code for the TrwC derivatives indicated in the second column.
Transfer frequencies are expressed as the number of transconjugants per donor cell. The results represent the means of three to nine independent experiments.
FIG. 5.
Yeast nuclear import/export assays. (a) Scheme of the yeast nuclear import assay. The test protein (green circle) is fused to transcriptional regulators, which turn on reporter genes if driven into the nucleus, thus allowing the expression of HIS3 and lacZ. Selection is illustrated in the right panels. HIS3 expression allows growth in histidine-deficient medium (−His), and lacZ is monitored by β-galactosidase activity in cells transferred to nitrocellulose filters with X-Gal (NC+X-Gal). (b) Results of the nuclear import (left two panels) and export (right two panels) assay with TrwC and derivatives. The numbers on the plates in panels a and b represent the following: 1, pNIA:VirD2; 2, pNIA:E2; 3, pNIA:N293; 4, pNIA:C774; 5, pNIA:TrwC; 6, pNIA:N600; 7, pNEA:VirE2; 8, pNEA:TrwC; and 9, pNEA:C774.
The cytoplasmic localization of TrwC could be due to the lack of a functional NLS, but this is surprising considering that the N-terminal derivatives locate to the nucleus. Another option would be the presence of a sequence acting as a nuclear export signal (NES) within the helicase domain of the protein, which would redirect the protein back into the cytoplasm. An analysis of the TrwC sequence using NetNES (CBS; Technical University of Denmark), which predicts leucine-rich NESs in eukaryotic proteins (25) (http://www.cbs.dtu.dk/services/NetNES/), rendered no putative NES candidates; however, there is no general consensus for NES, since export pathways other than the CMR1-mediated pathways are known to exist (20). We performed the related yeast nuclear export assay (36), implemented to detect the reverse protein transport into the cytoplasm. To this end, the pNEA vector, containing an NLS fused to LexA-Gal4AD, was used. By default, there will be nuclear localization of the transcriptional activators and HIS expression. Fusion of an NES expressing protein would result in the transport of the protein back into the cell cytoplasm and lack of expression of the reporter gene. We cloned both full-length TrwC and the helicase domain C774 in this plasmid. We checked that pCMS15 (pNEA:trwC) fully complemented the R388 TrwC-deficient plasmid in conjugation (Table 3). The results (Fig. 5b, right panels) show that fusion to the whole-length TrwC protein resulted in growth, meaning that the NLS from the vector targeted the fusion protein to the nucleus, where it remained; while fusion to the helicase domain impedes growth in DO-his plates, meaning this domain is driving the fusion protein out of the nucleus. Thus, it appears that the localization of TrwC could be at equilibrium between the nucleus (driven by the NLS in the relaxase domain) and the cytoplasm (driven by an NES present in the helicase domain).
Intragenic complementation of TrwC.
N600, the recombinase domain of TrwC, enters the nucleus of yeast and human cells and remains stably within it. From the point of view of its possible application as an integrase for mammalian genomic engineering, it would be interesting to determine whether N600 can be delivered to the recipient cell in vivo. If N600 can be delivered to recipient bacteria by conjugation, it can be assumed that it will be also delivered to recipient human cells by conjugation as described previously (45).
We know that C-terminal deletions of TrwC are not functional in conjugation. However, it was published elsewhere (32) that, when coexpressed in the donor bacteria, a relaxase fragment (N348) and the helicase domain C774 of TrwC could complement each other functionally to substitute TrwC in conjugation, with a frequency 104 lower than the wild-type protein. This N348 fragment is, however, rather unstable (7). We assayed the N600 fragment, together with other TrwC fragments known to be stable, N293 and N450 (7), for their capacity to complement an R388 TrwC-deficient plasmid in conjugation in the presence of the helicase domain. The results in Table 3 indicate that larger fragments N450 and N600 can functionally complement the helicase with the same efficiency as the relaxase fragment N293. It is possible that the N-terminal domain contains part of the secretion signal that would allow the protein to be delivered through the T4SS into the recipient cell but, as shown for most T4SSs, the main determinant for substrate recognition could reside in the C terminus (4). Hence, N600 can be transferred to a recipient cell by conjugation if the helicase domain is provided in the donor cell, albeit with very low efficiency.
TrwC mutants which enter the nucleus.
Since the recombinase domain of TrwC targets efficiently the human nucleus but can be transferred with very low efficiency by conjugation, it would be desirable to obtain a full-length TrwC protein which locates to the nucleus. The results from the yeast nuclear import/export assays suggested that a NES in the C-terminal domain of TrwC was responsible for the cytoplasmic localization of the protein. We reasoned that we could target TrwC to the nucleus in two ways: by adding an extra NLS or by getting rid of the NES.
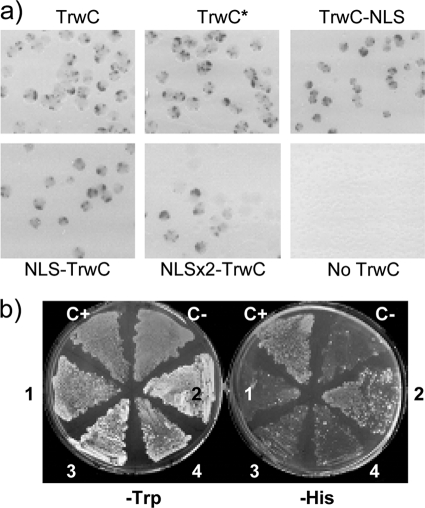
We constructed pNIA:TrwC derivatives carrying either an extra NLS at the C terminus or several NLSs at its N terminus by the addition of oligonucleotides coding for the peptide IPKKKRKV in phase with trwC (see the description of plasmid constructions in Materials and Methods and Table 2). Cloning of the insert gave rise to constructs containing one, two, or five NLSs in frame with the N terminus of TrwC. By Western blotting, we checked that NLS-TrwC levels were similar to the wild type, except for the construct with five NLSs, which was not detected (not shown), and thus we did not use it further. All plasmids complemented R388 (TrwC−) with high efficiency (Table 3). These TrwC derivatives were also tested for their ability to catalyze site-specific recombination. As can be observed in Fig. 6a, the LexA-Gal4AD-TrwC fusion protein is recombination proficient (top panel 1), and the addition of an NLS to the C terminus of TrwC does not affect its recombination capacity (compare the top panels 1 and 3; 100% colonies with blue sectors); however, progressive addition of an NLS to the N terminus of TrwC affects its recombinase activity (Fig. 6a, bottom panels), from a mild effect when a single NLS is added (more than 70% colonies showing blue sectors, albeit smaller than in the wild type) to a stronger effect with two N-terminal additional NLS (ca. 40% colonies showing small blue sectors). When these plasmids were introduced into L40 yeast cells to test their cellular localization, we found that none of them showed nuclear localization, as indicated by the lack of growth in DO-his plates (Fig. 6b and data not shown).
FIG. 6.
TrwC mutants for nuclear targeting. (a) Recombination assays. DH5α cells containing the recombination substrate pCIG1030 (including trwA) and the plasmid coding for the indicated TrwC fusion derivative were plated on X-Gal containing selective media. TrwC* refers to the TrwC mutant coded by plasmid pLA44. (b) Yeast nuclear import assays (as in Fig. 5). C+ and C− refer to pNIA:VirD2 and pNIA:VirE2, respectively. The numbers on the plates represent the following: 1, pNIA:TrwC; 2, pNIA:TrwC* (TrwC mutant coded by pLA44); 3, pNIA:TrwC-NLS; and 4, pNIA:NLS(x2)-TrwC.
Our second strategy was to obtain a TrwC mutant in the NES sequence which would show nuclear localization while probably maintaining its function in conjugation and recombination. We performed random mutagenesis on plasmid pCMS9 (pNIA:trwC), selecting for mutants which grew in DO-his plates, as explained in Materials and Methods. In total, of ca. 70,000 colonies screened, we obtained six His+ colonies. Three of the six plasmids had no trwC insert, so they represented recircularized pNIA3b molecules (which code for a LexA-Gal4AD fusion that enters the nucleus by passive diffusion [36]). The other three constructs were introduced in E. coli DH5α harboring pSU1445, a TrwC-deficient R388 derivative, to test TrwC function. Two of them were transfer negative and were no longer characterized. The remaining construct, named pLA44, complemented pSU1445 almost to the same level as pCMS9 (Table 3). The His+ phenotype of pLA44 was confirmed and compared to that of TrwC; it can be observed that the mutant protein allows growth on DO-his plates as the positive control VirD2 (Fig. 6b). We also checked its recombination activity: it can be observed (Fig. 6a, top of panel 2) that the mutant TrwC protein is as capable as wild-type TrwC in catalyzing oriT-oriT recombination.
The trwC insert was sequenced to determine the TrwC mutations responsible for the His+ phenotype: it carried a T-to-G and two G-to-A mutations at positions 1986, 2529, and 2710 of the trwC open reading frame, giving rise to two silent mutations and the missense mutation Ala904-Thr in TrwC, respectively. In addition, presumably due to a defective oligonucleotide molecule, a CG dinucleotide was missing at position 2893 of trwC, producing a sense mutation which added to the trwC open reading frame a tail from the vector sequence, giving rise to the extra peptide KVNSCSHGSSRSTRD after residue 964 of TrwC. In order to determine which of the two mutations was responsible for nuclear localization, a plasmid was constructed carrying only the missense mutation in TrwC (pLA66, Table 2); this protein did not enter the nucleus according to the lack of growth in DO-his plates (data not shown).
In conclusion, we obtained a TrwC mutant that is fully proficient in conjugation and recombination and targets the nucleus, which will presumably be a useful biotechnological tool for genomic engineering of human cells.
DISCUSSION
We previously reported that protein TrwC of the conjugative plasmid R388 is capable of promoting site-specific recombination between short oriT sequences (6, 7) and also to catalyze site-specific integration upon conjugative transfer into a resident oriT copy (10). In the present study, we describe the ability of TrwC to mediate recombination on specific putative human target sequences, while discriminating among more frequent ubiquitous sequences. We also report the competence of TrwC recombinase domain to enter the human nucleus and the possibility to transport this domain to the recipient cell when complemented intragenically with the helicase domain of the protein in conjugation assays. Finally, we succeeded in obtaining a full-length TrwC mutant that targets the nucleus while it maintains its function in conjugation and recombination. Overall, these results emphasize the features of TrwC as a site-specific integrase that could be of potential biotechnological use for the genetic manipulation of human cells.
TrwC is structurally related to AAV-Rep protein (17, 21), which catalyzes the integration of the single-stranded viral genome into a unique human sequence homologous to the viral origin of replication, containing the 16-bp Rep binding motif and the 6-bp nicking site (26). In a similar way, TrwC could catalyze the integration of incoming ssDNA into target sites present in the genome of the recipient cell; such putative targets would be short DNA sequences resembling the R388 nic site. We searched the human genome database, and we found no matches containing the precise R388 14+2 sequence. Sequences 12+3 or 13+1 were not targets for TrwC-mediated recombination with a wild-type R388 oriT copy (Fig. 3, bottom panels), underscoring the sequence specificity of TrwC. A substrate containing either sequences 15+3 (−7) on chromosome X or 15+3 (−10) on chromosome 5, with the mismatch lying outside of the essential nicking region, yielded a recombination efficiency similar to that of the R388 14+3 sequence (Fig. 3, top panels). TrwC-mediated recombination is affected by vector DNA replication and by local DNA topology (6, 7), so DNA packaging in the human genome may well affect TrwC-mediated integration of foreign DNA; it has been shown that the ability of Tn7 to transpose to a putative human target site is decreased in in vitro-assembled nucleosomes (24). Integration assays in human cells will be required to determine whether any of the above mentioned human sequences works as a target for TrwC-mediated integration of foreign DNA and whether this integration has any deleterious effect on human cell physiology; although the putative targets do not affect any known coding sequence, the Hu5 target lies in an intronic region, which could affect correct splicing of the MCC gene.
In order to integrate exogenous DNA into these specific sites of the human genome, the protein must reach the nucleus. Although the nuclear membrane is not a permanent barrier, it has been reported that the addition of an NLS to an integrase increases its integration frequency in eukaryotes (8). TrwC or its functional domains were expressed in human cell lines, and their localization was confirmed by immunofluorescence analysis (Fig. 4): the relaxase and recombinase domains of TrwC showed nuclear localization, supporting the existence of a functional NLS in the N-terminal domain of TrwC. Similar results were obtained in a yeast-based assay (Fig. 5), where TrwC fragments were expressed fused to transcriptional regulators, thus discarding nuclear entrance by passive diffusion. However, the whole-length protein showed cytoplasmic localization in both assays. The results of the yeast nuclear export assay suggested the presence of an NES-like sequence in the helicase domain of the protein (Fig. 5), which could transport TrwC back into the cytoplasm. We obtained by random mutagenesis a TrwC mutant that targets the nucleus while retaining full activity in conjugation and recombination, which is of potential interest for future experimentation on TrwC activity as a site-specific integrase in mammals. The mutant carried a missense A904T mutation close to the C terminus and a sense mutation which added a peptide tail to the protein KVNSCSHGSSRSTRD. Since a plasmid carrying only the A904T mutation did not enter the nucleus, the added tail is responsible for nuclear localization of TrwC, presumably by affecting an NES not belonging to any defined consensus.
The addition of NLSs to either the N or the C terminus of TrwC did not target the protein to the nucleus. Curiously, the progressive addition of the NLS to the N terminus of the protein did not alter its function in conjugation but affected its recombinase activity (Table 3 and Fig. 6). This is the first time that recombinase activity can be separated from TrwC function in conjugation. This different effect could be due to a higher demand of TrwC for recombination than conjugation; in conjugation, we have observed repeatedly that very small amounts of TrwC are enough to efficiently mobilize DNA. Fusion proteins could alter TrwC function, and yet this would not affect conjugation due to the high number of TrwC molecules in the cell, whereas recombination would be affected. A similar effect has been recently reported for conjugative coupling protein TrwB (9): to observe the effect of certain TrwB mutants in conjugation, a TrwB-limiting mating assay had to be used. Otherwise, mild phenotypes are masked by the high number of TrwB molecules present in the cell.
Recombinases of bacterial or viral origin have been widely used for mammalian genomic modification (42). Among them, the only integrases known to work on naturally existing human targets are the TrwC-related AAV-Rep protein and integrases of the phiC31 family; these proteins pose several problems for genome modification, such as their low sequence specificity, viral limited DNA packaging capacity, or toxicity of the expression of the integrase in human cells (35). These problems could be overcome by the system we propose, based on in vivo TrwC-DNA transfer to mammalian cells by bacterial conjugation (45). The obtained TrwC nuclear mutant could mediate site-specific integration of the incoming foreign DNA into specific human targets. Conjugation is a processive mechanism that imposes no limit on the DNA length that could be transferred into human cells. trwC overexpression does not seem to affect cell viability or morphology; even more importantly, since TrwC is delivered to the recipient cell attached to the DNA, there is no need for trwC expression in the recipient cell, thus avoiding toxicity problems. Finally, conjugative relaxases show high sequence specificity. In the future, a bank of mutant relaxases could be obtained that target different sequences. In fact, the first such TrwC mutants have already been obtained (13). We are also conducting a mutagenesis analysis of TrwC-related relaxases to obtain recombinase-proficient mutants that could be similarly used on other possible targets present in human and mammalian genomes.
Acknowledgments
We are grateful to Vitaly Citovsky for providing the yeast strain L40 and pNIA/pNEA plasmids for the nuclear import/export assays.
This study was supported by grant BIO2008-00133 from the Spanish Ministry of Education to M.L. and grant FIS08/29 from the Spanish Instituto de Salud Carlos III to M.D.D.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 2.Bravo-Angel, A. M., V. Gloeckler, B. Hohn, and B. Tinland. 1999. Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J. Bacteriol. 181:5758-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan-Wollaston, V., J. E. Passiatore, and F. Cannon. 1987. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328:172-175. [Google Scholar]
- 4.Cambronne, E. D., and C. R. Roy. 2006. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic 7:929-939. [DOI] [PubMed] [Google Scholar]
- 5.Campos-Olivas, R., J. M. Louis, D. Clerot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. U. S. A. 99:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.César, C. E., and M. Llosa. 2007. TrwC-mediated site-specific recombination is controlled by host factors altering local DNA topology. J. Bacteriol. 189:9037-9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.César, C. E., C. Machón, F. de la Cruz, and M. Llosa. 2006. A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol. Microbiol. 62:984-996. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., and S. L. Woo. 2005. Complete and persistent phenotypic correction of phenylketonuria in mice by site-specific genome integration of murine phenylalanine hydroxylase cDNA. Proc. Natl. Acad. Sci. U. S. A. 102:15581-15586. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.de Paz, H. D., D. Larrea, S. Zunzunegui, C. Dehio, F. de la Cruz, and M. Llosa. 2010. Functional dissection of the conjugative coupling protein TrwB. J. Bacteriol. 192:2655-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper, O., C. E. César, C. Machón, F. de la Cruz, and M. Llosa. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. U. S. A. 102:16385-16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontes, M. R., T. Teh, D. Jans, R. I. Brinkworth, and B. Kobe. 2003. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 278:27981-27987. [DOI] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and R. A. Woods. 2001. Genetic transformation of yeast. Biotechniques 30:816-828. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Perez, B., J. D. Carballeira, G. Moncalian, and F. de la Cruz. 2009. Changing the recognition site of a conjugative relaxase by rational design. Biotechnol. J. 4:554-557. [DOI] [PubMed] [Google Scholar]
- 14.Grandoso, G., P. Avila, A. Cayón, M. A. Hernando, M. Llosa, and F. de la Cruz. 2000. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 295:1163-1172. [DOI] [PubMed] [Google Scholar]
- 15.Grandoso, G., M. Llosa, J. C. Zabala, and F. de la Cruz. 1994. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur. J. Biochem. 226:403-412. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guasch, A., M. Lucas, G. Moncalián, M. Cabezas, R. Pérez-Luque, F. X. Gomis-Rüth, F. de la Cruz, and M. Coll. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 10:1002-1010. [DOI] [PubMed] [Google Scholar]
- 18.Guralnick, B., G. Thomsen, and V. Citovsky. 1996. Transport of DNA into the nuclei of Xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell 8:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinemann, J. A., and G. F. Sprague, Jr. 1989. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340:205-209. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 21.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus. Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 22.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard, E. A., J. R. Zupan, V. Citovsky, and P. C. Zambryski. 1992. The VirD2 protein of Agrobacterium tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68:109-118. [DOI] [PubMed] [Google Scholar]
- 24.Kuduvalli, P. N., R. Mitra, and N. L. Craig. 2005. Site-specific Tn7 transposition into the human genome. Nucleic Acids Res. 33:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527-536. [DOI] [PubMed] [Google Scholar]
- 26.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U. S. A. 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llosa, M., S. Bolland, and F. de la Cruz. 1991. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol. Gen. Genet. 226:473-483. [DOI] [PubMed] [Google Scholar]
- 28.Llosa, M., S. Bolland, G. Grandoso, and F. de la Cruz. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J. Bacteriol. 176:3210-3217. (Erratum, 176:6414.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Llosa, M., F.-X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Llosa, M., G. Grandoso, and F. de la Cruz. 1995. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J. Mol. Biol. 246:54-62. [DOI] [PubMed] [Google Scholar]
- 32.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 33.Lucas, M., B. Gonzalez-Perez, M. Cabezas, G. Moncalian, G. Rivas, and F. de la Cruz. 2010. Relaxase DNA binding and cleavage are two distinguishable steps in conjugative DNA processing that involve different sequence elements of the nic site. J. Biol. Chem. 285:8918-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelczar, P., V. Kalck, D. Gomez, and B. Hohn. 2004. Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic T-DNA complexes in mammalian cells. EMBO Rep. 5:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recchia, A., and F. Mavilio. 2006. Site-specific integration into the human genome: ready for clinical application? Rejuvenation Res. 9:446-449. [DOI] [PubMed] [Google Scholar]
- 36.Rhee, Y., F. Gurel, Y. Gafni, C. Dingwall, and V. Citovsky. 2000. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 18:433-437. [DOI] [PubMed] [Google Scholar]
- 37.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 38.Sadler, J. R., M. Tecklenburg, and J. L. Betz. 1980. Plasmids containing many tandem copies of a synthetic lactose operator. Gene 8:279-300. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Silby, M. W., G. C. Ferguson, C. Billington, and J. A. Heinemann. 2007. Localization of the plasmid-encoded proteins TraI and MobA in eukaryotic cells. Plasmid 57:118-130. [DOI] [PubMed] [Google Scholar]
- 41.Smith, R. H., and R. M. Kotin. 2002. Adeno-associated virus, p. 905-923. In N. L. Craig, R. Craigie, M. Gelert, and A. M. Lambowithz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 42.Sorrell, D. A., and A. F. Kolb. 2005. Targeted modification of mammalian genomes. Biotechnol. Adv. 23:431-469. [DOI] [PubMed] [Google Scholar]
- 43.Teramoto, H., P. Crespo, O. A. Coso, T. Igishi, N. Xu, and J. S. Gutkind. 1996. The small GTP-binding protein rho activates c-Jun N-terminal kinases/stress-activated protein kinases in human kidney 293T cells. Evidence for a Pak-independent signaling pathway. J. Biol. Chem. 271:25731-25734. [DOI] [PubMed] [Google Scholar]
- 44.Torrano, V., J. Navascues, F. Docquier, R. Zhang, L. J. Burke, I. Chernukhin, D. Farrar, J. Leon, M. T. Berciano, R. Renkawitz, E. Klenova, M. Lafarga, and M. D. Delgado. 2006. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J. Cell Sci. 119:1746-1759. [DOI] [PubMed] [Google Scholar]
- 45.Waters, V. L. 2001. Conjugation between bacterial and mammalian cells. Nat. Genet. 29:375-376. [DOI] [PubMed] [Google Scholar]
- 46.Williams, S. L., and J. F. Schildbach. 2006. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 34:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zupan, J., D. Ward, and P. Zambryski. 2002. Inter-kingdom DNA transfer decoded. Nat. Biotechnol. 20:129-131. [DOI] [PubMed] [Google Scholar]