Abstract
Tangential flow ultrafiltration (UF) was used to concentrate and recover bacterial indicators and enteric viruses from 100 liters of groundwater (GW; n = 10) and surface water (SW; n = 11) samples collected in Lower Yakima Valley, WA. Human and bovine enteric viruses were analyzed in SW and GW concentrates by real-time PCR by using integrated inhibition detection.
Microbial contamination of water poses a significant risk to public health. Pathogens associated with waterborne illnesses are excreted in the feces of humans and animals and transmitted via the direct consumption of water, recreational activities, consumption of shellfish (e.g., oysters), or ingestion of fresh market crops (i.e., fruits and vegetables) contaminated via irrigation water (22, 23). Water can become contaminated by pathogen-containing fecal pollution through point sources such as municipal sewage treatment plant discharges and concentrated animal feeding operations (CAFO) as well as nonpoint sources, including agricultural and urban runoff, wildlife, aging sewage collection infrastructure, and faulty septic systems (25).
The ability to reliably identify and track sources of fecal contamination to their origin is important for the management and mitigation of nonpoint source pollution. Water quality parameters that are traditionally assessed include nitrogen, phosphorus, and concentrations of fecal indicator bacteria (i.e., Escherichia coli and enterococci). However, elevated nutrient loads and fecal bacteria in surface water (SW) and groundwater (GW) do not indicate a single source of pollution, since numerous point and nonpoint sources, including animal wastes, poorly maintained septic systems, land application of biosolids, and discharge from municipal sewage treatment plants, may all be contributors to nutrient and fecal indicator bacteria levels (5). Recent strategies for tracking fecal contamination have expanded to encompass more than just these basic indicators. Of increasing interest is the assessment of host-specific enteric viruses for tracking fecal pollution. More than 100 types of pathogenic viruses are excreted in human and animal wastes and can enter the water environment and persist for extended periods of time (22). Enteric viruses are shed at extremely high concentrations in the feces of infected hosts and at somewhat lower concentrations in a proportion of healthy host populations (19). In addition, enteric viruses predominantly infect a single host species (i.e., swine, bovine, human). These characteristics of enteric viruses make them ideal candidates for source tracking of fecal contamination.
Prior to this study, an ultrafiltration (UF) method was optimized in our laboratory for the concentration and recovery of bacteria (E. coli and enterococci), protozoan surrogates (Clostridium perfringens spores), viral surrogates (murine norovirus 1 [MNV-1] and bacteriophages PRD1 and MS2), and human enteric viruses from 100-liter dechlorinated drinking water and SW samples (K. E. Gibson and K. J. Schwab, submitted for publication). The primary goal of the present study was to apply UF for the concentration of bovine enteric viruses, including bovine enterovirus (BoEV) and bovine norovirus (BoNoV), and human enteric viruses, including human norovirus (HuNoV), enterovirus (EV), human adenovirus (HuAdV), and human polyomavirus (HuPyV), from 100-liter SW and GW samples in Lower Yakima Valley, WA. The Lower Yakima Valley has a history of GW and SW pollution issues (i.e., elevated nitrate and nutrient levels) arising primarily from nonpoint source agricultural runoff of land-applied animal (bovine) wastes and inorganic fertilizers (27). In addition, MNV-1, a novel surrogate for the study of HuNoVs, was evaluated as an internal method control for assessing the recovery efficiency of enteric viruses by UF in a field setting. Sample inhibition was also systematically evaluated during molecular analyses to control for potential false negatives. To our knowledge, only one study has applied a comparable UF method for the recovery and concentration of indicator bacteria and viruses (adenovirus) from 100-liter GW samples in situ (14). However, no studies have reported the use of UF for the recovery and molecular detection of both animal and human enteric viruses from 100-liter GW and SW samples with inclusion of inhibition analysis and sample volume back-calculations.
Study area.
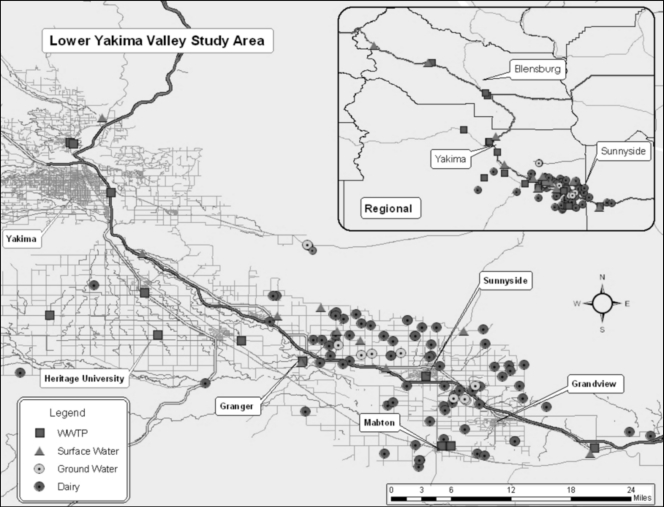
Lower Yakima Valley is located in the south central part of Washington. The primary economic activity in the region is agriculture, comprising 70 to 80% of land use (27). Sixty-one dairy CAFO containing approximately 290,000 animal units (214,600 milk cows) are concentrated in this region, largely in and around the cities of Sunnyside, Grandview, and Granger (Fig. 1) (27). There are 83 community water systems (CWS), which serve an estimated 47,000 people year-round within Yakima County. Each of the larger communities in Lower Yakima Valley, including Sunnyside, Granger, and Grandview, is served by a CWS that relies on treated groundwater for drinking water. The remaining population of about 34% resides in rural, unincorporated areas and relies on private groundwater wells for drinking water. In the entire Yakima River basin, there are more than 20,000 wells, of which 70% are shallow, 10- to 250-foot-deep, domestic wells (28).
FIG. 1.
Lower Yakima Valley study area and sampling sites. WWTP, wastewater treatment plant. (Courtesy of D'Ann L. Williams.)
Sample collection.
Water samples were collected between 24 July 2008 and 4 August 2008 from 10 different GW and 11 different SW sites. All GW and 9 of 11 SW sites were located within a 49-mile corridor between the cities of Prosser and Yakima. The two remaining SW samples were collected upstream of the City of Yakima, near the headwaters of the Yakima River at the Keechelus Dam just south of Snoqualmie Pass, and further downstream, toward the City of Yakima in South Cle Elum. Figure 1 shows the location of each GW and SW site as well as surrounding dairy operations and municipal sewage treatment plants. Site selection was based on convenience and ease of access. At each site 100 liters of water was collected in 25-liter high-density polyethylene water storage containers (Blitz U.S.A., Miami, OK), and water quality parameters were collected using a YSI multiparameter water quality sonde (model 6820 V2) and data system (model 650MDS; YSI Incorporated, Yellow Springs, OH). Groundwater samples were collected prior to any point-of-use household treatment (when present), primarily from outside water spigots. All GW sample sites were flushed for 10 min prior to collection. Samples were transported to local lab facilities at Heritage University in Toppenish, WA, and processed on the same day as collection.
MNV-1 stock production.
MNV-1 stocks were generated in monolayers of RAW 264.7 cells (ATCC TIB-71) as previously described (3). MNV-1 stock titers were determined by plaque assay as described by Bae and Schwab (3) with modifications. Briefly, six-well tissue culture plates were seeded with RAW 264.7 cells at a concentration of 2 × 106 viable cells per well and incubated for 24 h at 37°C with 5% CO2. Viral stock dilutions were prepared in complete Dulbecco's modified Eagle's medium with 2% low-endotoxin fetal bovine serum (Invitrogen) and 2.5 μg/ml amphotericin B antimycotic (Fungizone; Invitrogen), and 200 μl was inoculated into each well. Plates were incubated at 37°C and 5% CO2 for 1 h with continuous rocking followed by removal of the inoculum and application of 2 ml of prepared overlay medium (3). Plates were incubated at 37°C and 5% CO2 for 24 h, and then each well was stained with an additional 2 ml of overlay medium supplemented with 1% neutral red (3.3-g/liter stock solution) (Invitrogen) for visualization of plaques.
Specimens for bovine positive controls.
For BoNoV positive controls, bovine fecal specimens (n = 5) were collected from two separate locations, a dairy operation in Michigan (n = 3) and a calving operation in Virginia (n = 2). Prior to RNA extraction, fecal specimens were prepared in Dulbecco's phosphate-buffered saline (DPBS; Invitrogen) at 10% (wt/vol) and centrifuged at 3,000 × g for 5 min at 4°C in order to clarify the sample. RNA was extracted from 140 μl of the clarified fecal samples by using the QIAamp viral RNA mini kit (Qiagen) following the manufacturer's spin protocol. Total RNA was eluted from the Qiagen spin column by performing a double elution using 40 μl of 2× diethyl pyrocarbonate (DEPC)-treated water supplemented with a 0.01% dilution of a 500-U/μl solution of RNase inhibitor. For the BoEV positive control, BoEV type 7 (ATCC VR-744) RNA was extracted from 140 μl of cell lysate as described for clarified fecal samples. Eluted RNA was stored at −80°C in 20-μl aliquots until analysis. Total extracted RNA samples from the cell lysate and each bovine specimen were analyzed for BoEV and BoNoV, respectively, using the methods described below.
Preparation of water samples.
All four 25-liter water containers collected from each sampling site were combined into a sterilized, 120-liter polypropylene storage container (Sterilite, Townsend, MA). The chemical surfactant sodium polyphosphate (NaPP; Sigma, St. Louis, MO) was added to each sample to achieve a final concentration of 0.01% as previously described (9). Approximately 5 × 106 PFU of prepared MNV-1 stock were added to each sample as a positive control to evaluate the viral recovery efficiency of the UF method in a field setting. Each sample was allowed to equilibrate at room temperature for 30 min prior to UF.
Ultrafiltration setup and procedure.
The ultrafiltration setup was conducted as previously reported (21), with modifications. High-performance, platinum-cured LS/36 and LS/24 silicon tubing (Masterflex; Cole-Parmer Instrument Co., Vernon Hills, IL) was used in each experiment and then reused after disinfection. Disinfection consisted of submersion in a 50 ppm chlorine bleach solution followed by a 5-min rinse with deionized (DI) water. After rinsing with DI water, the tubing was soaked in a 3 M excess sodium thiosulfate (Sigma) solution for neutralization of remaining chlorine, followed by a final rinse with DI water. Polypropylene NS4 quick-disconnect couplings (Colder Products Company, St. Paul, MN), screw clamps, brass fittings, rubber stoppers, and polypropylene storage containers (Sterilite) were disinfected in the same way prior to use in the UF setup and between each water sample. Exceltra Plus 210 dialysis filters (Baxter International, Deerfield, IL) were utilized during UF. New filters were used for each experiment. A Cole-Parmer model 7524-40 peristaltic pump and Masterflex model 77800-52 pump heads were used for processing all samples. Before filtration, ultrafilters were blocked with 0.1% NaPP, and filtration was conducted as described by Polaczyk et al. (21). UF concentrates were stored at 4°C and shipped on ice to Johns Hopkins Bloomberg School of Public Health laboratories in Baltimore, MD, for additional sample processing and analysis.
Analysis of indicator bacteria.
For detection and enumeration of total coliforms and E. coli, the Colilert Quanti-tray system (IDEXX Laboratories, Westbrook, ME) was used to determine the most probable number (MPN) in each sample before and after UF. The Enterolert Quanti-tray system was used to determine the MPN for enterococci in each sample before and after UF. Duplicate 101-ml and 10.1-ml samples were collected from prepared samples (i.e., after addition of NaPP and MNV-1) of GW and SW, respectively, prior to filtration in order to analyze for the presence of total coliforms, E. coli, and enterococci. Following filtration, duplicate 10.1-ml and 1.1-ml samples were collected from GW and SW UF concentrates, respectively. Sample volumes of less than 100 ml (10 ml, 1 ml, or 100 μl) were added to 0.1% peptone (Invitrogen) to bring the total volume to 100 ml. A negative control containing 100 ml of 0.1% peptone was analyzed by using Colilert and Enterolert for each batch of samples.
Secondary processing of UF concentrates.
Using Centricon Plus-70 (Millipore) centrifugal filtration devices with molecular weight cutoffs (MWCO) of 30 or 100, 70-ml aliquots of UF concentrates were further concentrated following the manufacturer's protocol. Before secondary concentration, SW samples were preclarified by centrifugation at 5,000 × g for 5 min at 4°C. Supernatant was removed and applied to the Centricon filter unit. The pellet was archived and processed separately during total viral nucleic acid (NA) extraction as described below. Aliquots (200 μl) of the secondary concentrates and pellets were processed by using QIAamp MinElute virus spin kits (Qiagen, Valencia, CA) following the manufacturer's protocol. Total viral NA was eluted from the Qiagen spin column by a double elution using 50 μl of 2× DEPC-treated water supplemented with a 0.01% dilution of a 500-U/μl solution of RNase inhibitor (Applied Biosystems, Foster City, CA). Eluted NA was aliquoted and archived at −80°C until analysis. During total viral NA extraction, a negative-control extraction containing 200 μl DEPC-treated water was also processed to verify that no cross-contamination had occurred.
Real-time PCR and RT-PCR.
Amplification of viral DNA and RNA targets was performed using a Prism 7300 sequence detection system (Applied Biosystems). Total viral NA extracted from UF secondary concentrates was analyzed for BoEV, BoNoV, HuAdV, HuNoV, pan-EV, and HuPyV, including JC virus and BK virus, by real-time PCR or reverse transcription-PCR (RT-PCR). All assays were performed in a 96-well plate format. The sequences and sources of the primers and probes utilized in this study are shown in Table 1 (7, 11-13, 20, 30). All assays were validated using positive controls and negative controls consisting of nontarget NA and DEPC-treated water.
TABLE 1.
Virus primers and probes
| Virus | GenBank accession no. | Primer or probe name | Primer/probe final concn (nM) | Probe label | Sequencec | Product size (bp) | Product region | Reference |
|---|---|---|---|---|---|---|---|---|
| Murine norovirus | AY228235 | MNVKS1 | 400 | 5′-AGGTCATGCGAGATCAGCTT-3′ | 159 | ORF1 | Bae and Schwab (3) | |
| MNVKS2 | 400 | 5′-CCAAGCTCTCACAAGCCTTC-3′ | ||||||
| MNVKS3 | 200 | FAMa | 5′-CAGTCTGCGACGCCATTGAGAA-3′ | |||||
| Human norovirus GI | M87661 | COG1F | 1,000 | 5′-CGYTGGATGCGNTTYCATGA-3′ | 85 | ORF1-ORF2 junction | Kageyama et al. (13) | |
| COG1R | 1,000 | 5′-CTTAGACGCCATCATCATTYAC-3′ | ||||||
| RING 1A | 100 | FAMa | 5′-AGATYGCGATCYCCTGTCCA-3′ | |||||
| RING 1B | 100 | FAMa | 5′-AGATCGCGGTCTCCTGTCCA-3′ | |||||
| Human norovirus GII | AF145896 | COG2F | 1,000 | 5′-CARGARBCNATGTTYAGRTGGATGAG-3′ | 88 | ORF1-ORF2 junction | Kageyama et al. (13) | |
| COG2R | 1,000 | 5′-CGACGCCATCTTCATTCACA-3′ | ||||||
| RING2-TP | 200 | FAMa | 5′-TGGGAGGGCGATCGCAATCT-3′ | |||||
| Human adenovirus | AC_000008 | JTVXF | 400 | 5′-GGACGCCTCGGAGTACCTGAG-3′ | 96 | Hexon region | Jothikumar et al. (12) | |
| (types A to F) | JTVXR | 400 | 5′-ACIGTGGGGTTTCTGAACTTGTT-3′ | |||||
| JTVXP | 150 | FAMa | 5′-CTGGTGCAGTTCGCCCGTGCCA-3′ | |||||
| Human polyomaviruses | AB092584 | SM2 | 500 | 5′-AGTCTTTAGGGTCTTCTACCTTT-3′ | 173 (JC) 176 (BK) | Partial T antigen | McQuaig et al. (20) | |
| (JC and BK) | P6 | 500 | 5′-GGTGCCAACCTATGGAACAG-3′ | |||||
| KGJ3 | 400 | FAMb | 5′-TCATCACTGGCAAACAT-3′ | |||||
| Human enteroviruses | AJ293918 | EV1R | 700 | 5′-TGTCACCATAAGCAGCCA-3′ | 143 | 5′ untranslated region | Gregory et al. (7) | |
| EV1F | 700 | 5′-CCCTGAATGCGGCTAAT-3′ | ||||||
| EV probe | 120 | FAMa | 5′-ACGGACACCCAAAGTAGTCGGTTC-3′ | |||||
| Hepatitis G virus | U44402 | HepG-F | 400 | 5′-CGGCCAAAAGGTGGTGGATG-3′ | 185 | 5′ untranslated region | Lambertini et al. (15) | |
| (internal standard) | HepG-R | 400 | 5′-CGACGAGCCTGACGTCGGG-3′ | |||||
| HepG probe | 200 | FAMa | 5′-AGGTCCCTCTGGCGCTTGTGGCGAG-3′ | |||||
| Bovine enteroviruses | PS87 isolate | BEV-5FL | 500 | 5′-GCCGTGAATGCTGCTAATCC-3′ | 92 | 5′ noncoding region | Jimenez-Clavero et al. (11) | |
| BEV-3FL | 500 | 5′-GTAGTCTGTTCCGCCTCCACCT-3′ | ||||||
| BEV-SON | 250 | FAMa | 5′-CGCACAATCCAGTGTTGCTACGTCGTAAC-3′ | |||||
| Bovine noroviruses | AJ011099 | SW GIII forw | 400 | 5′-CGCTCCATGTTYGCBTGG-3′ | 91 | ORF1-ORF2 junction | Wolf et al. (30) | |
| SW GIII rev | 400 | 5′-TCAGTCATCTTCATTTACAAAATC-3′ | ||||||
| SW GIII probe | 200 | FAMa | 5′-TGTGGGAAGGTAGTCGCGACRYC-3′ |
The 6-carboxyfluorescein (FAM) quencher was BHQ-1 (black hole quencher 1).
The FAM quencher was MGBNFQ (minor groove binder nonfluorescent quencher).
Mixed bases in degenerate primers and probes were as follows: Y = C, T; R = A or G; B = C, T, or G; N = A, C, T, or G.
For viral RNA amplification, each 25-μl reaction mixture contained 12.5 μl of 2× master mix (QuantiTect probe RT-PCR kit; Qiagen), 5 U RNase inhibitor (Applied Biosystems), custom primers (Invitrogen), dually labeled TaqMan probes (Biosearch Technologies, Novato, CA) at the final concentrations listed in Table 1, 5 μl of prepared sample, and DEPC-treated water for the remaining volume. Real-time RT-PCR amplification for five of the assays (for MNV-1, HuNoV GI and GII, pan-EV, and BoEV) was performed under the following conditions: reverse transcription for 30 min at 50°C and then denaturation for 15 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 60 s. Real-time RT-PCR amplification for BoNoV RNA was the same as for the above assay except the annealing/extension was performed at 57°C.
For viral DNA amplification, each 25-μl reaction mixture contained 10 μl of 2× master mix (QuantiTect probe PCR kit; Qiagen), custom primers (Invitrogen), dually labeled TaqMan probes (Biosearch Technologies) at the concentrations listed in Table 1, 5 μl of prepared sample, and DEPC-treated water for the remaining volume. Real-time PCR amplification for HuAdV was performed under the following conditions: denaturation for 15 min at 95°C, followed by 40 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 60 s. Real-time PCR amplification for HuPyV was performed under the following conditions: denaturation for 15 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 60°C for 60 s. Dilutions of sample NA extracts were prepared in DEPC-treated water. All real-time PCR analyses were performed utilizing a positive control for each target and DEPC-treated water as the negative control with each thermocycler run to ensure reagent and cycling efficiencies.
Detection of sample inhibition.
An internal standard for the identification of inhibition in real-time PCR and RT-PCR assays was prepared using hepatitis G virus (HGV) armored RNA (Asuragen, Austin, TX). RNA was extracted from 140 μl HGV armored RNA by using a QIAamp viral RNA mini kit (Qiagen) following the manufacturer's spin protocol. The extracted RNA was then amplified by real-time RT-PCR using a Prism 7300 sequence detection system (Applied Biosystems). Primers and probes for the HGV assay are shown in Table 1, and amplification was performed as described previously (15), with modifications. Each 25-μl reaction mixture was prepared as described above for viral RNA, with the inclusion of 2 μl of a 100-fold dilution of internal standard HGV RNA. Real-time RT-PCR amplification for HGV was performed under the same conditions described above for MNV-1, HuNoV, pan-EV, and BoEV. Each batch of samples assayed for inhibition included a negative control of HGV master mix containing no HGV RNA and at least three positive-control reaction mixtures containing only HGV RNA and no sample. For controls, 5 μl DEPC-treated water was added to bring the reaction volume to 25 μl.
Volume back-calculations and statistical analysis.
For indicator bacteria, the percent recovery efficiency was calculated as the number of microbes recovered after UF divided by the number of microbes before UF, multiplied by 100. Correlation analyses were performed using the Pearson product-moment correlation. Strong correlations were defined as a correlation with a Pearson r value of greater than or equal to 0.6 or less than or equal to −0.6. Volume back-calculations were performed for molecular data in order to estimate the volume of preconcentrated sample analyzed during a given assay.
Water quality.
A total of 21 100-liter SW (n = 11) and GW (n = 10) samples were collected. Water quality parameters were obtained for these samples at the time of sample collection. Tables 2 and 3 display the values for GW and SW, respectively, for each parameter by sample collected.
TABLE 2.
Water quality and bacterial indicator data for the 100-liter GW samplesa
| GW sample no. | Water quality parameters |
Bacterial indicators (log10 MPN/100 ml) |
% RE |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Conductivity (μS/cm) | Turbidity (NTU) | DO (mg/liter) | Nitrate (mg/liter) | Ammonium (mg/liter) | Before UF |
After UF |
||||||
| TC | EC | Entero | TC | EC | Entero | TC | |||||||
| 1 | 7.63 | 4.4 | 6.58 | 7.03 | 0.394 | 1.61 | 1.48 | ||||||
| 2 | 7.63 | 527 | 6.53 | 9.16 | 0.689 | 1.00 | |||||||
| 3 | 7.80 | 334 | 0.3 | 6.66 | 4.42 | 0.733 | 0.93 | 3.27 | 1.48 | >100 | |||
| 4 | 7.41 | 1225 | 0.4 | 7.08 | 59.20 | 0.808 | 2.17 | ||||||
| 5 | 7.63 | 343 | 0.1 | 7.07 | 7.52 | 0.570 | |||||||
| 6 | 7.57 | 617 | 0.4 | 6.77 | 9.57 | 0.451 | 2.76 | ||||||
| 7 | 7.67 | 151 | 0.6 | 9.26 | 0.09 | 0.375 | 1.00 | 1.00 | |||||
| 8 | 7.39 | 305 | 1.0 | 11.55 | 6.19 | 0.485 | 1.72 | ||||||
| 9 | 7.66 | 746 | 0.6 | 9.96 | 13.26 | 0.451 | 0.30 | 2.58 | 51.5 | ||||
| 10 | 7.83 | 857 | 0.2 | 10.42 | 18.99 | 0.532 | 0.72 | 3.61 | 1.49 | >100 | |||
Abbreviations: NTU, nephelometric turbidity unit; DO, dissolved oxygen; TC, total coliforms; EC, E. coli; Entero, enterococci; RE, recovery efficiency.
TABLE 3.
Water quality and bacterial indicator data for the 100-liter SW samplesa
| SW sample no. | Water quality parameters |
Bacterial indicators (log10 MPN/100 ml) |
% RE |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Conductivity (μS/cm) | Turbidity (NTU) | DO (mg/liter) | Nitrate (mg/liter) | Ammonium (mg/liter) | Before UF |
After UF |
||||||||
| TC | EC | Entero | TC | EC | Entero | TC | EC | Entero | |||||||
| 1 | 8.11 | 91 | 10.16 | 0.432 | 0.141 | 3.19 | 1.75 | 1.24 | 5.38 | 3.71 | 3.08 | 50.5 | 29.9 | 22.7 | |
| 2 | 7.19 | 82 | 3.1 | 10.61 | 0.219 | 0.137 | 3.54 | 1.49 | 1.30 | 5.79 | 3.74 | 3.93 | >100 | >100 | >100 |
| 3 | 7.67 | 93 | 7.5 | 10.79 | 0.271 | 0.191 | 3.24 | 1.80 | 2.12 | 5.99 | 4.01 | 4.56 | >100 | 38.5 | 66.2 |
| 4 | 7.69 | 89 | 5.8 | 11.17 | 0.179 | 0.187 | 3.71 | 1.00 | 1.88 | 5.84 | 3.59 | 3.88 | 66.4 | >100 | 50.0 |
| 5 | 7.41 | 48 | 2.7 | 10.38 | 0.416 | 0.119 | 3.27 | 1.30 | 1.30 | 5.91 | 4.14 | 4.77 | 79.9 | >100 | >100 |
| 6b | 7.77 | 96 | 9.6 | 11.18 | 0.399 | 0.120 | 3.66 | 1.87 | 2.62 | 5.59 | 3.67 | 3.64 | 97.2 | 72.7 | 12.2 |
| 7 | 7.80 | 49 | 6.6 | 11.41 | 0.391 | 0.120 | 3.39 | 1.55 | 1.38 | 5.89 | 3.88 | 3.71 | 95.9 | 65.0 | 65.1 |
| 8 | 6.58 | 81 | 4.2 | 11.84 | 0.270 | 0.125 | No test | No test | 1.49 | 5.30 | 3.80 | 3.38 | >100 | ||
| 9c | 6.71 | 38 | 3.1 | 13.49 | 0.087 | 0.048 | 1.16 | 0.30 | >4.38 | 2.72 | 2.36 | >100 | 55.0 | ||
| 10c | 6.95 | 47 | 1.3 | 13.33 | 0.044 | 0.054 | 2.02 | 0.80 | 0.00 | 4.42 | 3.13 | 3.10 | 48.5 | 41.7 | >100 |
| 11 | 7.20 | 238 | 73.7 | 10.53 | 1.71 | 0.336 | 3.00 | 2.08 | 1.72 | 5.91 | 4.34 | 4.09 | >100 | 24.1 | 31.2 |
Abbreviations: NTU, nephelometric turbidity unit; DO, dissolved oxygen; TC, total coliforms; EC, E. coli; Entero, enterococci; RE, recovery efficiency.
The elution step was not completed on this sample.
Upstream surface water sample.
Recovery of bacterial indicators.
Grab samples (i.e., before concentration with UF) of the GW samples (n = 10) revealed that three GW sites were positive for total coliforms and none was positive for either E. coli or enterococci (Table 2). Following UF concentration, Table 2 shows that total coliforms and enterococci were present in nine and four GW UF concentrates, respectively. All SW samples (n = 11) were positive for the presence of total coliforms, E. coli, and fecal enterococci before and after UF concentration (Table 3). None of the water quality parameters collected was found to be correlated with reported UF recovery efficiencies based on a Pearson r value of greater than or equal to 0.6 or less than or equal to −0.6 (data not shown).
Evaluation of inhibition.
Inhibition was evaluated in viral NA extracts from GW, SW, and SW pellets by using an HGV RNA internal standard. Five-microliter viral NA extract samples were analyzed undiluted or at 10-fold and/or 100-fold dilutions to determine the level of inhibition. Sample inhibition occurred when the sample HGV RNA cycle threshold (CT) value deviated from the average CT value of the positive-control HGV RNA by 1 CT value. Inhibition was detected in 4 of 10 GW samples and all SW samples. Analysis for additional target viral NA was then determined based on the dilutions where inhibition occurred. For example, if total inhibition (i.e., a CT output of “undetermined”) was detected when the sample was undiluted but not at 10-fold or 100-fold dilutions, then target NA would be analyzed at the 10-fold and 100-fold dilutions but not in the undiluted sample. If the samples had partial or no inhibition, then undiluted and 10-fold portions were analyzed.
Real-time PCR and RT-PCR analyses.
Total viral NA extracted from GW and SW UF secondary concentrates and SW pellets was analyzed for MNV-1, HuAdV (types A to F), EV (pan-EV), HuPyV (JC and BK), HuNoV (GI and GII), BoEV, and BoNoV. MNV-1 positive-control RNA was detected in all GW samples (n = 10) and 9 of 11 SW samples (data not shown). Table 4 displays results for human and bovine enteric viruses detected in surface water and groundwater samples.
TABLE 4.
Detection of human and bovine enteric viruses in total viral NA extracts by real-time PCR or RT-PCR
| Source water | n | No. of positive samples for virus target by real-time PCR or RT-PCR |
||||||
|---|---|---|---|---|---|---|---|---|
| pan-EV | HuNoV |
HuAdV (types A to F) | HuPyV (JC or BK) | BoNoV | BoEV | |||
| GI | GII | |||||||
| GWa | 10 | NDc | 1 | 1 | 1 | 1 | ND | ND |
| SWb | 11 | 1 | 1 | 1 | 2 | 2 | 2 | 1 |
None of the GW samples positive for human enteric viruses were from the same sample.
One of the SW samples was positive for all human and bovine enteric viruses.
ND, not detected.
Volume back-calculations.
To determine the volume of initial sample analyzed, back-calculations from the total viral NA extracts were completed for each GW and SW sample. Table 5 shows the average volumes per sample processing step as they relate to the initial sample volumes. GW and SW samples were grouped separately for clarity. The average sample volumes analyzed in a 5-μl real-time PCR or RT-PCR mixture were 836 ml and 201 ml for GW and SW, respectively. These volumes decreased by 10- or 100-fold as the sample was diluted to overcome the effects of inhibition.
TABLE 5.
Volume back-calculations for average final sample volumes in UF concentrates, secondary concentrates, and total viral NA extracts
| Sample type | n | Total sample vol (liters) | Avg (range) concentrate vol (ml) | Total sample vol (ml)/avg concentrate vol (ml) | Avg total secondary concentrate vol (ml)a | Avg total sample vol (ml)/secondary concentrate vol (liters) | Avg total viral NA extract vol (μl)b | Avg total sample vol (ml) calculated for 5 μl of NA extract |
|---|---|---|---|---|---|---|---|---|
| GW | 10 | 100 | 286 (80-550) | 465 | 0.39 | 96 | 94 | 836 |
| SW | 11 | 100 | 380 (131-1,160) | 373 | 3.1 | 19 | 93 | 201 |
Total secondary concentrate from 70 ml of UF concentrate, except for one GW UF concentrate, for which a 35-ml volume was used.
Total viral NA extract from 200 μl of secondary concentrate, except for five GW secondary concentrates, for which volumes of 135, 193, 195, 117, and 132 μl were used.
In the present study, a combined tangential flow UF and real-time PCR methodology was applied for the assessment of human and animal enteric viruses in 100-liter SW and GW samples potentially impacted by animal and human waste in Lower Yakima Valley, WA. In addition, MNV-1, a model surrogate for the investigation of norovirus, was utilized in this study to evaluate the viral recovery efficiency of the UF system during application to GW and SW samples in a field setting. A standardized system for the identification of sample inhibition during molecular analysis was also utilized in this study (Gibson and Schwab, submitted).
During the collection of GW and SW samples, water quality parameters were collected at each site. Average values for all parameters, except pH, differed between SW and GW sources, although not unexpectedly. Nitrate levels either exceeded the maximum contaminant level (MCL; >10 mg/liter N) or were elevated (5 to 9.9 mg/liter N) in 8 out of 10 GW samples. The association of agricultural processes (i.e., excess use of fertilizers, irrigation practices, and land application of animal waste) with excess groundwater nitrate levels has been demonstrated throughout the United States (4, 17). Ammonium levels in all GW samples were also slightly higher than the natural levels of less than 0.2 mg/liter. The presence of ammonium in GW sources is often an indicator of potential human and animal waste pollution (24).
The analysis of bacterial indicators in GW and SW samples before and after UF provided information on microbial water quality at each source and enabled the assessment of recovery efficiency for bacteria of each sample. The detection of bacterial indicators prior to UF in GW was limited to total coliform concentrations less than 10 MPN/100 ml for three samples. After UF, total coliforms were detected in six additional GW samples. E. coli was not detected in any GW samples before or after filtration. Conversely, enterococci were present in four GW samples after UF at concentrations ranging from 10 to 31 MPN/100 ml UF concentrate. Indicator microorganisms are intended to act as sentinels for the potential presence of human pathogens of fecal origin, including enteric viruses and protozoa. However, previous studies have demonstrated that these bacteria, especially total coliforms, are poorly correlated with the presence of human pathogenic bacteria, enteric viruses, and protozoa; thus, the overall public health implications of the presence of total coliforms and enterococci in concentrated GW samples are unclear (5, 8).
Molecular methods utilized in this study focused on the detection of both RNA and DNA human and animal enteric viruses. The selection of human enteric viruses was primarily based on the U.S. EPA Drinking Water Contaminant Candidate List 3 (CCL 3) (26). Human enteric viruses included in this study and identified in the CCL 3 include enterovirus, adenovirus, and calicivirus (e.g., norovirus). Human polyomavirus was also included, as recent research suggested that HuPyV could be utilized as a reliable viral indicator of human fecal contamination in water (2, 10, 20). Human enteric viruses were detected in 4 of the 10 private GW wells sampled. The presence of human enteric viruses in groundwater has been associated with unsanitary wellhead conditions, breaks or leaks in service lines, local sources of fecal contamination in the immediate area around the well, improper disinfection after construction and repairs, substandard well construction, and groundwater aquifers under the influence of surface water (i.e., alluvial and sand gravel aquifers) (1). Although this study reports detection of human enteric viral RNA and DNA in groundwater, the public health implications are not known. Unlike cell culture systems, real-time PCR and RT-PCR do not detect infectious virus particles. Insufficient evidence exists regarding the stability of viral NA when inactivation occurs due to environmental stressors. However, the detection of viral NA should not be considered to be detection of noninfectious particles, because although infectivity cannot be determined, the potential for those microorganisms to be infectious prior to NA extraction cannot be excluded (16).
The bovine enteric viruses (BoEV and BoNoV) were also selected for analysis, as animal wastes in the area are predominantly from bovine sources due to intensive dairy CAFO and scattered cattle feedlots. Two previous studies looked at BoEV in surface waters under the influence of agricultural activities (dairy and cattle) (6, 11). BoEV was detected in one SW sample in the present study. To our knowledge, no previous studies investigated the presence of BoNoV in environmental water sources. However, several studies have investigated BoNoV in fecal samples from both asymptomatic and diarrheic bovine (18, 29, 30). In the present study, BoNoV was detected in 2 of 11 SW samples.
A simple method for identifying sample inhibition within real-time PCR and RT-PCR assays was reported here. This method uses a commercially available RNA (HGV) as an internal standard. Even though the sample processing method was optimized for the elimination of sample inhibitors, 4 of 10 and 11 of 11 GW and SW viral NA extracts, respectively, were determined to be inhibited. Evaluation of sample inhibition is often missing during real-time PCR and RT-PCR evaluations of environmental water samples, which may contain molecular inhibitors, including humus, complex polysaccharides, bacterial debris, metal ions, and nucleases. If inhibition analysis is not included, the estimated risk of exposure to a particular pathogen in drinking water or recreational water could be underestimated.
Volume back-calculations for each sample were also determined in this study. These calculations were conducted to determine original sample volumes that were analyzed during molecular analyses, and we found that the GW and SW volumes differed by more than 4-fold. This difference was primarily due to higher levels of turbidity in SW than in GW sources. The ability to analyze 100 liters of water concentrated to 300 ml by UF provides a better understanding of true water quality than does a smaller volume grab sample. This distinction is important when developing microbial risk assessment approaches for determination of health-based standards for individual microorganisms. For example, a risk estimate based on the presence of a given microorganism in 100 ml of water would likely provide less protection to public health than an estimate based on a 100-liter composite sample, as large volumes are frequently required for the direct detection of low levels of pathogens in ambient waters.
Conclusions.
Overall, this work demonstrates the use of optimized UF and real-time PCR systems to assess large volumes of groundwater and surface water for the presence of bacterial indicators and enteric viruses simultaneously. In addition, this study reports the use of MNV-1 as a potential viral positive control throughout each step of the described method. The importance of evaluating each sample for inhibition during PCR has also been demonstrated.
Acknowledgments
We thank Sarah Ehmer at Heritage University in Toppenish, WA, for allowing us to utilize laboratory space for sample processing; D'Ann L. Williams for guidance on sampling site locations, assistance with collection and processing, and for providing the map of the Lower Yakima Valley Study Area; and James Schissler for his laboratory assistance and support.
This study was supported by a U.S. EPA STAR grant (R833002), the Johns Hopkins University Global Water Program, and the Johns Hopkins University Bloomberg School of Public Health's Center for a Livable Future. K.E.G. is a Center for a Livable Future predoctoral fellow.
The views expressed herein have not been subjected to U.S. EPA review and therefore do not necessarily reflect the views of the agency, and no official endorsement should be inferred.
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1.Abbaszadegan, M., M. W. LeChevallier, and C. P. Gerba. 2003. Occurrence of viruses in US groundwaters. J. Am Water Resour. Assoc. 95:107-120. [Google Scholar]
- 2.Albinana-Gimenez, N., P. Clemente-Casares, S. Bofill-Mas, A. Hundesa, F. Ribas, and R. Girones. 2006. Distribution of human polyomaviruses, adenoviruses, and hepatitis E virus in the environment and in a drinking-water treatment plant. Environ. Sci. Technol. 40:7416-7422. [DOI] [PubMed] [Google Scholar]
- 3.Bae, J., and K. Schwab. 2008. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74:477-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkholder, J., B. Libra, P. Weyer, S. Heathcote, D. Kolpin, P. Thorne, and M. Wichman. 2007. Impacts of waste from concentrated animal feeding operations on water quality. Environ. Health Perspect. 115:308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colford, J. M., T. J. Wade, K. C. Schiff, C. C. Wright, J. F. Griffith, S. K. Sandhu, S. Burns, M. Sobsey, G. Lovelace, and S. B. Weisberg. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27-35. [DOI] [PubMed] [Google Scholar]
- 6.Fong, T. T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory, J. B., R. W. Litaker, and R. T. Noble. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 72:3960-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill, V. R., A. L. Polaczyk, D. Hahn, J. Narayanan, T. L. Cromeans, J. M. Roberts, and J. E. Amburgey. 2005. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 71:6878-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hundesa, A., C. Maluquer de Motes, S. Bofill-Mas, N. Albinana-Gimenez, and R. Girones. 2006. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 72:7886-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiménez-Clavero, M. A., E. Escribano-Romero, C. Mansilla, N. Gómez, L. Córdoba, N. Roblas, F. Ponz, V. Ley, and J. C. Sáiz. 2005. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jothikumar, N., T. Cromeans, V. Hill, X. Lu, M. Sobsey, and D. Erdman. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knappett, P. S., A. Layton, L. D. McKay, D. Williams, B. J. Mailloux, M. R. Huq, M. J. Alam, K. Matin Ahmed, Y. Akita, M. L. Serre, G. S. Sayler, and A. van Geen. 2010. Efficacy of hollow-fiber ultrafiltration for microbial sampling in groundwater. Ground Water [Epub ahead of print.] doi: 10.1111/j.1745-6584.2010.00712.x. [DOI] [PubMed]
- 15.Lambertini, E., S. Spencer, P. Bertz, F. Loge, B. Kieke, and M. Borchardt. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limsawat, S., and S. Ohgaki. 1997. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 63:2932-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallin, M. A., and L. B. Cahoon. 2003. Industrialized animal production: a major source of nutrient and microbial pollution to aquatic ecosystems. Popul. Environ. 24:369-385. [Google Scholar]
- 18.Mauroy, A., A. Scipioni, E. Mathijs, C. Saegerman, J. Mast, J. C. Bridger, D. Ziant, C. Thys, and E. Thiry. 2009. Epidemiological study of bovine norovirus infection by RT-PCR and a VLP-based antibody ELISA. Vet. Microbiol. 137:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McQuaig, S. M., T. M. Scott, V. J. Harwood, S. R. Farrah, and J. O. Lukasik. 2006. Detection of human-derived fecal pollution in environmental waters by use of a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuaig, S. M., T. M. Scott, J. O. Lukasik, J. H. Paul, and V. J. Harwood. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polaczyk, A., J. Narayanan, T. Cromeans, D. Hahn, J. Roberts, J. Amburgey, and V. Hill. 2008. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods 73:92-99. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds, K., K. Mena, and C. Gerba. 2008. Risk of waterborne illness via drinking water in the United States. Rev. Environ. Contam. Toxicol. 192:117-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sair, A. I., D. H. D'Souza, and L. A. Jaykus. 2002. Human enteric viruses as causes of foodborne disease. Compr. Rev. Food Sci. Food Saf. 1:73-89. [DOI] [PubMed] [Google Scholar]
- 24.Sell, R., and L. Knutson. 2002. Quality of ground water in private wells in the Lower Yakima Valley, 2001-02. Valley Institute for Research and Education, Yakima, WA.
- 25.U.S. Environmental Protection Agency. 2005. Protecting water quality from agricultural runoff. EPA 841-F-05-001. U.S. Environmental Protection Agency, Office of Water, Washington, DC.
- 26.U.S. Environmental Protection Agency. 2009. Fact sheet: final third drinking water Contaminant Candidate List (CCL 3), EPA 815-F-09-001. U.S. Environmental Protection Agency, Office of Water, Washington, DC.
- 27.U.S. Environmental Protection Agency, Washington State Departments of Agriculture, Ecology, and Health, and Yakima County Public Works Department. 2010. Lower Yakima Valley groundwater quality: preliminary assessment and recommendations document. Publication no. 10-10-009. Department of Ecology, Olympia, WA. http://www.ecy.wa.gov/biblio/1010009.html.
- 28.Vaccaro, J. J., M. A. Jones, D. M. Ely, M. E. Keys, T. D. Olsen, W. B. Welch, and S. E. Cox. 2009. Hydrogeologic framework of the Yakima River Basin aquifer system, Washington. 2009-5152. U.S. Geological Survey, Reston, VA.
- 29.Wise, A., S. Monroe, L. Hanson, D. Grooms, D. Sockett, and R. Maes. 2004. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 100:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf, S., W. Williamson, J. Hewitt, M. Rivera-Aban, S. Lin, A. Ball, P. Scholes, and G. Greening. 2007. Sensitive multiplex real-time reverse transcription-PCR assay for the detection of human and animal noroviruses in clinical and environmental samples. Appl. Environ. Microbiol. 73:5464-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]