Abstract
Lactobacillus helveticus can possess one or two cell envelope proteinases (CEPs), called PrtH2 and PrtH. The aim of this work was to explore the diversity of 15 strains of L. helveticus, isolated from various origins, in terms of their proteolytic activities and specificities on pure caseins or on milk casein micelles. CEP activity differed 14-fold when the strains were assayed on a synthetic substrate, but no significant differences were detected between strains possessing one or two CEPs. No correlation was observed between the proteolytic activities of the strains and their rates of acidification in milk. The kinetics of hydrolysis of purified αs1- and β-casein by L. helveticus whole cells was monitored using Tris-Tricine sodium dodecyl sulfate (SDS) electrophoresis, and for four strains, the peptides released were identified using mass spectrometry. While rapid hydrolysis of pure β-casein was observed for all strains, the hydrolysis kinetics of αs1-casein was the only criterion capable of distinguishing between the strains based on the number of CEPs. Fifty-four to 74 peptides were identified for each strain. When only PrtH2 was present, 22 to 30% of the peptides originated from αs1-casein. The percentage increased to 41 to 49% for strains in which both CEPs were expressed. The peptide size ranged from 6 to 33 amino acids, revealing a broad range of cleavage specificities, involving all classes of amino acids (Leu, Val, Ala, Ile, Glu, Gln, Lys, Arg, Met, and Pro). Regions resistant to proteolysis were identified in both caseins. When strains were grown in milk, a drastic reduction in the number of peptides was observed, reflecting changes in accessibility and/or peptide assimilation during growth.
Lactobacillus helveticus is used as a starter component in many fermented dairy products, such as fermented milks, Swiss-type cheeses (Emmental, Comté), and long-ripened Italian cheeses (Parmigiano Reggiano, Grana Padano, Provolone). With a high nutritional requirement for numerous amino acids, L. helveticus strains possess an efficient proteolytic system capable of producing short peptides and amino acids from the casein matrix. Cell envelope proteinases (CEPs) located in the cell wall are the first bacterial enzymes that hydrolyze milk caseins into peptides, which are then transported into the cell, where they are further hydrolyzed by numerous intracellular peptidases (30, 39). L. helveticus strains are therefore capable of generating a diverse range of interesting textural or bioactive properties in dairy products (4, 11, 33). Despite the multiple technological applications of L. helveticus, little is known regarding the activity and specificity of its CEPs (23, 24, 26, 42, 45, 46).
Many species of lactic acid bacteria possess only one CEP, such as PrtP for Lactococcus lactis (27), PrtS for Streptococcus thermophilus (10), PrtR for Lactobacillus rhamnosus (35), and PrtB for Lactobacillus bulgaricus (17). In L. helveticus, a minimum of two genes encoding the CEPs, named prtH and prtH2, have been reported for strain CNRZ32 (43), and two others, named prtH3 and prtH4, remain putative (3). In a previous study the distribution of prtH and prtH2 was shown to be strain dependent (16), with prtH2 present and expressed in all of the 29 strains studied, while prtH was present and expressed in only 18. Even though all of the strains tested possess a prtH2 gene, sequencing of an internal fragment revealed the existence of intraspecific diversity, which could be related either to two alleles of prtH2 or to another, homologous gene, such as prtH3 (3, 26). Although the prtH and prtH2 genes were both shown to be expressed in dairy matrices (16) and in the absence of respective mutants, it is still difficult to distinguish the activity and specificity of each CEP. The aim of this work was to explore the diversity of strains of L. helveticus containing one or two CEPs with regard to their global proteolytic activities on a synthetic substrate and to identify the peptides produced from the main caseins, either purified or in milk, in which caseins are present as micelles.
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
Fifteen strains of L. helveticus were chosen based on their technological use in dairy products, as well as their geographic origins (Table 1). All strains came from the International Centre of Microbial Resources, a resource center dedicated to bacteria for fermented products (CIRM-BIA, INRA, Rennes, France). According to the results of Genay et al. (16), these strains could be divided into two groups: strains containing one CEP (PrtH2) and strains containing two CEPs (PrtH and PrtH2) (Table 1). L. helveticus strains were propagated three times without shaking at 43°C in MRS growth culture medium (Difco, Detroit, MI) from a cryoball or cryotube stored at −80°C. The strains were then grown in milk, either in 10 ml of commercial ultrahigh-temperature (UHT)-treated milk or in 10% (wt/vol) reconstituted low-low-heat (LLH)-dried milk (INRA-STLO, France) (41). An overnight MRS broth culture was subsequently inoculated at different concentrations (1%, 0.5%, 0.1%, 0.05%, 0.01%, 0.005%, and 0.001%) at 43°C without shaking. The first dilution in which no casein precipitation was visible (i.e., nonclotted milk) was used to inoculate at 5% (i) 20 ml of UHT milk, which was then incubated without shaking at 43°C until the pH reached 4.6, and (ii) 150 ml of LLH milk incubated at 43°C until an optical density at 480 nm (OD480) of 1 was reached. The absorbance at 480 nm was measured after clarification of the milk with 0.2% EDTA (pH 12).
TABLE 1.
L. helveticus strains used in this work
| Straina | CIRM-BIA no.b | Biotope or peculiaritiesc | Origin | Reference or source | Presence or absenced of: |
|
|---|---|---|---|---|---|---|
| PrtH | PrtH2 | |||||
| ITGLH1 | CIRM-BIA 107 | Whey | France | Deutsch et al. (7) | + | + |
| ITGLH3 | CIRM-BIA 453 | Aminopeptidase-deficient mutant strain derived from ITGLH1 | France | Blanc et al. (1) | + | + |
| CNRZ32 | CIRM-BIA 103 | Artisanal starter used for Comté | France | INRA, Jouy-en-Josas, France | + | + |
| CNRZ32A | CIRM-BIA 454 | prtH mutant derived from CNRZ32 | United States | J. Steele, University of Wisconsin, Madison | + | + |
| CNRZ241 | CIRM-BIA 679 | Artisanal starter used for Comté | France | + | + | |
| ROSELL5088 | CIRM-BIA 797 | ND | Canada | + | + | |
| ROSELL5089 | CIRM-BIA 798 | ND | Canada | + | + | |
| ITGLH77 | CIRM-BIA 99 | Artisanal starter, commercial strain | France | Deutsch et al. (6) | − | + |
| CNRZ303 | CIRM-BIA 680 | Artisanal starter used for Comté | France | Deutsch et al. (7) | − | + |
| CNRZ328 | CIRM-BIA 110 | Artisanal starter used for Emmental; Prt− phenotype | Finland | Hebert et al. (22) | − | + |
| CNRZ414 | CIRM-BIA 681 | Cow milk koumiss | Russia | − | + | |
| ISLC5 | CIRM-BIA 104 | Artisanal lactic starter used for Grana Padano | Italy | Valence and Lortal (44) | − | + |
| CP615 | CIRM-BIA 105 | ND | Japan | − | + | |
| CIP103146T | CIRM-BIA 101 | Artisanal starter used for Emmental | France | − | + | |
| DPC4571 | CIRM-BIA 796 | Commercial Swiss cheese isolate | Ireland | T. Beresford, Teagasc | − | + |
ITG, Institut Technique du Gruyère, France (renamed Actilait); CNRZ, Centre National de Recherche Zootechnique, INRA, Jouy-en-Josas, France; ISLC, Instituto Sperimentale Lattiero-Caesario, Italy; CIP, Collection of the Institut Pasteur, France.
CIRM-BIA, Centre International de Ressources Microbiennes-Bactéries d'Intérêt Alimentaire, INRA, Rennes, France.
ND, not determined.
+, presence; −, absence.
Acidification rate.
Milk acidification was monitored using CINAC software (5) until a pH of 4.6 was reached. The acidification rate (in pH units [UpH] per hour) was calculated from the slope of the pH curve as a function of time. The bacterial cells and residual caseins were pelleted by centrifugation of acidified UHT milk at 6,000 × g for 10 min at 4°C, and the supernatants were filtered through 0.45-μm-pore-size polyvinylidene difluoride (PVDF) filters (Millex; Millipore Corporation, MA), aliquoted, and frozen at −20°C until further use.
Assessment of CEP activity.
Cells grown in LLH milk were harvested by centrifugation (10,000 × g for 10 min at 4°C) at the end of the exponential-growth phase (OD480, 1), washed three times with 50 mM Tris-HCl buffer (pH 7.5) supplemented with 2 mM CaCl2, and resuspended to a final OD of approximately 10 in 100 mM sodium phosphate buffer (pH 7.0). The CEP activity of the whole-cell suspension was measured as described previously (8, 20) by using the chromogenic substrate succinyl-alanyl-alanyl-prolyl-phenylalanine-p-nitroanilide derivative (Sigma). The released nitroaniline was measured at 410 nm by using a spectrophotometer (UVICON922; Kontron Instruments, Serlabo Technologies, Bonneuil sur Marne, France). One unit of CEP activity was expressed as the release of 1 nmol p-nitroanilide·ml−1·min−1. Specific activity was expressed as units per milligram (dry weight) of cells.
Casein purification.
The αs1- and β-caseins were purified as follows. First, caseins were precipitated in the presence of 6.6 M urea and sulfuric acid to pH 1.5, according to the method of Zittle and Custer (47), and αs- and β-caseins were then separated from κ-casein. αs1-casein was further separated from β-, γ-, and αs2-caseins by dilution with urea to 3.3 M at pH 4.6, followed by ethanol precipitation according to the method of Brignon et al. (2) and Fox and Guiney (12). The enriched αs1- and β-casein fractions were further purified by anion-exchange chromatography using 210 ml of the Q Sepharose fast-flow separating phase (Pharmacia, St. Quentin Yvelines, France), according to the method of Guillou et al. (19). Two grams of each casein was dissolved in buffer A, containing 5 mM Tris-HCl (pH 8), 4.5 M urea, and 6.4 × 10−5 M dithiothreitol (DTT), and was injected onto the column. The flow rate was 3 ml/min. Elution was performed by using a gradient consisting of 0 to 25% 1 M NaCl for 1 h, 25 to 30% 1 M NaCl for 100 min, 30 to 60% 1 M NaCl for 600 min, and 60% 1 M NaCl for 60 min (19).
All peaks were collected; those corresponding to αs1- or β-casein were dialyzed and lyophilized; and their purity was verified by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% SDS-PAGE) (31).
Assessment of casein hydrolysis.
After growth in LLH milk (OD480, 1), cells were harvested by centrifugation (10,000 × g for 10 min at 4°C), washed three times with 50 mM Tris-HCl buffer (pH 7.5) and 2 mM CaCl2, and resuspended to a final OD of approximately 10 in the same buffer (21). Two hundred microliters of the αs1- or β-casein solution at 5 mg/ml in 50 mM Tris-HCl buffer (pH 7.5) was added to 3 ml of the cell suspension, and the mixture was incubated at 40°C in a shaking water bath. Samples (300 μl) were withdrawn at 0, 5, 15, 30, 60, 120, and 180 min of hydrolysis, and the reaction was stopped with 3 μl of 100 mM phenylmethylsulfonyl fluoride (PMSF). Cells were eliminated by centrifugation (6,000 × g for 10 min at 4°C), and the supernatant was filtered through a 0.45-μm-pore-size PVDF filter and was stored at −20°C until further use.
SDS-PAGE.
Hydrolysis of purified β- and αs1-caseins by strains of L. helveticus grown in LLH milk was monitored by SDS-PAGE at different times (from 5 to 180 min). Electrophoresis was performed as described by Schägger and von Jagow (40) using a Protean II system (16 by 16 by 0.1 cm; Bio-Rad, Marnes-la-Coquette, France) with SDS-Tris-Tricine buffer and a concentration gradient from 12 to 18% acrylamide. Samples were solubilized in a denaturing buffer at pH 6.8 (vol/vol) containing 1.25 M Tris, 138 mM SDS, 38 mM DTT, 2.17 M glycerol, and 0.28 mM bromophenol blue in distilled water according to the method of Laemmli (31). Gels were run overnight at a constant voltage of 90 V. Proteins and oligopeptides were stained with Coomassie brilliant blue R-250 (Serva Electrophoresis, Heidelberg, Germany). After scanning of the gels (Image Scanner II; Amersham Biosciences), the molecular weights and volumes of the visualized bands were determined using ImageQuant TL (GE Healthcare) 1-dimensional (1D) densitometry software.
Peptide identification by LC-ESI-MS-MS.
Following the growth of L. helveticus strains in milk, the peptides released into the pH 4.6 supernatant or during the hydrolysis of the purified αs1- and β-caseins were identified by online liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS-MS). Prior to injection, pH 4.6 supernatant samples were half-diluted with 200 μl of 8.75 M urea and 50 μl of 10% trifluoroacetic acid (TFA) (vol/vol).
MS experiments were performed using an LC-Packings NanoLC system (Dionex) fitted to a QStar XL mass spectrometer (MDS Sciex, Ontario, Canada) equipped with a nano-ESI source (Proxeon Biosystems A/S, Odense, Denmark). The instrument was calibrated with multipoint calibration using fragment ions that resulted from the collision-induced decomposition of a peptide from β-casein, β-CN(193-209). A preliminary sample concentration step was performed on a reverse-phase PepMap 100 column (C18; particle size, 5 μm; inner diameter [i.d.], 300 μm; length [L], 5 mm) (LC-Packings, Dionex, Amsterdam, Netherlands). Separation was performed on a reverse-phase PepMap 100 column (C18; particle size, 3 μm; i.d., 75 μm; L, 150 mm) (LC-Packings, Dionex, Amsterdam, Netherlands) at room temperature using solvent A (2% [vol/vol] acetonitrile, 0.08% [vol/vol] formic acid, and 0.01% [vol/vol] TFA in deionized water) and solvent B (95% [vol/vol] acetonitrile, 0.08% [vol/vol] formic acid, and 0.01% [vol/vol] TFA in deionized water). A linear gradient of solvent B from 10 to 70% in 90 min was applied for the elution at a flow rate of 0.2 μl/min. Eluted peptides were electrosprayed directly into the mass spectrometer operated in positive mode. A full continuous MS scan was carried out, followed by three data-dependent MS-MS scans. Spectra were collected in the selected mass range of m/z 300 to 2,000 for MS and m/z 60 to 2,000 for MS-MS. The first, second, and third most intense ions from the MS scan were selected individually for collision-induced dissociation (2+ to 4+ charged ions were selected for the MS-MS analysis). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS-MS acquisition, using Analyst QS software, version 1.1.
To identify peptides, all data (MS and MS-MS) were submitted to MASCOT (version 2.2). The search was performed against a homemade database that deals with the major milk proteins and that represents a portion of the SwissProt database (http://www.expasy.org). No specific enzymatic cleavage was used, and the peptide mass tolerance was set to 0.3 Da for MS and 0.2 Da for MS-MS. Three variable modifications (phosphorylation on serine and threonine, oxidation of methionine, and deamidation of asparagine and glutamine residues) were selected. For each peptide identified, a minimum MASCOT score corresponding to a P value below 0.05 was considered a prerequisite for peptide validation with a high degree of confidence.
Estimation of bacterial cell lysis.
Cell lysis was estimated by measurement of the intracellular aminopeptidase activity of L. helveticus, released in the pH 4.6 culture supernatants or in the hydrolysates of purified caseins. As described by Goldbarg and Rutenberg (18), the following β-naphthylamide (β-Na) derivatives were used: Lys β-Na and Phe-Pro β-Na, for lysine aminopeptidase (PepN) and X-prolyl-dipeptidyl aminopeptidase (PepX), respectively.
Statistical analyses.
Unilateral Student t tests were performed to compare the mean values of the proteolytic activities of PrtH2 and PrtH/PrtH2 strains for each incubation time. Principal-component analysis (PCA) was applied to the standardized data in order to map the individual strains, while hierarchical-clustering analysis (HCA) procedures were used to build homologous groups (Statbox, version 6.3; Grimmersoft, Paris, France).
Molecular modeling.
Three-dimensional representations of casein models were constructed using the life science modeling and simulation suite of Discovery Studio, version 2.1, 2008 (Accelrys Inc., San Diego, CA). These representations were deduced from the data of Kumosinski et al. for αs1-casein (28) and β-casein (29) by using molecular-modeling tools integrating their physicochemical properties and secondary-structure predictions. For αs2-casein, a model was produced by threading the backbone sequence of the protein onto a homologous protein according to the method of Farrell et al. (9) (http://www.arserrc.gov/CaseinModels/).
RESULTS
Acidification rate.
Acidification rates differed depending on the strain, ranging from 0.11 to 0.44 UpH/h, as shown in Table 2. Most strains acidified the milk to pH 4.6 in 5 to 10 h. No significant differences (P, >0.05) in the acidification rates were detected between the groups possessing one versus two CEPs. Strains ROSELL5088 and ROSELL5089 were the fastest-acidifying strains, while strain ITGLH3 was the slowest. In fact, strain ITGLH3, an aminopeptidase-deficient mutant derived from ITGLH1, acidified milk at half the rate of the parent strain. Strain CNRZ328, which was considered CEP negative in a previous study (22), also acidified milk slowly; pH 4.6 was reached only after 17 h, due to a longer lag phase (15 h versus about 3 h) and not to a lower acidification rate.
TABLE 2.
Acidification rates and specific activities of cell envelope proteinases of L. helveticus strainsa
| Strain | Acidification rate (UpH/h) | Acidification time (h)b | Sp act of CEP (U/mg [dry wt] of bacteria) |
|---|---|---|---|
| ITGLH1 | 0.28 ± 0.01 | 8.0 | 2.73 ± 0.99 |
| ITGLH3 | 0.11± 0.00 | 17.0 | 3.28 ± 0.94 |
| CNRZ32 | 0.33 ± 0.00 | 7.2 | 4.38 ± 1.65 2.88 ± 0.77,c 0.35 ± 0.19d |
| CNRZ32A | 0.26 ± 0,02 | 9.4 | 2.42 ± 0.57 |
| CNRZ241 | 0.28 ± 0.00 | 8.3 | 2.75 ± 0.00 |
| ROSELL5088 | 0.43 ± 0.01 | 5.6 | 5.82 ± 0.44 |
| ROSELL5089 | 0.44 ± 0.02 | 5.6 | 3.60 ± 0.79 |
| ITGLH77 | 0.34 ± 0.01 | 6.8 | 7.33 ± 0.47 4.95 ± 1.30,c 0.74 ± 0.06d |
| CNRZ303 | 0.23 ± 0,00 | 9.4 | 3.27 ± 0.04 |
| CNRZ328 | 0.24 ± 0.01 | 17.3 | 0.05 ± 0.07 |
| CNRZ414 | 0.23 ± 0.00 | 9.9 | 5.66 ± 0.54 |
| ISLC5 | 0.29 ± 0.01 | 8.2 | 5.28 ± 1.21 |
| CP615 | 0.25 ± 0.03 | 10.8 | 0.53 ± 0.13 |
| CIP103146T | 0.26 ± 0.04 | 8.6 | 5.29 ± 0.57 |
| DPC4571 | 0.30 ± 0.01 | 8.5 | 3.61 ± 0.88 |
Values are means ± standard deviations of results from three or two independent experiments. Numbers in boldface are the highest and the lowest values for each column.
Time necessary to acidify milk to pH 4.6.
Measured after growth of cells in low-low-heat milk supplemented with 1% tryptic and chymotryptic β-casein hydrolysates.
Measured after growth of cells in MRS broth.
CEP activity on a synthetic substrate.
The CEP activities of L. helveticus strains were initially measured by using the synthetic substrate succinyl-alanyl-alanyl-prolyl-phenylalanine-p-nitroanilide to characterize overall activity during growth in milk. The activities of the strains ranged from 0.05 to 7.33 U/mg (dry weight) of cells (Table 2). The highest CEP activity was observed for strain ITGLH77, while the CEP activities of strains CNRZ328 and CP615 were 146 and 14 times lower, respectively. Significant differences were observed between the strains tested (P, <0.05) but not between groups containing one versus two CEPs. The finding that strain CNRZ328 had the lowest CEP activity was in agreement with the use of this strain (also classified as CRL1176) as a negative control for proteolytic activity by Hébert et al. (23, 24).
For two strains, ITGLH77 (with one CEP) and CNRZ32 (with two CEPs), the CEP activities were reduced 10 and 12 times, respectively, when the bacterial cells were grown in MRS broth, and 1.5 times when the cells were grown in milk supplemented with 1% tryptic and chymotryptic hydrolysates (Table 2).
Kinetics of purified β- and αs1-casein hydrolysis.
The activities of the CEPs on purified β- and αs1-caseins were subsequently determined, from the same ratio of bacterial cells to caseins, and were evaluated by Tris-Tricine SDS-PAGE. Ten strains were analyzed.
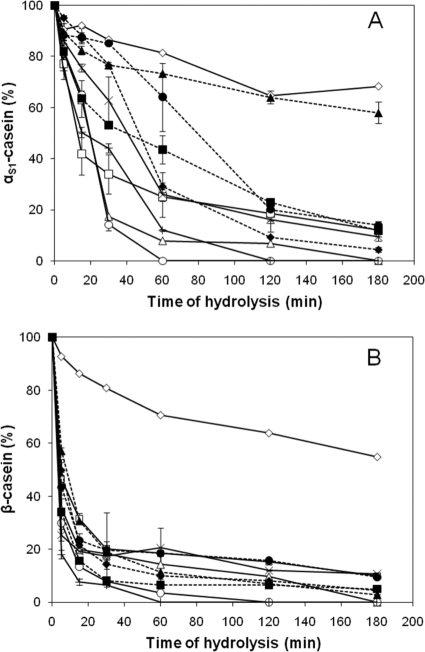
Regarding casein hydrolysis (Fig. 1), 60% of the β-casein was hydrolyzed after 5 min of incubation with the bacterial cells. The rate of hydrolysis was lower for αs1-casein: only 10 to 20% of αs1-casein was hydrolyzed after 5 min of incubation. When αs1-casein was hydrolyzed, significant differences were observed between the strains for each of the time periods analyzed. Low rates of hydrolysis were observed on both caseins for CNRZ32A, a prtH mutant derived from CNRZ32 (Fig. 1). At 3 h, 70 and 60% of αs1- and β-caseins, respectively, remained intact for the mutant strain. In the case of strain ISLC5, only 40% of αs1-casein was hydrolyzed, while β-casein was completely hydrolyzed, after 3 h of incubation. The level of bacterial lysis during hydrolysis was monitored by measuring the intracellular aminopeptidase activity released into the hydrolysates. No bacterial lysis had occurred over the incubation period except for two strains: DPC4571, where lysis had started during incubation in the presence of the casein substrates (20% lysis after 5 min), and ITGLH1 (23% after 2 h).
FIG. 1.
Kinetics of hydrolysis of purified αs1-casein (A) and β-casein (B) by whole-cell suspensions of L. helveticus strains at 0, 5, 15, 30, 60, 120, and 180 min. The strains used were ITGLH1 (□), ITGLH3 (▵), CNRZ32 (○), CNRZ32A (⋄), ROSELL5088 (×), ROSELL5089 (+), ITGLH77 (▪), ISLC5 (▴),CIP103146T (•), and DPC4571 (♦). The percentage of casein degradation was estimated by densitometry (n = 2).
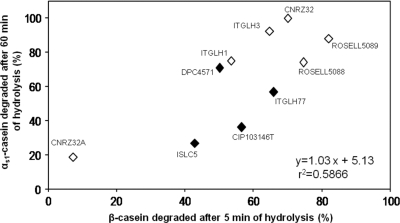
Using the percentage of αs1-casein hydrolysis, strains were classified in terms of the number of proteinases at any incubation time point analyzed (P, <0.05). In contrast, for β-casein, similar classification was possible only for the 5-min incubation time point. Strains with one or both proteinases were better distinguished when the degradation of β-casein was reported as a function of αs1-casein degradation (Fig. 2). Similar discrimination was obtained by PCA and HCA (data not shown). However, the strains possessing one or two CEPs were not clearly distinguished when the acidification rate was reported as a function of the global CEP activity (data not shown). Moreover, no correlation was shown between these two phenotypic criteria, in agreement with the previous results of Gatti et al. (15).
FIG. 2.
Relationships of L. helveticus strains with regard to the percentages of casein degradation, i.e., αs1-casein degraded after 1 h of hydrolysis versus β-casein degraded after 5 min of hydrolysis. Symbols: ⋄, strains with two CEPs, PrtH2 and PrtH; ♦, strains with PrtH2 only.
The most representative patterns of hydrolysis by Tris-Tricine SDS-PAGE are shown in Fig. S1 in the supplemental material. For β-casein, the degradation profile led to the production of two common bands at the end of the hydrolysis time. For αs1-casein, one intermediate peptide (indicated by an arrow in Fig S1A) was produced by most strains and remained intact for at least 60 min. Thereafter, the production of subsequent peptides with lower apparent sizes was strain dependent. Most of the large peptides produced had disappeared by the end of hydrolysis for both caseins.
Identification of peptides from the hydrolysis of αs1- and β-caseins. (i) Hydrolysis of purified caseins.
The peptides identified after 2 h of incubation for four strains of L. helveticus with either one CEP (ITGLH77 and ISLC5) or two CEPs (ITGLH1 and ROSELL5088) are presented in Tables S1 (for purified αs1-casein) and S2 (for purified β-casein) in the supplemental material. The total number of peptides identified for both caseins was as follows (in decreasing order): 74 for ISLC5, 70 for ROSELL5088, 59 for ITGLH77, and 54 for ITGLH1. In accordance with the kinetics studies, the number of αs1-casein-derived peptides was lower than that of β-casein-derived peptides, regardless of the strain. The total number of peptides was not significantly related to the presence of one or two CEPs (P, <0.05). However, the proportion of αs1-casein-derived peptides was significantly higher (P, <0.05) for strains possessing two CEPs (ITGLH1 and ROSELL5088) than for strains possessing one CEP (ITGLH77 and ISLC5): 41 and 49% versus 29 and 22%, respectively.
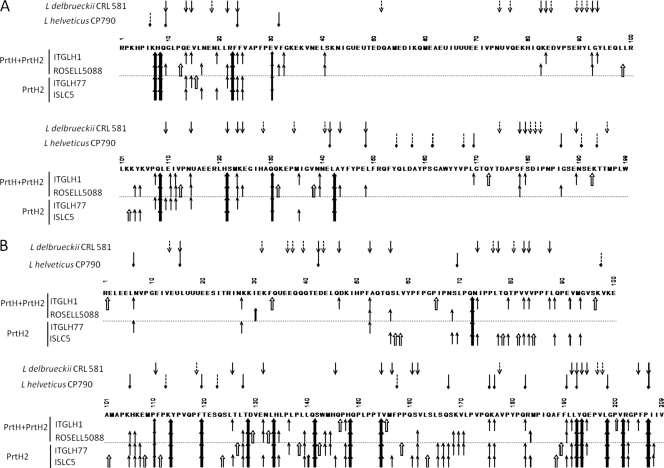
The number of cleavage sites was 17 to 34 on αs1-casein and 40 to 61 on β-casein (Fig. 3). The cleavage specificity was rather broad in that various classes of amino acids were involved: aliphatic (Leu, Val, Ala, and Ile), dicarboxylic (Glu and its amide form Gln), basic (Lys and Arg), aromatic (Phe, Tyr, and Trp), hydroxylated (Ser and Thr), sulfur containing (Met), and heterocyclic (Pro). Even though some sites were found only for strains possessing two CEPs (ITGLH1 and ROSELL5088), it was difficult to determine definitely the particular amino acids that were specific to PrtH.
FIG. 3.
Profiles of degradation of the purified αs1-casein (A) and β-casein (B) by L. helveticus strains. The arrows correspond to sites of cleavage by cell envelope proteinases of L. helveticus ITGLH1, ROSELL5088, ITGLH77, and ISLC5 (↑, ); L. helveticus CP790 (45) (
); L. helveticus CP790 (45) ( ); and L. delbrueckii subsp. lactis CRL581 (20) (↓). Cleavage sites specific to one strain (⇑,
); and L. delbrueckii subsp. lactis CRL581 (20) (↓). Cleavage sites specific to one strain (⇑, ,
, ) are also shown. U represents a phosphoserine residue.
) are also shown. U represents a phosphoserine residue.
Cleavage sites were observed all along the β-casein sequence. A high number of cleavage sites were common to the different strains: 14 cleavage sites were common to all strains, and another 14 cleavage sites were observed in three out of the four strains. However, more than two-thirds of the sites were located in the C-terminal end of β-casein, regardless of the strain or the presence of one or two CEPs.
Not surprisingly, the cleavage sites were distributed differently on αs1-casein. For strains that possessed only PrtH2, two regions of αs1-casein were hydrolyzed, one at the N-terminal end (between residues 1 and 30) and another in the middle of the sequence (between residues 100 and 148). Eight cleavage sites were common to the four strains, and seven sites were common to three out of the four strains. Some additional cleavage sites were observed only when both PrtH and PrtH2 were present: sites between residues 80 and 100 and sites in the C-terminal end of the molecule, between residues 169 and 199. These regions were hydrolyzed only by strains ITGHLH1 and ROSELL5088.
For both caseins, regions containing a cluster of three phosphoserine residues were resistant to hydrolysis, in agreement with the previous results of Deutsch et al. (6).
(ii) Hydrolysis of casein micelles in milk.
The pH 4.6 supernatants obtained after bacterial growth in UHT milk were analyzed by mass spectrometry, and the peptides generated were identified. Sterile milk was used as a control and was incubated under the same conditions as those used for the growth of the lactobacilli. Peptides identified in the sterile milk were produced by endogenous enzymes in the milk, plasmin and cathepsin D. The well-characterized cleavage sites of both proteinases were considered in order to characterize the cleavage specificities corresponding to those of the L. helveticus CEPs.
The number of casein-derived peptides identified was severely reduced when lactobacilli were grown in milk, as shown in Tables S1 and S2 and in Fig. S2 in the supplemental material. The total numbers of peptides produced from αs1-, αs2-, and β-caseins were 38, 26, 38, and 5 for strains ITGLH1, ROSELL5088, ITGLH77, and ISLC5, respectively. Only 9, 1, and 4 αs1-casein-derived peptides were identified in the milk pH 4.6 supernatants of ITGLH1, ROSELL5088, and ITGLH77, respectively, while no peptides were identified for ISLC5. The reduction in the number of peptides identified was less significant for β-casein, where 26, 21, and 27 peptides were identified for ITGLH1, ROSELL5088, and ITGLH77, respectively, and only 3 for ISLC5. Only a few peptides (between 1 and 7) were identified from αs2-casein and none from κ-casein. Even though the peptides identified were not identical to those from purified caseins and from milk caseins (see Tables S1 and S2 in the supplemental material), they belonged to the same regions of the casein sequence, regardless of the casein tertiary structure.
Surprisingly, there was also a great reduction in the number of sites that were cleaved. The cleavage sites that remained corresponded to those that were common for all strains (Fig. 3), as clearly shown for αs1-casein. The hydrolysis of β-casein was not as constrained for strains with two CEPs, since peptides were found throughout the sequence (see Fig. S2 in the supplemental material). In contrast, an uncleaved region was observed for strains with only one CEP; no peptides were identified between the Gln141 and Ser161 amino acid residues, except for strain ITGLH77.
The sizes of the peptides identified from the casein micelles ranged from 6 to 27 amino acid residues for β-casein, from 6 to 22 amino acid residues for αs1-casein, and from 12 to 20 amino acid residues for αs2-casein. All strains produced more medium-sized peptides (between 10 and 20 amino acid residues) than small peptides (fewer than 10 amino acid residues). Such a reduction in the number of cleavage sites suggests that some regions are no longer accessible for hydrolysis in milk and/or that some small peptides are assimilated into the bacterial cell during the growth of lactobacilli. Only common cleavage sites, such as Phe52-Ala53, Phe119-Thr120,Thr128-Asp129, Gln141-Ser142, Leu163-Ser164, Gln182-Arg183, and Tyr193-Gln194, were hydrolyzed in β-casein. For αs1-casein, there were His8-Gln9, Gln9-Gly10, Leu16-Asn17, and Glu30-Val31. For αs2-casein, there were Met4-Glu5,Glu5-His6, Tyr98-Leu99, Arg114-Asn115, and Asn115-Ala116.
Interpretation of hydrolyzed regions by casein 3D modeling.
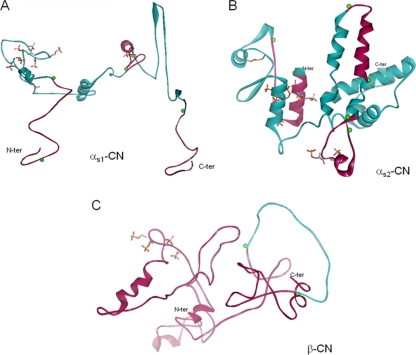
3D molecular-modeling representations of the caseins, deduced from the data of Kumosinski et al. (28) for αs1-casein, Farrell et al. (9) for αs2-casein, and Kumosinski et al. (29) for β-casein, are shown in Fig. 4.
FIG. 4.
3D models of αs1-casein (A), αs2-casein (B), and β-casein (C) represented as a solid ribbon backbone for defined secondary structures (coiled ribbons represent α-helices, and broad ribbons represent β-sheets). Colored green balls indicate the beginnings and ends of some hydrolyzed regions. Pink areas correspond to hydrolyzed regions and green areas to regions not cleaved by cell wall proteinases of L. helveticus strains in milk. Side chains of phosphorylated serine residues and of cysteine residues are shown as sticks and are colored by atom type (orange, phosphate; gray, carbon; yellow, sulfur). The representations were deduced from the data of Kumosinski et al. (28) for αs1-casein, Farrell et al. (9) for αs2-casein, and Kumosinski et al. (29) for β-casein.
The N- and C-terminal ends of αs1-casein had less secondary structure than the other parts of the molecule and were more susceptible to hydrolysis in milk (Fig. 4A; the pink areas correspond to hydrolyzed regions). In contrast, the carboxyl-terminal region of αs1-casein (amino acid residues Ile136 to Pro160), which exhibits a high degree of hydrophobicity, was not hydrolyzed in milk, probably due to interactions enhancing the pronounced self-association of the casein monomer in aqueous solution (28). Obviously, one α-helix was hydrolyzed on both ends by 7 of the 12 strains regardless of the number of CEPs present, and therefore, this α-helix may not be involved in intermolecular interactions (see Fig. S2 in the supplemental material). The interactions between caseins and minerals appear to reduce the level of access, as reflected by a reduction in the level of hydrolysis of the phosphoserine cluster (14) (represented by the phosphate side chain in Fig. 4). A low level of hydrolysis on the phosphorylated region was also observed in purified casein and was considered likely to be due to the presence of calcium in the buffer, which allowed mineral binding and refolding of the αs1-casein.
Regarding αs2-casein (Fig. 4B), the 3D model shown is based on more recently developed and refined model calculations (9). Again, the N-terminal end of the molecule, corresponding to a phosphorylated region, was hydrolyzed from either end, and the other phosphorylated region present in the α-helix (Glu50 to Glu60) was weakly accessible. Moreover, the α-helix was surrounded by cysteine residues (Cys36 and Cys40). which mainly formed intramolecular disulfide bridges (85%) and, to a lesser extent, intermolecular dimers (15%) (37, 38). Among the six regions that could be located in the interior of the molecule—the regions from amino acid residues 46 to 49, 65 to 68, 97 to 100, 148 to 151, 171 to 174, and 191 to 194 (9)—-four (residues 46 to 49, 65 to 68, 97 to 100, and 191 to 194) were not hydrolyzed in milk by any of the CEPs of L. helveticus.
Holt and Sawyer (25) and Kumosinski et al. (29) found that β-casein was more unstructured than the αs-caseins, as shown on Fig. 4C, rendering this molecule more accessible to cleavage and subsequently more hydrolyzed than the other caseins. The easier access to β-casein by CEPs seems to occur whether β-casein is in the purified or the assembled state, as in milk. With the exception of strain ITGLH77, the C-terminal end (Leu140 to Val162) was not hydrolyzed by strains that possessed only one CEP (PrtH2) (shown in green in Fig. 4C). This region, rich in proline and glutamine, is likely involved in interactions with the other caseins (25).
DISCUSSION
In contrast to many strains of lactic acid bacteria, L. helveticus possesses several genes encoding CEPs, in particular prtH and prtH2, both of which have been well characterized. prtH2 is ubiquitous, whereas the presence of prtH has been shown to be strain dependent (16). The overall proteolytic efficiencies of the 15 strains investigated in this study were shown to differ 14-fold when a synthetic substrate was used, but no statistical correlation with the number of CEP genes present was observed. However, while the extended and rapid hydrolysis of purified β-casein did not distinguish between strains possessing one or two CEPs, these strains could be distinguished both quantitatively and qualitatively by measuring the hydrolysis of purified αs1-casein.
Moreover, strains with only one CEP hydrolyzed αs1-casein slowly, produced a smaller proportion of αs1-casein-derived peptides (expressed as a percentage of the total number of peptides identified), and did not produce any peptides in the His80-to-Arg100 and Leu169-to-Trp199 regions of αs1-casein. The absence of hydrolysis was not observed in previous studies, likely because the αs1-casein peptide f(1-23), on which both CEPs are active and have at least three common cleavage sites, was used as a marker of specificity (32, 34, 36, 42) rather than native whole αs1-casein as in this study. Interestingly, the proteinase of Lactobacillus delbrueckii subsp. lactis CRL 581 (20) had a large number of cleavage sites throughout the αs1-casein sequence, which could explain differences in final texture and biological properties between dairy products produced with that species and those produced with L. helveticus.
The cleavage sites of L. helveticus proteinases were numerous when purified αs1- and β-caseins were used but were not specifically related to one type of amino acid residue, in agreement with other studies (26, 45, 46). The question is how the active sites of the CEPs, either PrtH or PrtH2, can interact with such a variety of amino acid residues, since they both belong to the serine-type family. Although in milk the cleavage sites were more restricted to sites common to the strains, they remained diverse, thus conferring an advantage on the lactobacilli by supplying all amino acids essential for growth.
For strains grown in milk, the number of peptides identified was much lower than the number identified when the cells were incubated with purified caseins. The same overall peptide fingerprint was observed for the 13 strains tested regardless of the number of CEPs. Most of the cleavage sites found were common to all strains when purified αs1- or β-casein was used as a substrate. Accessibility, therefore, seems altered when caseins are assembled into micelles, as previously suggested for trypsin activity (14). A fortiori, most of the hydrolyzed regions in milk were also detected during the ripening of Swiss-type cheese, where a higher degree of casein organization is shown in the curd (13) for all caseins except for the region between the Thr128 and Phe189 of β-casein and the C-terminal end of αs1-casein.
The activities of the CEPs decreased when the bacterial cells were grown in MRS broth or in milk supplemented with 1% tryptic and chymotryptic hydrolysates. It is still difficult to know whether such a decrease concerned only one or both CEPs. Indeed, both genes have previously been reported to be transcribed in MRS broth (16, 43). The genes encoding CEPs were shown to be downregulated in MRS broth compared to milk for strain CNRZ32 (43). However, a decrease in transcription may partly explain the observed change in activity. The regulation of proteinase activity by the nature and amount of peptides present in milk or MRS broth may be another factor that requires further study for strains of L. helveticus.
The 3D modeling of casein molecules allows regions with different levels of organization within each casein type to be identified. When purified caseins were used as substrates, the regions that possessed less secondary structure were highly hydrolyzed by the proteinases of L. helveticus: (i) the N- and C-terminal ends of αs1-casein and (ii) most of the β-casein sequence, which is highly unstructured in comparison to the three other caseins. In milk, only some regions in the middle of the αs2-casein sequence were hydrolyzed. The phosphorylated regions of the caseins were poorly hydrolyzed, in agreement with previous studies (6). When strains were grown in milk, a drastic reduction in the number of peptides was observed, highlighting the parts of the casein sequence that were more resistant to proteolysis when αs1-, αs2,- and β-caseins were assembled into micelles. This study provides a first insight into the change in the accessibility of cleavage sites to their CEPs for a more complex casein organization, such as that in cheese curd. Further study is required to explain fully why cheese texture and functionalities differ so greatly when different proteolytic systems are involved.
Supplementary Material
Acknowledgments
This work was supported by research grants from INRA and the Brittany Region, reference 08008390.
We are grateful to John A. Hannon (Teagasc Food Research Centre, Moorepark, Fermoy, Ireland) for careful reading of the manuscript and fruitful discussions.
Footnotes
Published ahead of print on 29 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Blanc, B., P. Laloi, D. Atlan, C. Gilbert, and R. Portalier. 1993. Two cell-wall-associated aminopeptidases from Lactobacillus helveticus and the purification and characterization of APII from strain ITGL1. J. Gen. Microbiol. 139:1441-1448. [DOI] [PubMed] [Google Scholar]
- 2.Brignon, G., B. Ribadeau-Dumas, and J. C. Mercier. 1976. Premiers éléments de structure primaire des caseines αs2 bovines. FEBS Lett. 71:111-116. [DOI] [PubMed] [Google Scholar]
- 3.Broadbent, J. R., R. L. Thompson, J. E. Hughes, D. L. Welker, J. L. Steele, H. Cai, Y. Ardo, F. K. Vogensen, T. A. Thompkins, K. Hagen, and E. Altermann. 2008. Comparative genome analysis of the obligately homofermentative lactic acid bacterium Lactobacillus helveticus, poster A 063. 9th Int. Symp. Lactic Acid Bacteria, Egmond aan Zee, Netherlands, 31 August to 4 September 2008.
- 4.Bütikofer, U., J. Meyer, R. Sieber, B. Walther, and D. Wechsler. 2008. Occurrence of the angiotensin-converting enzyme inhibiting tripeptides Val-Pro-Pro and Ile-Pro-Pro in different cheese varieties of Swiss origin. J. Dairy Sci. 91:29-38. [DOI] [PubMed] [Google Scholar]
- 5.Corrieu, G., D. Picque, B. Perret, and P. Quemener. 1992. CINAC, an automated system for control of starters. Process (Rennes) 1068:24?-25. [Google Scholar]
- 6.Deutsch, S. M., D. Mollé, V. Gagnaire, M. Piot, D. Atlan, and S. Lortal. 2000. Hydrolysis of sequenced β-casein peptides provides new insight into peptidase activity from thermophilic lactic acid bacteria and highlights intrinsic resistance of phosphopeptides. Appl. Environ. Microbiol. 66:5360-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutsch, S. M., A. Neveu, S. Guezenec, P. Ritzenthaler, and S. Lortal. 2003. Early lysis of Lactobacillus helveticus CNRZ 303 in Swiss cheese is not prophage-related. Int. J. Food Microbiol. 81:147-157. [DOI] [PubMed] [Google Scholar]
- 8.Exterkate, F. A. 1990. Differences in short peptide-substrate cleavage by two cell-envelope-located serine proteinases of Lactococcus lactis subsp. cremoris are related to secondary binding specificity. Appl. Microbiol. Biotechnol. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 9.Farrell, H. M., Jr., E. L. Malin, E. M. Brown, and A. Mora-Gutierrez. 2009. Review of the chemistry of αs2-casein and the generation of a homologous molecular model to explain its properties. J. Dairy Sci. 92:1338-1353. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Espla, M. D., P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flambard, B. 2002. Role of bacterial cell wall proteinase in antihypertension. Sci. Aliment. 22:209-222. [Google Scholar]
- 12.Fox, P. F., and J. Guiney. 1972. A procedure for the partial fractionation of the αs1-casein complex. J. Dairy Res. 39:49-53. [Google Scholar]
- 13.Gagnaire, V., D. Mollé, M. Herrouin, and J. Léonil. 2001. Peptides identified during Emmental cheese ripening: origin and proteolytic systems involved. J. Agric. Food Chem. 49:4402-4413. [DOI] [PubMed] [Google Scholar]
- 14.Gagnaire, V., A. Pierre, D. Mollé, and J. Léonil. 1996. Phosphopeptides interacting with colloidal calcium phosphate isolated by tryptic hydrolysis of bovine casein micelles. J. Dairy Res. 63:405-422. [DOI] [PubMed] [Google Scholar]
- 15.Gatti, M., M. E. Fornasari, A. Perrone, and E. Neviani. 1999. Relationship between acidification capacity in milk, presence of yeast extract, proteolytic and peptidase activities in Lactobacillus helveticus species. Microbiol. Aliment. Nutr. 17:23-31. [Google Scholar]
- 16.Genay, M., L. Sadat, V. Gagnaire, and S. Lortal. 2009. prtH2, not prtH, is the ubiquitous cell wall proteinase gene in Lactobacillus helveticus. Appl. Environ. Microbiol. 75:3238-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, C., B. Blanc, J. Frot-Coutaz, R. Portalier, and D. Atlan. 1997. Comparison of cell surface proteinase activities within the Lactobacillus genus. J. Dairy Res. 64:561-571. [Google Scholar]
- 18.Goldbarg, J. A., and A. M. Rutenberg. 1958. The colorimetric determination of leucine aminopeptidase in urine and serum of normal subjects and patients with cancer or diseases. Cancer 11:283-291. [DOI] [PubMed] [Google Scholar]
- 19.Guillou, H., G. Miranda, and J. P. Pelissier. 1987. Analyse quantitative des caseines dans le lait de vache par chromatographie liquide rapide d'échange d'ions (FPLC). Lait 67:135-148. [Google Scholar]
- 20.Hébert, E. M., G. Mamone, G. Picariello, R. R. Raya, G. S. De Giori, P. Ferranti, and F. Addeo. 2008. Characterization of the pattern of αs1- and β-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Environ. Microbiol. 74:3682-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hébert, E. M., R. R. Raya, and G. S. De Giori. 1997. Characterization of a cell membrane-associated proteinase from Lactobacillus helveticus CRL 581. Curr. Microbiol. 35:161-164. [Google Scholar]
- 22.Hébert, E. M., R. R. Raya, and G. S. De Giori. 2002. Modulation of the cell-surface proteinase activity of thermophilic lactobacilli by the peptide supply. Curr. Microbiol. 45:385-389. [DOI] [PubMed] [Google Scholar]
- 23.Hébert, E. M., R. R. Raya, G. Oliver, and G. S. De Giori. 1996. Characterization of proteolytic activity of Lactobacillus helveticus strains in milk. Microbiol. Aliment. Nutr. 14:65-72. [Google Scholar]
- 24.Hébert, E. M., R. R. Raya, P. Tailliez, and G. S. De Giori. 2000. Characterization of natural isolates of Lactobacillus strains to be used as starter cultures in dairy fermentation. Int. J. Food Microbiol. 59:19-27. [DOI] [PubMed] [Google Scholar]
- 25.Holt, C., and L. Sawyer. 1993. Caseins as rheomorphic proteins: interpretation of primary and secondary structures of the αs1-, β- and κ-caseins. J. Chem. Soc. Faraday Trans. 89:2683-2692. [Google Scholar]
- 26.Jensen, M. P., F. K. Vogensen, and Y. Ardo. 2009. Variation in caseinolytic properties of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 19:661-668. [Google Scholar]
- 27.Kok, J., K. Leenhouts, A. J. Haandrikman, A. Ledeboer, and G. Venema. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumosinski, T. F., E. M. Brown, and H. M. Farrell, Jr. 1991. Three-dimensional molecular modeling of bovine caseins: αs1-casein. J. Dairy Sci. 74:2889-2895. [DOI] [PubMed] [Google Scholar]
- 29.Kumosinski, T. F., E. M. Brown, and H. M. Farrell, Jr. 1993. Three-dimensional molecular modeling of bovine caseins: an energy-minimized β-casein structure. J. Dairy Sci. 76:931-945. [DOI] [PubMed] [Google Scholar]
- 30.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Hernandez, M. C., A. C. Alting, and F. A. Exterkate. 1994. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89. Appl. Microbiol. Biotechnol. 40:828-834. [Google Scholar]
- 33.Matar, C., J. C. Valdez, M. Medina, M. Rachid, and G. Perdigon. 2001. Immunomodulating effects of milks fermented by Lactobacillus helveticus and its non-proteolytic variant. J. Dairy Res. 68:601-609. [DOI] [PubMed] [Google Scholar]
- 34.Oberg, C. J., J. R. Broadbent, M. Strickland, and D. J. McMahon. 2002. Diversity in specificity of the extracellular proteinases in Lactobacillus helveticus and Lactobacillus delbrueckii subsp. bulgaricus. Lett. Appl. Microbiol. 34:455-460. [DOI] [PubMed] [Google Scholar]
- 35.Pastar, I., I. Tonic, N. Golic, M. Kojic, R. Kranenburg, M. Kleerebezem, L. Topisirovic, and G. Jovanovic. 2003. Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl. Environ. Microbiol. 69:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pederson, J. A., G. J. Mileski, and B. C. Weimer. 1999. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen, L. K., P. Hojrup, and T. E. Petersen. 1992. Localization of two interchain disulfide bridges in dimers of bovine αs2-casein. Parallel and antiparallel alignments of the polypeptide chains. J. Biochem. 203:381-386. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen, L. K., P. Hojrup, and T. E. Petersen. 1994. Disulphide arrangement in bovine caseins: localization of intrachain disulphide bridges in monomers of κ- and αs2-casein from bovine milk. J. Dairy Res. 61:485-493. [DOI] [PubMed] [Google Scholar]
- 39.Savijoki, K., H. Ingmer, and P. Varmanen. 2006. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71:394-406. [DOI] [PubMed] [Google Scholar]
- 40.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 41.Schuck, P., M. Piot, S. Mejean, J. Fauquant, G. Brulé, and J. L. Maubois. 1994. Déshydratation des laits enrichis en caséine micellaire par microfiltration; comparaison des propriétés des poudres obtenues avec celles d'une poudre de lait ultra-propre. Lait 74:47-63. [Google Scholar]
- 42.Scolari, G., M. Vescovo, C. Zacconi, and F. Vescovi. 2006. Extraction and partial characterization of proteolytic activities from the cell surface of Lactobacillus helveticus Zuc2. J. Dairy Sci. 89:3800-3809. [DOI] [PubMed] [Google Scholar]
- 43.Smeianov, V. V., V. P. Wechter, J. R. Broadbent, J. E. Hughes, B. T. Rodriguez, T. K. Christensen, Y. Ardo, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valence, F., and S. Lortal. 1995. Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl. Environ. Microbiol. 61:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, N., A. Akino, and T. Takano. 1993. Purification and specificity of a cell-wall-associated proteinase from Lactobacillus helveticus CP790. J. Biochem. 114:740-745. [DOI] [PubMed] [Google Scholar]
- 46.Zevaco, C., and J.-C. Gripon. 1988. Properties and specificity of a cell-wall proteinase from Lactobacillus helveticus. Lait 68:393-408. [Google Scholar]
- 47.Zittle, C. A., and J. H. Custer. 1963. Purification and some of the properties of αs-casein and κ-casein. J. Dairy Sci. 46:1183-1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.