Abstract
Widespread fecal pollution of surface waters in developing countries is a threat to public health and may represent a significant pathway for the global dissemination of antibiotic resistance. The Minjiang River drainage basin in Fujian Province is one of China's most intensive livestock and poultry production areas and is home to several million people. In the study reported here, Escherichia coli isolates (n = 2,788) were sampled (2007 and 2008) from seven surface water locations in the basin and evaluated by PCR for carriage of selected genes encoding virulence factors, primarily for swine disease. A subset of isolates (n = 500) were evaluated by PCR for the distribution and characteristics of class 1 integrons, and a subset of these (n = 200) were evaluated phenotypically for resistance to a range of antibiotics. A total of 666 (24%) E. coli isolates carried at least one of the virulence genes elt, fedA, astA, fasA, estA, stx2e, paa, and sepA. Forty-one percent of the isolates harbored class 1 integrons, and these isolates had a significantly higher probability of resistance to tobramycin, cefoperazone, cefazolin, ciprofloxacin, norfloxacin, azitromycin, and rifampin than isolates with no class 1 integron detected. Frequencies of resistance to selected antibiotics were as high as or higher than those in fecal, wastewater, and clinical isolates in published surveys undertaken in China, North America, and Europe. Overall, E. coli in the Minjiang River drainage basin carry attributes with public health significance at very high frequency, and these data provide a powerful rationale for investment in source water protection strategies in this important agricultural and urban setting in China.
It is estimated that 3.2% of deaths globally are attributable to unsafe water caused by poor sanitation and hygiene, a problem that is particularly acute in rural areas in the developing world (45). In China, 89% of the population use drinking water from improved sources, a significant increase from the 67% estimated in 1990 (44). Nevertheless, increasing urbanization and expanding livestock and poultry production in many areas of the country pose significant challenges for maintaining or improving source water quality (39).
Escherichia coli is a ubiquitous commensal member of the gastrointestinal tract flora of warm-blooded animals. Given its ubiquity in fecal material and its relatively short persistence in environmental matrices, E. coli is the gold standard for detection of fecal pollution in water, and the presence and density of E. coli in water is widely used to measure and regulate water quality (28, 43). E. coli is also an important cause of disease in humans and agricultural livestock, and various pathotypes that cause specific enteric or extraintestinal disease syndromes are distinguished by the complements of virulence genes they carry (4, 5, 19, 27, 40). Furthermore, the emergence and spread of antimicrobial-resistant E. coli and other pathogenic bacteria have become serious global public health threats (34). Water contaminated with effluents from livestock farms, aquaculture, hospitals, municipal wastewater treatment, or pharmaceutical manufacturing can be enriched for enteric bacteria resistant to one or more antibiotics (2, 9, 14, 16, 23, 24, 26, 33, 38, 42, 54). Drinking or recreational use of compromised water represents a pathway for human exposure to antibiotic-resistant commensal and pathogenic bacteria, underscoring the wisdom of mandating practices to mitigate agricultural and human fecal pollution within source water protection strategies (39).
Fujian Province is one of the most important areas of livestock production in China and has a significant resident rural and urban population that draws upon the Minjiang River for drinking water. Although the Minjiang River basin water is of sufficient quality to be potable after treatment, fecal contamination is routinely detected (18). The purpose of the work reported here was to characterize isolates of E. coli obtained over a period of several months from several locations in the Minjiang River basin for attributes with significance for public and animal health, namely, resistance to various antibiotics, the carriage of selected virulence genes, and the distribution of class 1 integrons and associated gene cassettes.
MATERIALS AND METHODS
Sampling-site descriptions.
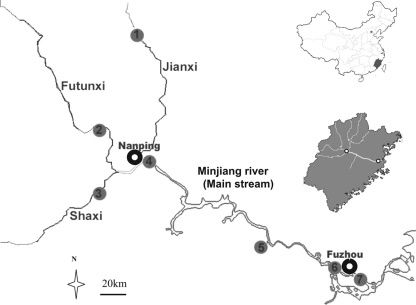
The Minjiang River is the largest river (577 km in length) in Fujian Province; known as the Mother River of Fujian, it is located in the southeast coastal area of China (Fig. 1, upper inset map). The river originates in the Wuyi Mountains, flows through 36 counties and districts, and then flows through the urbanized outskirts and inner city of Fuzhou, the capital of Fujian Province, before discharging into the East China Sea (Fig. 1, main and lower inset maps). The Minjiang River is a significant source of drinking and recreational water and of irrigation water for crop production. The entire river basin drains about 61,000 km2, representing about half of the total area of Fujian Province (Fig. 1, lower inset map). Approximately 16 million people reside within the area, including 4 million people in urban areas.
FIG. 1.
The Minjiang River study area is located in Fujian Province on the east coast of China (upper inset map). Within the province, the drainage basin discharges into the East China Sea (lower inset map). The water-sampling locations on tributaries (1 to 3) and the main reach (4 to 7), as well as the locations of the cities of Nanping and Fuzhou, are detailed in the main map. The two cities are also shown in the lower inset map as white dots.
All of the sampling sites (Fig. 1, main map) are routinely used for water quality measurements by the local environmental authorities. Sites 1, 2, and 3 are situated on the three major tributaries, the Jianxi, Futunxi, and Shaxi, which represent the upper reaches of the Minjiang River. Site 4 is downstream of Nanping City (population, 350,000), where the three main tributaries meet and the main reach of the Minjiang River starts. Site 5 is on the mainstream Minjiang River, and sites 6 and 7 represent, respectively, the upper and lower points of the river flowing through the urban area of Fuzhou City (population, 2 million). Site 6 is adjacent to the western drinking water treatment plant, about 10 km away from site 7, which is close to the eastern drinking water treatment plant of Fuzhou.
Surface water (0.5-m depth) samples were collected directly into triplicate sterile 500-ml glass bottles at each sampling time. Water samples were taken from sites 6 and 7 in February, May, and August 2007 and in January 2008. In September 2007, water samples were collected from each of the 7 sampling locations. All samples (n = 45) were kept in the dark, transported to the laboratory on ice, and analyzed for E. coli within 24 h. Water temperature and pH were measured at the individual sampling sites at sampling time.
Isolation and confirmation of E. coli.
Bacteria in the water were enumerated by membrane filtration as described previously (25). Briefly, surface water samples were passed through a sterile 47-mm-diameter cellulose ester disk filter (average pore size, 0.45 μm; TianHe, Hangzhou, China). The filters were placed on mFC-BCIG agar plates (BD Diagnostics, Franklin Lakes, NJ) and incubated at 44.5°C overnight. After 24 h of growth, the blue colonies appearing on each plate were counted. Single blue colonies were picked and streaked onto LB agar (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom). A total of 300 colonies were taken from each plate, or when fewer than 300 were available, all colonies were picked. The picked colonies were grown at 37°C overnight, and the isolates were purified by restreaking them twice on LB agar. The purified colonies were inoculated into sterile 96-well microplates containing 100 μl fresh LB broth per well and grown statically overnight at 37°C. For confirmation, the isolated cultures were replica plated (10 μl per well) into sterile 96-well microplates containing 100 μl of lactose broth (TianHe, Hangzhou, China) or 100 μl of tryptone broth (TianHe, Hangzhou, China) and were incubated overnight at 37°C. Positive confirmation was indicated by a color change from purple to yellow in lactose broth and by the formation of a red-pink color upon addition of 40 μl of Kovac's reagent to the tryptone broth wells, indicating indole production. Isolates were considered to be E. coli if they grew at 44.5°C, had a positive reaction for β-glucuronidase (blue color on mFC-BCIG agar), fermented lactose, and produced indole. Confirmed isolates were inoculated into sterile 96-well microplates containing 100 μl per well of LB broth and incubated overnight at 37°C. Sterile glycerol was then added to each well to a final concentration of 15% (vol/vol), and the plates were stored at −70°C.
Phenotypic evaluation of antibiotic resistance.
Antibiotic resistance was determined by the disk diffusion method using BBL Sensi-Disc Susceptibility Test Discs (BD Diagnostics, Franklin Lakes, NJ) as described by the Clinical and Laboratory Standards Institute (3) with E. coli ATCC 25922 as a control. One hundred class 1 integron-positive and 100 integron-negative E. coli isolates from seven sampling sites were tested for resistance to ampicillin (AM) (10 μg), tetracycline (TE) (30 μg), streptomycin (STR) (300 μg), gentamicin (GM) (10 μg), chloramphenicol (CHL) (30 μg), norfloxacin (NOR) (10 μg), sulfamethoxazole (SXT) (23.7 μg), ciprofloxacin (CTP) (5 μg), cefoperazone, (CFP) (75 μg), azithromycin (AZM) (15 μg), cefazolin (CZ) (30 μg), ceftriaxone (CRO) (30 μg), nitrofurantoin (F/M) (300 μg), polymyxin B (PB) (300 μg), imipenem (IPM) (10 μg), tobramycin (NN) (10 μg), and rifampin (RA) (5 μg).
PCR detection of virulence genes and class 1 integrons in E. coli isolates.
Nucleic acid for PCR was obtained from cell suspensions by using commercial DNA extraction kits (BioFlux, China) according to the manufacturer's instructions. E. coli isolates were evaluated for the presence of 12 virulence genes: stx2e (Shiga toxin variant), eae (intimin), paa (attaching and effacing protein), sepA (a secreted serine protease of the autotransporter family), astA (heat-resistant toxin), estA (heat-stable toxin A), estB (heat-stable toxin B), elt (heat-labile toxin), fedA (F18 fimbriae), faeG (F4 fimbriae), fanA (F5 fimbriae), and fasA (F6 fimbriae) (1, 19). The primer sequences, PCR conditions, and product sizes are described in Table 1. The 12 primer pairs for amplifying the target virulence factors from E. coli isolates were multiplexed into three groups (1, 7). The first targeted the genes estB, estA, elt, and faeG. The second targeted fanA, fedA, and stx2e, and the third targeted astA, paa, fasA, and sepA. All primers were synthesized by Sangong Inc. (Shanghai, China). E. coli strains RO8, B44, F107, P16M, AMR-472, and JG280 were used as positive controls for the detection of the virulence genes (1). Selected PCR products were sequenced as described below to confirm their identities. The multiplex PCR was performed with a Model 2720 thermocycler (ABI, Foster City CA). A reaction volume of 25 μl of PCR mixture contained PCR buffer, 2.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate (dNTP) (Solarbio, Beijing, China), 10 μM primers, 1.5 U DNA polymerase (Fermentas, Burlington, ON, Canada), and 2 μl of DNA extract (7, 40). The programs used for multiplex PCR are described by Duriez et al. (7).
TABLE 1.
Sequences of primers, PCR conditions, and E. coli strains used as positive controls for virulence genes
| Multiplex group | Gene | Primer sequence (5′-3′)a | Annealing temp (°C) | Fragment size (bp) | Positive control |
|---|---|---|---|---|---|
| 1 | estB | TGCCTATGCATCTACACAAT | 55 | 113 | RO8 |
| CTCCAGCAGTACCATCTCTA | |||||
| estA | CAACTGAATCACTTGACTCTT | 55 | 158 | RO8 | |
| TTAATAACATCCAGCACAGG | |||||
| elt | GGCGTTACTATCCTCTCTAT | 55 | 272 | RO8 | |
| TGGTCTCGGTCAGATATGT | |||||
| faeG | GAATCTGTCCGAGAATATCA | 55 | 499 | RO8 | |
| GTTGGTACAGGTCTTAATGG | |||||
| 2 | fanA | AATACTTGTTCAGGGAGAAA | 55 | 230 | B44 |
| AACTTTGTGGTTAACTTCCT | |||||
| fedA | TGGTAACGTATCAGCAACTA | 55 | 313 | F107 | |
| ACTTACAGTGCTATTCGACG | |||||
| fasA | GTAACTCCACCGTTTGTATC | 55 | 409 | P16M | |
| AAGTTACTGCCAGTCTATGC | |||||
| stx2e | AATAGTATACGGACAGCGAT | 55 | 733 | AMR-472 | |
| TCTGACATTCTGGTTGACGC | |||||
| 3 | astA | TCGGATGCCATCAACACAGT | 62 | 125 | JG280 |
| GTCGCGAGTGACGGCTTTGTAAG | |||||
| paa | GGCCCGCATACAGCCTTG | 62 | 282 | JG280 | |
| TCTGGTCAGGTCGTCAATACTC | |||||
| sepA | TAAAACCCGCCGCCTGAGTA | 62 | 611 | JG280 | |
| TGCCGGTGAACAGGAGGTTT |
Sequences were taken from reference 1.
Class 1 integrons were detected using specific primers (TGATGGCGACGCACGAC and TTGGGCAGCAGCGAAGT) developed with the Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA) software program (48). To evaluate the distribution of class 1 integrons within the E. coli collection, a subset of 500 E. coli isolates were selected from seven sampling locations for detailed analysis. At least 12 isolates from each sampling site were selected, and all of the selected isolates harbored at least one virulence gene (determined by PCR). To characterize the integron gene cassette content, 50 isolates that were class 1 integron positive based on PCR were randomly selected for a conserved-segment PCR (CS-PCR) based on the amplicon containing inserted gene cassettes flanked on both sides by small parts of the CSs (22).
PCR amplifications for class 1 integrons and gene cassettes were performed in a PTC-100 thermocycler (MJ Research Inc., Ramsey, MN). For class 1 integrons, the final 25-μl reaction mixture consisted of 10× Taq buffer with KCl, 2.5 μl; 2.5 mM MgCl2, 2 μl; 10 mM dNTP, 0.5 μl; Taq DNA polymerase, 0.7 μl; 5 μM (each) forward and reverse primers, 0.5 μl; and double-distilled H2O (ddH2O), 16.3 μl. The PCR program was as follows: an initial denaturation at 94°C for 15 min and 30 cycles of denaturation at 94°C for 45 s, annealing at 58°C for 45 s, and extension at 72°C for 45 s, followed by a final extension at 72°C for 10 min. For gene cassettes, the total 50-μl reaction mixture contained 10× buffer, 5 μl; 25 mM MgCl2, 2 μl; 10 mM dNTP, 1 μl; 10 pM each of forward and reverse primers; Taq polymerase, 1.5 μl; template, 4 μl; and ddH2O, 32.5 μl. The program of Lévesque et al. (22) was modified as follows: predenaturation at 95°C for 10 min and 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 40 s, and extension at 72°C for 90 s, followed by a final extension at 72°C for 10 min.
Ten microliters of PCR product were resolved electrophoretically through a 2% horizontal agarose gel in Tris-acetate-EDTA (TAE) buffer. The gel was stained with ethidium bromide solution and visualized with a Gel Doc 2000 (Bio-Rad, Hercules, CA). The banding patterns were digitally captured, and products were sized by reference to the Promega 100- to 2,000-bp DNA ladder.
Characterization of integrons.
Class 1 integron-positive E. coli isolates from different water samples were further characterized by using PCR with the primer pair 5′-CS and 3′-CS (22). The PCR products were then compared by restriction fragment length polymorphism (RFLP) typing with EcoRI according to the method of Gu et al. (12). Amplicons of similar size that yielded the same RFLP pattern were considered to be identical. Representative PCR products that had distinct RFLP patterns were sequenced after being cloned into the pGEM-T Takara D101A Easy vector following the manufacturer's instruction and transformation into E. coli DH5α. Transformants were selected on LB agar plates containing 50 μg/ml ampicillin, 0.5 mM isopropylthiogalactosidase (IPTG), and 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The presence of the required insert in the transformants was confirmed by PCR with the individual primers. Recombinant plasmid DNA extracted from the transformants was sequenced by Sangon Bioengineering Inc. (Shanghai, China) using the Sanger dideoxy method of DNA sequencing with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit, and the reaction mixtures were analyzed on an ABI 3730 (ABI, Foster City, CA). The inserts were sequenced using SP6 and T7 promoter primers. Using the BLASTN program (http:/www.ncbi.gove/blastn), the sequences were compared with those in the GenBank database.
RESULTS
Occurrence of virulence genes in water isolates of E. coli.
Overall, 2,788 water isolates were obtained from seven sampling locations between February 2007 and January 2008 and confirmed phenotypically as E. coli. Levels of E. coli contamination in the water samples obtained ranged from 185 to 1,295 CFU/100 ml. By means of PCR, 666 (24%) of these isolates were found to carry at least 1 of the 11 virulence genes evaluated. The gene astA was carried by 299 isolates (10.7% of the collection), estA by 102 isolates, (3.7%), elt by 85 isolates (3.0%), sepA by 66 isolates (2.4%), estB by 52 isolates (1.9%), fanA by 27 isolates (1%), and paa by 20 isolates (0.7%). The genes faeG (8 isolates), stx2e (5 isolates), and fedA (2 isolates) were each carried by less than 0.4% of the isolates, and fasA was not detected. The majority (568; 85%) of the 666 isolates carried only one virulence gene, whereas 98 isolates (15%) carried either two or three virulence genes. Notably, estA and estB were detected in 78 isolates either alone (n = 52) or in combination with elt (n = 26).
The spatial distribution of these virulence genes within the study area was determined with E. coli isolates (n = 339) obtained from each of the seven sampling sites in September 2007 (Table 2). The gene astA was detected in isolates from all 7 sampling locations. The genes elt, estA, and estB were detected in the majority of the isolates from site 4, whereas they were not detected together elsewhere. Three genes were detected at only one site, faeG at site 3, fedA at site 5, and stx2e at site 6. Altogether, astA was widely distributed within the study area, most genes were detected more frequently downstream of site 3 than above it, and isolates from site 4 were distinguished by the high frequency of carriage of elt, estA, and estB.
TABLE 2.
Distribution of virulence genes in collections of E. coli isolates recovered from each of seven sites sampled in September 2007
| Virulence gene | Frequency in E. coli isolates from sitea: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 (n = 32) | 2 (n = 44) | 3 (n = 12) | 4 (n = 88) | 5 (n = 48) | 6 (n = 49) | 7 (n = 66) | Total | |
| elt | 2 | 58 | 2 | 2 | 6 | 70 | ||
| astA | 3 | 10 | 4 | 5 | 20 | 10 | 18 | 70 |
| estA | 3 | 52 | 1 | 56 | ||||
| estB | 52 | 52 | ||||||
| faeG | 8 | 8 | ||||||
| sepA | 1 | 3 | 2 | 15 | 21 | |||
| paa | 1 | 1 | 1 | 3 | ||||
| fedA | 1 | 1 | ||||||
| stx2e | 1 | 1 | ||||||
| Total | 5 | 13 | 12 | 169 | 28 | 25 | 40 | 282 |
Indicated are the numbers of isolates from each site that carried the indicated gene. The site numbers correspond to those in Fig. 1.
The pathogenic potential of E. coli can be inferred on the basis of virulence gene profiles. Isolates that carry one or more enterotoxin-encoding genes (e.g., elt, estA, and estB) can be considered presumptive enterotoxigenic E. coli (ETEC) (27). On this basis, 538 of the 2,788 virotyped isolates (19.3%) were ETEC. Isolates that carry at least one enterotoxin-encoding gene (e.g., elt, estA, or estB) and one fimbrial gene (e.g., faeG, fasA, or fedA) may be considered potential ETEC for swine (5). On this basis, no swine ETEC isolates were detected. Among the 2,788 virotyped E. coli isolates recovered from the Minjiang River watershed in 2007 and 2008, only 10 possessed faeG, fasA, or fedA, and none of these isolates possessed a toxin gene, indicating that the frequency of swine ETEC isolates by this criterion was below the detection limit of about 0.1% (1/2,788).
The PCR products (n = 11) of elt (272 bp), astA (125 bp), and sepA (611 bp) were sequenced and analyzed with BLASTN. The sequences of the genes elt and astA examined in the present study had 97 to 100% identity with those in GenBank (accession no. EU11352.1 and AF161002) (data not shown). Sequences of sepA amplified from 11 E. coli isolates had 95 to 97% identity with those of the gene sepA in GenBank (accession no. AF386526) (data not shown).
Class 1 integron distribution and variation.
Five hundred representative E. coli isolates were evaluated by PCR for the presence of class 1 integrons. A total of 206 isolates (41% of those evaluated) yielded the expected 587-bp PCR product. A representative subset of 50 of these 206 class 1 integron-positive isolates (isolatesint+) were further analyzed by PCR using the 5′-CS and 3′-CS primers (22). Fifteen isolates yielded larger amplicons of 1,010 bp, 1,406 bp, and 1,587 bp (8 of 15) in various patterns, and 35 isolates did not produce an amplicon. The RFLP (EcoRI) patterns of the three kinds of amplicons were identical, indicating that they each comprised homologous fragments. Upon sequencing, 3 different gene cassettes were identified, including genes encoding resistance to aminoglycosides (aadA1; 1,010 bp), to trimethoprim and aminoglycosides (dfrA1 and aadA1; 1,587 bp), and to rifampin ADP-ribosylating transferase and dihydrofolate reductase (arr-3 and dfrA27; 1,406 bp). The aadA1 sequence shared 99% identity with that reported for another strain of E. coli (GenBank accession no. AB188267; direct deposit). The dhfr1-aadA1 sequence presented 99% identity with those found in E. coli isolates recovered from cattle, pigs, and chickens in Kenya (20) (GenBank accession no. AJ884723). The arr-3 and dfrA27 sequences shared 100% identity with those found in E. coli isolates from China (41) (GenBank accession no. EU675686). The distributions of the gene cassettes and the antimicrobial resistance patterns of the 15 isolates are shown in Table 3. All isolates carried aadA1, and eight also carried dfrA1. Two isolates carried two gene cassettes (dfrA1 and aadA1/arr-3 and dfrA27). The cassette dfrA1 and aadA1 was detected in isolates from sites 1, 2, 4, 5, and 7; the gene addA1 was carried by the isolates from 6 sites; and dfrA1 and aadA1/arr-3 and dfrA27 were detected only in isolates from site 5 (Table 3).
TABLE 3.
Antibiotic resistance phenotypes of E. coli isolates carrying class 1 integrons with the indicated gene cassettes
| Isolate no. | Sampling site | Gene cassette | GenBank accession no. with closest similarity | Size (bp) | Antibiotic resistance profile |
|---|---|---|---|---|---|
| BCHS-16 | 6 | addA1 | AB188267 | 1,010 | CIP, NOR, RA |
| BCMQ-11 | 5 | addA1 | AB188267 | 1,010 | NN, AM, CHL, TE, RA, SXT |
| BCHS-31 | 6 | addA1 | AB188267 | 1,010 | AM, CZ, AZM, F/M, C, RA |
| BC3JF-14 | 7 | addA1 | AB188267 | 1,010 | AM, CRO, CFP, CZ, NOR, AZM, CHL, TE, RA |
| BCJF-60 | 7 | addA1 | AB188267 | 1,010 | AM, CFP, CZ, CHL, TE, RA, PB |
| BC3HS-10 | 6 | addA1 | AB188267 | 1,010 | NN, AM, CRO, CFP, CZ, F/M, CHL, TE, RA |
| BC3JF-10 | 7 | addA1 | AB188267 | 1,010 | GM, STR, NN, AM, CRO, CFP, CZ, NOR, F/M, TE, RA, SXT |
| BCMQ-07 | 5 | dfrA1 and aadA1/arr-3 and dfrA27 | AJ884723/EU675686 | 1,587/1,406 | AM, CRO, CFP, CZ, AZM, CHL, RA |
| BCSC-12 | 2 | dfrA1 and aadA1 | AJ884723 | 1,587 | AM, CFP, CZ, CIP, AZM, CHL, RA |
| BCSC-13 | 2 | dfrA1 and aadA1 | AJ884723 | 1,587 | AM, CRO, CZ, CIP, AZM, F/M, CHL, TE, RA, SXT |
| BCJF-29 | 7 | dfrA1 and aadA1 | AJ884723 | 1,587 | GM, AM, CIP, NOR, AZM, TE, RA, SXT |
| BC3JF-07 | 7 | dfrA1 and aadA1 | AJ884723 | 1,587 | AM, CFP, CZ, CIP, NOR, AZM, F/M, TE, RA |
| BCNP-21 | 4 | dfrA1 and aadA1 | AJ884723 | 1,587 | GM, NN, AM, AZM, TE, RA, SXT |
| BCJY-01 | 1 | dfrA1 and aadA1 | AJ884723 | 1,587 | GM, NN, AM, CRO, CFP, CZ, CIP, NOR, AZM,CHL, TE, RA, SXT |
| BCMQ-36 | 5 | dfrA1 and aadA1/arr-3 and dfrA27 | AJ884723/EU675686 | 1,587/1,406 | NN, AM, CRO, CFP, CZ, AZM, F/M, CHL, RA, SXT |
Antimicrobial resistance phenotypes of class 1 integron-positive and -negative isolates.
Using the K-L disk diffusion method, 100 isolates carrying class 1 integrons (isolatesint+) and 100 isolates for which class 1 integrons were not detected (isolatesint−) were examined for their resistance to 17 antibiotics (Table 4). One hundred ninety-nine isolates (99.5%) were resistant to four or more antimicrobial agents tested. More than half the isolates were resistant to ampicillin, tetracycline, rifampin, and chloramphenicol. Less than a quarter of the isolates were resistant to gentamicin, tobramycin, cefoperazone, norfloxacin, and polymyxin B. Not a single isolate was resistant to carbapenem. Isolates that carried a class 1 integron had a higher frequency (P < 0.05) of resistance to tobramycin, cefoperazone, cefazolin, ciprofloxacin, norfloxacin, azithromycin, and rifampin (Table 4). One hundred and ninety-nine of 200 randomly selected isolates were resistant to at least 3 and up to 13 antimicrobial agents with a total of 175 multiple-antibiotic resistance phenotypic patterns (data not shown).
TABLE 4.
Comparison of antimicrobial resistance between integron-positive (n = 100) and integron-negative (n = 100) E. coli isolatesa
| Antimicrobial agent | Resistance frequency (%) |
Mean % | χ2 | P value | |
|---|---|---|---|---|---|
| Integron-positive isolates (n = 100) | Integron-negative isolates (n = 100) | ||||
| Aminoglycosides | |||||
| GM | 20 | 25 | 22.5 | 0.1 | >0.05 |
| STR | 50 | 37 | 43.5 | 2.93 | >0.05 |
| NN | 15 | 6 | 10.5 | 4.5 | <0.05 |
| Cephalosporins | |||||
| CFP | 31 | 15 | 23 | 6.35 | <0.05 |
| CRO | 26 | 27 | 26.5 | 0 | >0.05 |
| CZ | 37 | 54 | 40.5 | 5.16 | <0.05 |
| β-Lactams | |||||
| AM | 75 | 71 | 73 | 0.23 | >0.05 |
| Quinolones | |||||
| CIP | 36 | 19 | 27.5 | 6.42 | <0.05 |
| NOR | 36 | 6 | 21 | 27.12 | <0.05 |
| Macrolides | |||||
| AZM | 36 | 22 | 29 | 4.1 | <0.05 |
| Tetracyclines | |||||
| TE | 71 | 67 | 69 | 0.21 | >0.05 |
| Sulfonamides | |||||
| SXT | 46 | 36 | 41 | 1.67 | >0.05 |
| Rifamycins | |||||
| RA | 96 | 79 | 87.5 | 11.7 | <0.05 |
| Phenicols | |||||
| CHL | 79 | 84 | 81.5 | 0.67 | >0.05 |
| Carbapenem | |||||
| IPM | 0 | 0 | 0 | ||
| Lipopeptide | |||||
| PB | 2 | 8 | 5 | 2.63 | >0.05 |
| Nitrofuran | |||||
| F/M | 33 | 46 | 39.5 | 3.01 | >0.05 |
Differences in resistance between the two groups were established using the χ2 test and were considered significant at a P value of <0.05.
DISCUSSION
Overall, about a quarter of the E. coli isolates carried at least 1 of the 11 virulence genes evaluated. These 11 genes are a small fraction of the virulence genes that would be required to detect the many known enteric and extraintestinal pathotypes within the E. coli collection (14). With the exception of astA, which was detected throughout the study area, virulence genes were detected less frequently in isolates from the more pristine sites 1 to 3 than in those from sites with more urban and agricultural development (sites 4 to 7) (Table 2). In particular, isolates obtained from site 4, downstream from Nanping, carried the toxin genes elt, estA, and estB at high frequency. We isolated E. coli on the basis of growth at 44.5°C and the presence of β-glucoronidase activity. These conditions would not allow the growth or detection of isolates of E. coli O157:H7 (32). Taken together, the data obtained in this study most likely underrepresent the pathogenic potential of waterborne E. coli in this drainage basin and should be interpreted conservatively.
Class 1 integrons are crucial elements in the worldwide problem of antibiotic resistance and are often embedded in promiscuous plasmids, transposons, and chromosomes, as well, facilitating their lateral transfer into a wide range of pathogens (10, 11, 41). In the present study, 41% of the 500 E. coli isolates screened harbored class 1 integrons. This compares with 13% of E. coli isolates from the Rio Grande River (35), 3.6% of Gram-negative bacteria isolated from an estuary in the United Kingdom (36), and 3% of E. coli isolates from tap and well water in Turkey (29). Up to 70% of Gram-negative bacteria isolated from humans or livestock carry class 1 integrons (8, 11, 13, 49, 53). Fifteen percent of bacteria in wastewater from a penicillin-manufacturing plant harbored class 1 integrons, whereas over 97% did so in wastewater from an oxytetracycline-manufacturing plant (23, 24). An estimated 2.7% of the total bacteria in creek sediments carried class 1 integrons, based on quantitative-PCR (qPCR) evaluation of extracted DNA (37). Taken together, the frequency of class 1 integrons in E. coli isolates from the Minjiang River is more characteristic of human and livestock fecal isolates or isolates from water exposed to massive antibiotic concentrations than of isolates that have been obtained from other freshwater surveys.
Isolates that carried class 1 integrons were more likely to be resistant to tobramycin, cefoperazone, cefazolin, ciprofloxacin, norfloxacin, azithromycin, and rifampin than isolates that did not carry integrons (Table 5). However, the occurrence of gene cassettes found in this study could not account for resistance to most of the antimicrobial agents investigated, except for arr-3 and dfrA27 (rifampin ADP-ribosylating transferase and dihydrofolate reductase), conferring resistance to RA and SXT. For example, among 15 isolates carrying aadA, only one isolate (BC3JF-10) was phenotypically resistant to streptomycin. Likewise, isolates resistant to AM, TE, and CHL, as well as CRO, CFP, and CZ, had no corresponding resistance genes, such as β-lactamases (bla), tetracycline (tet), and chloramphenicol (cat), detected in the variable regions of the integrons analyzed. The discordance between genotypes and phenotypes in this limited study is not surprising, since several genes may confer a given phenotype.
TABLE 5.
Comparison of resistances to selected antibiotics of Minjiang River basin E. coli isolates (this study) with those of E. coli isolates obtained in other studies from surface water, fecal material, retail meat, or clinical specimens
| Source of isolate | % of isolates resistant |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|
| Gentamicin | Streptomycin | Ampicillin | Ciprofloxacin | Norfloxacin | Tetracycline | Chloramphenicol | ||
| Minjiang River surface water (n = 200) | 22 | 44 | 73 | 27 | 21 | 69 | 81 | This study |
| Animal feces (United States) (n = 1,037) | 0.8 | 13.2 | 5.6 | 28 | 1 | 15 | ||
| Livestock waste lagoons (United States) (n = 2,008) | 16 | 39 | 36 | 30 | ||||
| Retail meat [(beef; Canada) (n = 572)] | 2 | 7 | 4 | 1 | 20 | 3 | 31 | |
| Chicken abattoir (Canada) (n = 170) | 8 | 45 | 38 | 1 | 50 | 4 | 31 | |
| Swine on farm (n = 1,425) | 2 | 37 | 38 | 0.1 | 80 | 20 | 31 | |
| Seine River downstream of STPa (France) (n = 104) | 5.8 | 9.6 | 43.6 | 38 | 21 | |||
| STP effluent (Austria) (n = 767) | 0 | 16 | 0 | 0 | 35 | 8 | 34 | |
| Clinical isolates (China) (n = 732) | 60 | 65 | 49 | |||||
| Wenyu River basin (China) (n = 388) | 0 | 7.9 | 36 | 16 | ||||
STP, sewage treatment plant.
The frequency of resistance to a variety of antibiotics observed in this study was generally at least as high as those reported in other surveys of antibiotic-resistant E. coli in source water, food, or animals (Table 5). The high level of antibiotic resistance found in this survey was most probably due to the widespread and heavy use of numerous antimicrobial agents in human therapy, livestock, poultry, and fish production in the area. An estimated 30% of drugs sold in Chinese hospitals and medical stores are antibiotics, while in the developed world, the proportion is only 10% (17). China had the highest levels of antibiotic resistance (mean prevalences of resistance, 41% in hospital-acquired infections and 26% in community-acquired infections) in a comparative analysis with Kuwait and the United States (52). Furthermore, China had a higher rate of resistance development (22% average annual growth from 1994 to 2000) than Kuwait (17% from 1999 to 2003) or the United States (6% from 1999 to 2002) (52). Overall, antibiotic resistance is a public health issue of great significance in China (47, 53).
Like many other parts of China, Fujian Province is now experiencing extraordinary environmental pressures accompanying rapid economic growth, increasing urbanization, and expansion of the agricultural sector. Although some progress has been made in creating a healthy environment, investments in the water treatment infrastructure have not been sufficient to mitigate widespread water pollution (45, 46, 51). An estimated 600 million tons of poultry and livestock manure was produced in Fujian Province in 2004, most of which was discharged directly into the Minjiang River and other waterways without any treatment (6, 50). The highest density of livestock and crop farms in Fujian Province is in proximity to Nanping City, including two of the biggest dairy farms. In recent years, only 82 of the 793 farms in the Nanping area have launched pollution control programs, and only 10% of the farms are estimated to participate. Although livestock are the major source of aquatic pollution in the area, municipal domestic sewage is likewise a major source of the surface water pollution in the Fuzhou region (18, 55).
In conclusion, this study has detected virulence genes, antibiotic resistance, and class 1 integrons at very high frequency in waterborne E. coli bacteria within the Minjiang drainage basin. The widespread occurrence of these attributes of public health and zoonotic concern underscores the critical need to improve the management of fecal wastes within the area for the protection of human and animal health and with respect to managing the spread of antibiotic resistance.
Acknowledgments
This study was funded in part by the Fujian Science and Technology Foundation for Environmental Protection through grant no. 2006-12 to W.Z., the Fujian Natural Science Foundation through grant no. 2010J01097 to B.C., and Agriculture and Agri-Food Canada.
We are thankful to three anonymous reviewers whose comments greatly improved this paper.
Footnotes
Published ahead of print on 5 November 2010.
REFERENCES
- 1.Boerlin, P., R. M. Travis, C. L. Gyles, R. Reid-Smith, N. Janecko, L. Heather, V. Nicholson, S. A. McEwen, R. M. Friendship, and M. Archambault. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabello, F. C. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8:1137-1144. [DOI] [PubMed] [Google Scholar]
- 3.Clinical Laboratory Standards Institute. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: approved standard, 3rd ed. CLSI document M31-A3. Clinical Laboratory Standards Institute, Wayne, PA.
- 4.Croxen, M. A., and B. B. Finlay. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26-38. [DOI] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 68:7028-7038. [DOI] [PubMed] [Google Scholar]
- 6.Duan, Y., Y. Z. Zhang, Y. F. Li, and Z. Y. Niu. 2007. Pollution load and environmental risk assessment of livestock manure in Minjiang River Valley. J. Ecol. Rural Environ. 23:55-59. [Google Scholar]
- 7.Duriez, P., Y. Zhang, Z. Lu, A. Scott, and E. Topp. 2008. Loss of virulence genes in Escherichia coli populations during manure storage on a commercial swine farm. Appl. Environ. Microbiol. 74:3935-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebner, P., K. Garner, and A. Mathew. 2004. Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Meleagridis. J. Antimicrob. Chemother. 53:1004-1009. [DOI] [PubMed] [Google Scholar]
- 9.Edge, T. A., and S. Hill. 2005. Occurrence of antibiotic resistance in Escherichia coli from surface waters and fecal pollution sources near Hamilton, Ontario. Can. J. Microbiol. 51:501-505. [DOI] [PubMed] [Google Scholar]
- 10.Gillings, M., Y. Boucher, M. Labbate, A. Holmes, S. Krishnan, M. Holley, and H. W. Stokes. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190:5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu, B., S. Pan, T. Wang, W. Zhao, Y. Mei, P. Huang, and M. Tong. 2008. Novel cassette arrays of integrons in clinical strains of Enterobacteriaceae in China. Int. J. Antimicrob. Agents 32:529-533. [DOI] [PubMed] [Google Scholar]
- 13.Gu, B., M. Tong, W. Zhao, G. Liu, M. Ning, and S. Pan. 2008. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J. Clin. Microbiol. 45:241-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamelin, K., G. Bruant, A. El-Shaarawi, S. Hill, T. A. Edge, J. Fairbrother, J. Harel, C. Maynard, L. Masson, and R. Brousseau. 2007. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl. Environ. Microbiol. 73:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwicke, S. A., H. W. Stokes, F. Sophia, T. Mark, and R. G. Michael. 2008. Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 278:207-212. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J. Y., J. C. Shi, H. Chang, D. Li, M. Yang, and Y. C. Kamagata. 2008. Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 42:3415-3420. [DOI] [PubMed] [Google Scholar]
- 17.Jia, H. P. 2008. Antibiotic resistance and the developing world. Science and Development Network. http://www.scidev.net/en/features/antibiotic-resistance-and-the-developing-world.html.
- 18.Jiang, Y., L. Jin, and T. Lin. 2010. Higher water tariffs for less river pollution—evidence from Min River and Fuzhou City, People's Republic of China. ADB economics working paper series no. 201. Asian Development Bank, Manila, Philippines.
- 19.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 20.Kikuvi, G. M., S. Schwarz, J. N. Ombui, E. S. Mitema, and C. Kehrenberg. 2007. Streptomycin and chloramphenicol resistance genes in Escherichia coli isolates from cattle, pigs, and chicken in Kenya. Microb. Drug Resist. 13:62-68. [DOI] [PubMed] [Google Scholar]
- 21.Laroche, E., B. Pawlak, T. Berthe, D. Skurnik, and F. Petit. 2009. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol. Ecol. 68:118-130. [DOI] [PubMed] [Google Scholar]
- 22.Lévesque, C., L. Piche, C. Larose, and P. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, D., M. Yang, J. Hu, J. Zhang, R. Liu, X. Gu, Y. Zhang, and Z. Wang. 2009. Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ. Microbiol. 11:1506-1517. [DOI] [PubMed] [Google Scholar]
- 24.Li, D., T. Yu, Y. Zhang, M. Yang, Z. Li, M. Liu, and R. Qi. 2010. Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river. Appl. Environ. Microbiol. 76:3444-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyautey, E., Z. Lu, D. R. Lapen, G. Wilkes, A. Scott, T. E. Berkers, T. A. Edge, and E. Topp. 2010. Distribution and diversity of Escherichia coli populations in the South Nation River drainage basin, Eastern Ontario, Canada. Appl. Environ. Microbiol. 76:1486-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan, S. L., E. J. Hollis, M. M. Depas, M. Van Dyke, J. Harris, and C. O. Scopel. 2007. Distribution and fate of Escherichia coli in Lake Michigan following contamination with urban stormwater and combined sewer overflows. J. Great Lakes Res. 33:566-580. [Google Scholar]
- 27.Müller, D., L. Greune, G. Heusipp, H. Karch, A. Fruth, H. Tschape, and M. A. Schmidt. 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl. Environ. Microbiol. 73:3380-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Research Council and Committee on Indicators for Waterborne Pathogens (ed.). 2004. Indicators for waterborne pathogens. National Academies Press, Washington, DC.
- 29.Ozgumus, O. B., E. Celik-Sevim, S. Alpay-Karaoglu, C. Sandalli, and A. Sevim. 2007. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J. Microbiol. 45:379-387. [PubMed] [Google Scholar]
- 30.Parveen, S., J. Lukasik, T. M. Scott, M. L. Tamplin, K. M. Portier, S. Sheperd, K. Braun, and S. R. Farrah. 2006. Geographical variation in antibiotic resistance profiles of Escherichia coli isolated from swine, poultry, beef and dairy cattle farm water retention ponds in Florida. J. Appl. Microbiol. 100:50-57. [DOI] [PubMed] [Google Scholar]
- 31.Public Health Agency of Canada. 2010. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2007. Public Health Agency of Canada, Guelph, Ontario, Canada. [DOI] [PubMed]
- 32.Raghubeer, E. V., and J. R. Matches. 1990. Temperature range for growth of Escherichia coli serotype O157:H7 and selected coliforms in E. coli medium. J. Clin. Microbiol. 28:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinthaler, F. F., J. Posch, G. Feierl, G. Wüst, D. Haas, G. Ruckenbauer, F. Mascher, and E. Marth. 2003. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37:1685-1690. [DOI] [PubMed] [Google Scholar]
- 34.Rice, L. B. 2009. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 12:476-481. [DOI] [PubMed] [Google Scholar]
- 35.Roe, M. T., E. Vega, and S. D. Pillai. 2003. Antimicrobial resistance markers of class 1 and class 2 integron-bearing Escherichia coli from irrigation water and sediments. Emerg. Infect. Dis. 9:822-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosser, S. J., and H. K. Young. 1999. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 44:11-18. [DOI] [PubMed] [Google Scholar]
- 37.Sayah, R. S., J. B. Kaneene, Y. Johnson, and R. Miller. 2005. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szczepanowski, R., B. Linke, I. Krahn, K. H. Gartemann, T. Gutzkow, W. Eichler, A. Pühler, and A. Schlüter. 2009. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155:2306-2319. [DOI] [PubMed] [Google Scholar]
- 39.Wang, X. 2006. Management of agricultural nonpoint source pollution in China: current status and challenges. Water Sci. Technol. 53:1-9. [DOI] [PubMed] [Google Scholar]
- 40.Watterworth, L., E. Topp, H. Schraft, and K. T. Leung. 2005. Multiplex PCR-DNA probe assay for the detection of pathogenic Escherichia coli. J. Microbiol. Methods 60:93-105. [DOI] [PubMed] [Google Scholar]
- 41.Wei, Q. H., X. F. Jiang, and Y. G. Lü. 2008. Advances in integrons of bacteria. Chin. J. Antibiot. 33:1-5, 40. [Google Scholar]
- 42.Whitman, R. L., M. B. Nevers, and M. N. Byappanahalli. 2006. Examination of the watershed-wide distribution of Escherichia coli along southern Lake Michigan: an integrated approach. Appl. Environ. Microbiol. 72:7301-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization. 2004. Guidelines for drinking-water quality. World Health Organization, Geneva, Switzerland.
- 44.World Health Organization. 2009. Global health risks: mortality and burden of disease attributable to selected major risks. WHO Press, Geneva, Switzerland.
- 45.World Health Organization and United Nations Children's Fund. 2010. Progress on sanitation and drinking-water: 2010 update report. World Health Organization, Geneva, Switzerland.
- 46.Wu, C., C. Maurer, Y. Wang, S. Xue, and D. L. Davis. 1999. Water pollution and human health in China. Environ. Health Perspect. 107:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, Y., J. Wang, Y. Li, and MOH National Antimicrobial Resistance Investigation Net. 2008. Bacterial resistance surveillance in China: a report from Mohnarin 2004-2005. Eur. J. Clin. Microbiol. Infect. Dis. 27:697-708. [DOI] [PubMed] [Google Scholar]
- 48.Xiulei, C., W. Qiang, C. Yanshun, B. Guolian, H. Cuimei, J. Quanan, L. Jianliang, and X. Zhang. 2007. Drug sensitivity test and detection of class I integron in clinical isolates of Riemerella anatipestifer. Chin. J. Prev. Vet. Med. 9:727-731. [Google Scholar]
- 49.Xu, X., F. Kong, X. Cheng, B. Yan, X. Du, J. Gai, H. Ai, L. Shi, and J. Iredell. 2008. Integron gene cassettes in Acinetobacter spp. strains from South China. Int. J. Antimicrob. Agents 32:441-445. [DOI] [PubMed] [Google Scholar]
- 50.Ye, X.-M., Z.-Z. Chang, X. Chen, H.-Y. Huang, Y. Ma, and J.-Y. Zhang. 2007. Hazard of pathogenic microorganisms in discharge from livestock and poultry breeding farms. J. Ecol. Rural Environ. 23:66-70. [Google Scholar]
- 51.Zhang, J., D. L. Mauzerall, T. Zhu, S. Liang, M. Ezzati, and J. V. Remais. 2010. Environmental health in China: progress towards clean air and safe water. Lancet 375:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, R., K. Eggleston, V. Rotimi, and R. J. Zeckhauser. 2006. Antibiotic resistance as a global threat: evidence from China, Kuwait and the United States. Global Health 7:2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, X. Y., L. J. Ding, and J. Yue. 2009. Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in Northeastern China. Microb. Drug Resist. 15:323-328. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., F. Guo, W. Meng, and X. Q. Wang. 2009. Water quality assessment and source identification of Daliao river basin using multivariate statistical methods. Environ. Monitor. Assess. 152:105-121. [DOI] [PubMed] [Google Scholar]
- 55.Zhong, S. F. 2004. Current status of the drainage of municipal domestic sewage in Fuzhou and counter measures. J. Fujian Univ. Technol. 2:320-325. [Google Scholar]