Abstract
Improved understanding of the ecology and epidemiology of Campylobacter in the poultry farm environment is key to developing appropriate farm-based strategies for preventing flock colonization. The sources of Campylobacter causing broiler flock colonization were investigated on one poultry farm and its environment, from which samples were obtained on three occasions during each of 15 crop cycles. The farm was adjacent to a dairy farm, with which there was a shared concreted area and secondary entrance. There was considerable variation in the Campylobacter status of flocks at the various sampling times, at median ages of 20, 26, and 35 days, with 3 of the 15 flocks remaining negative at slaughter. Campylobacters were recoverable from various locations around the farm, even while the flock was Campylobacter negative, but the degree of environmental contamination increased substantially once the flock was positive. Molecular typing showed that strains from house surroundings and the dairy farm were similar to those subsequently detected in the flock and that several strains intermittently persisted through multiple crop cycles. The longitudinal nature of the study suggested that bovine fecal Campylobacter strains, initially recovered from the dairy yard, may subsequently colonize poultry. One such strain, despite being repeatedly recovered from the dairy areas, failed to colonize the concomitant flock during later crop cycles. The possibility of host adaptation of this strain was investigated with 16-day-old chickens experimentally exposed to this strain naturally present in, or spiked into, bovine feces. Although the birds became colonized by this infection model, the strain may preferentially infect cattle. The presence of Campylobacter genotypes in the external environment of the poultry farm, prior to their detection in broiler chickens, confirms the horizontal transmission of these bacteria into the flock and highlights the risk from multispecies farms.
Case control studies indicate that the handling or consumption of poultry meat is an important source of campylobacteriosis, accounting for about 20 to 40% of cases in those countries for which data are available, including the Netherlands (18), United Kingdom (40), and Australia (37). However, source attribution studies using multilocus sequence typing (MLST) suggest that Campylobacter strains associated with poultry may account for up to 80% of cases (35). In a recent European Food Safety Authority opinion, this difference has been in part explained by the contamination of the general environment by “poultry-associated” campylobacters (http://www.efsa.europa.eu/en/scdocs/scdoc/1437.htm). Thus, reduction of the prevalence of broiler flocks colonized with Campylobacter is considered key to the control of contamination along the whole food production chain, including the farm environment, and subsequently control of campylobacteriosis in humans (28).
Campylobacter is rarely recovered from intensively reared broiler chicks until 14 to 21 days of age (12, 36, 39), and vertical transmission is now generally dismissed as an important source of flock infection (7, 28). Campylobacters are ubiquitous in most environments, and horizontal transmission is considered the major route for colonization of housed broilers. Nevertheless, the implementation of generic biosecurity measures effective at preventing flock infection has proven extremely difficult, indicating that targeted biosecurity intervention strategies will be required. However, before such measures can be introduced, the key farm-level environmental sources of flock infection must be identified.
Farm-level epidemiological survey-based studies have identified a number of important risk factors for infection of broiler flocks (4, 15, 22, 24) and in recent systematic reviews (1; http://www.foodbase.org.uk/results.php?f_category_id=&f_report%20id=384) of available literature, the most important risk factors identified included the age of the flock, the use of staggered slaughter, the presence of multiple broiler houses, farmworkers, and other livestock on the farm. Molecular epidemiological investigations have provided supporting evidence for the role of farm surrounds and on-farm puddles (5, 20), flies (16), transport crates (17), and broiler house water systems (30) as sources. However, the direction of this environmental contamination is uncertain and may be both from and into the poultry house. The presence of other livestock on poultry farms is of particular interest because such animals can constitute a substantial amplification reservoir for campylobacters. Campylobacter jejuni or Campylobacter coli is frequently isolated from the feces of livestock, including dairy and beef cattle (10, 27, 29, 38), and strains matching those found in poultry flocks have been identified in cattle housed on or near broiler farms (14). However, once again the direction of transmission is unclear.
Investigation of the molecular epidemiology of campylobacters on poultry farms is fraught with difficulties. These organisms are fastidious to recover and maintain in culture, particularly from environmental sources. In order to optimize the chance of identifying the source of flock infection and establishing the direction of transmission, longitudinal studies involving multiple samples with a structured sampling strategy are necessary. Such studies can result in large numbers of isolates which need to be appropriately typed, using a strategy that allows sufficient discriminatory power to identify identical strains but minimize resource usage (41). The latter requirement is confounded by the genetic instability of campylobacters under various environmental conditions, including chicken colonization (32).
The primary objective of our study was to investigate the detailed molecular epidemiology of Campylobacter on one broiler farm contiguous with a dairy farm to determine the impact of rearing two species of animals within the same vicinity. A structured longitudinal sampling strategy was adopted over a 2-year period, including 15 flocks to investigate temporal trends. Strains were genotyped by a layered approach. The transmission of Campylobacter strains from the dairy farm to the poultry farm was studied and further investigated with experimental challenge studies of chickens.
MATERIALS AND METHODS
Rearing of the birds.
The farm investigated was managed under contract for a large United Kingdom poultry producer and was located in a rural area surrounded by fields. The farm was relatively new and comprised of four paired poultry sheds located adjacent to a dairy farm with an adjoining concrete farmyard. The farmer managing the dairy farm also reared young calves housed in pens in this adjoining yard.
One target house was selected for sampling on the poultry farm. The back doors of the target poultry house were contiguous with the concreted area (including drains) of the calf pens. The distance between the calf pens and these doors was approximately 15 m. Overall biosecurity measures were considered by visiting researchers to be relatively good with regard to the changing of footwear, provision and use of foot dips, and presence of hand washing and sanitization facilities in each anteroom. However, biosecurity relative to vehicle access to the poultry farm was relatively poor in so much as the gate was not closed during the day, the wheel wash was not operational for most of the study period, and there was no defined parking area.
The dairy and poultry farms shared a drive adjacent to some of the poultry houses, which was used by tractors going to the dairy farm and vehicles exiting the poultry farm. The poultry farm office was situated in the middle of the four poultry houses, so farmer and visitor vehicles tended to park in this area. With regard to the poultry houses themselves, these had a shared anteroom/control room between two houses. The poultry farm sourced its water through a borehole, and the water was treated with chlorine dioxide via an automatic dosing system. The ventilation fans in all the poultry houses were reversible and were situated at a low elevation on the side walls. In practice, the fans were only reversed when the houses were being depopulated. Following clearance, the houses were extensively cleaned; the litter was removed, and the houses were washed down with Hyperox (DuPont Animal Health Solutions, Sudbury, United Kingdom) and disinfected with a formaldehyde-based product.
Collection of samples.
In all, 15 broiler crops were studied on this farm. Standardized sampling, according to a predefined schedule, usually took place during three visits per crop cycle, between March 2006 and April 2008. The timing of the sampling visits varied (Table 1). Partial depopulation (i.e., the practice of the early removal of a portion of the birds) was practiced for crops 1 and 2 only, and the second visit coincided with this event. For the remaining flocks, visit 1 was at 20 to 24 days, visit 2 was at 24 to 35 days, and visit 3 was at 34 to 36 days, which coincided with clearance. Additional sampling was conducted on the day of fill for the first crop cycle only, to determine the general level of contamination on the farm and to confirm that Campylobacter was not detectable from chicks and crates arriving from the hatchery prior to placement of the birds. At each visit, the flock was sampled by gathering pools of six freshly voided fecal or cecal droppings from each of six predefined zones in the target house. Two overshoe samples were also collected at each visit from a walk-through of the whole house, again following a predetermined pattern. In addition, 16 pairs of ceca were collected in the processing plant at slaughter of the target flock.
TABLE 1.
Age of birds on the days in which crop cycle visits were undertaken and flock infection status at each visit
| Crop no. | Age of birds in days (infection status) at: |
||
|---|---|---|---|
| Visit 1 | Visit 2 | Visit 3 | |
| 1 (thinned) | 28 (−)a | 36 (+)b | 45 (+) |
| 2 (thinned) | 27 (−) | 35 (+) | 45 (+) |
| 3 | 23 (+) | No visit | 35 (+) |
| 4 | 24 (+) | No visit | 35 (+) |
| 5 | 20 (−) | 25 (−) | 35 (+) |
| 6 | 20 (−) | 24 (−) | 36 (+) |
| 7 (affected by AI) | 20 (−) | No visit | 35 (+)b |
| 8 | 20 (−) | 26 (+) | 35 (+) |
| 9 | 21 (+) | 25 (+) | 35 (+) |
| 10 | 20 (+) | 25 (+) | 34 (+) |
| 11 | 20 (+) | 28 (+) | 36 (+) |
| 12 (affected by AI) | 20 (−) | 24 (−) | 35 (−)b |
| 13 (affected by AI) | No visit | No visit | 34 (−) |
| 14 | 20 (−) | 26 (−) | 35 (+)b |
| 15 | 22 (−) | 28 (−) | 34 (−) |
Crop 1 was also sampled at fill to check that the birds (chick papers) and target house were negative at the point of fill.
From ceca at processing plant.
At each visit, a set of samples (n = 58) were always collected from the environment surrounding the broiler houses. These samples were collected by a standardized protocol and were categorized as follows: surroundings of the monitored shed and track, which was used by personnel from a neighboring dairy farm; main entrance area and wheel-wash; water from an effluent tank and trough; dead birds stored on site; litter scattered or stored outside; anteroom doors and floor areas; and water from the mains and in-house water. Additional samples were collected when feasible. These included samples from on-farm vehicles and equipment (including vehicles owned by either farmer, tractors, trailers, and brushes or footwear present in farm office and/or anteroom areas); air in and outside the poultry house; and feces of other wild or domestic animals, such as wild birds and rabbits, in the near vicinity of the target house. Beetles and flies recovered from the house were also sampled when present. Other potential Campylobacter reservoirs, such as vehicles and equipment belonging to the catching crews, entering the farm, were also sampled. In practice, some of these samples were inconsistently collected because they were not always present: i.e., puddles, farm-associated vehicles and equipment, and wildlife feces.
Surfaces were sampled by swabbing an area of approximately 100 cm2 from each site with a sterile Readiwipe (Robinson Healthcare, Ltd., Chesterfield, United Kingdom), while boot swabs in the form of gauze overshoes (Mike Bowden Livestock Service, Attleborough, Norfolk, United Kingdom) were used for sampling large areas of grass or concrete, as previously described (6). Fecal droppings from calves, wildlife, and the flock were collected by inverting sterile plastic bags using sterile disposable gloves (2, 6). Insects were crushed and placed in 28 ml of modified Exeter broth (mEB) (19) before being transported to the laboratory (2).
Samples of water (10 ml) were taken from the nipples on the drinker lines and swabs taken from the nipple drinkers and cups by pushing the swab onto the nipple until wet and then wiped around the cup. Each water sample was added to 10 ml double-strength mEB. In addition, during the final two crop cycles, water (1 liter) was taken from the blind end of one drinker line at each sampling visit. In addition, air sampling was performed during eight visits from six crop cycles, as previously described (6).
Culture and identification of Campylobacter.
Fecal and cecal samples from broiler chickens were cultured on selective agar (sheep blood agar containing Skirrow's supplement, plus cycloheximide [ActiDione] and cefoperazone [BASAC]) at 42°C under microaerobic conditions (5% O2, 10% CO2, 85% N2), as previously described (2, 6). All environmental samples were transported to the laboratory, under chilled conditions, within 24 h of collection. Swabs, overshoes, and nonpoultry fecal samples were placed in mEB before transportation. All nonwater samples were then enriched for Campylobacter in mEB for 48 h at 37°C prior to being streaked onto modified charcoal cefoperazone deoxycholate agar (mCCDA) and incubated at 41.5°C for 48 h. The method used for analysis of water was performed as described in EN ISO 17995 “Water quality—detection and enumeration of thermotolerant Campylobacter species” (http://www.iso.org/iso/catalogue_detail.htm?csnumber=42082).
Where available, three colonies per sample were subcultured onto blood agar base no. 2 Oxoid CM0271 and incubated microaerobically at 41.5°C for 24 h. The following confirmatory tests were performed: cell morphology with a wet preparation or a Gram stain (with dilute carbol fuchsin as the counterstain), oxidase test, and lack of growth in air at 25°C after 48 h. A selection of isolates were examined by Oxoid Campy Dry Spot DR0150M (Basingstoke, United Kingdom). Confirmed isolates were stored at −80°C in cryovials containing porous beads and glycerol, as previously described (2).
Statistical analysis.
Information such as date, flock age, and downtime between flocks (number of days between flocks) was collected routinely and was entered together with microbiological results into a Microsoft Access database. The Campylobacter status of each potential reservoir and the flocks was described at the sample, visit, and flock levels. Comparative analyses were carried out to assess associations between campylobacters in the flock and in potential environmental sources. Student's t tests and logistic regression were used, depending on the nature of the data. All data were cleaned and analyzed using STATA 10 (StataCorp, College Station, TX).
Molecular characterization.
A single colony was picked from a BASAC plate and cultured on blood agar at 42°C microaerobically for 24 h. Up to 3 isolates from culture-positive environmental samples and ceca were genotyped by pulsed-field gel electrophoresis (PFGE) using SmaI with pulse times increasing from 5 to 40 s with standardized parameters as proposed by CAMPYNET (http://www.medvetnet.org/cms/templates/doc.php?id=99&searchstring=CAMPYNET). Digital gel images of SmaI digests were compared by using Bionumerics software (Applied Maths, Kortrijk, Belgium), and cluster analyses were performed by the unweighted-pair group method with arithmetic averages (UPGMA).
Flagellin gene typing of selected strains was undertaken by sequencing of the PCR product of the flaA short variable region (SVR) with the primers FLA242FU and FLA625RU (25). A 321-bp sequence containing the flaA SVR nucleotide sequence was then compared with the database at http://pubmlst.org/campylobacter/flaA/ (8). Multilocus sequence typing (MLST) was performed by the previously published method (9). The PCR-based method of Best et al. (3) was used to identify to the species level a representative selection of strains belonging to each of the different flock-colonizing PFGE genotypes.
Experimental challenge of chickens with Campylobacter in cattle feces.
All animal studies were conducted under the Animals Scientific Procedures Act (1986) and were approved by the local ethical review committee. Two naturally contaminated cattle fecal samples (fecal sample 1, 211 g; fecal sample 2, 230 g) were collected from the concrete surface surrounding the calf pens. These samples were transported to the laboratory, and serial 10-fold dilutions were made in duplicate in phosphate-buffered saline (PBS). Aliquots were plated on BASAC agar to obtain viable counts of Campylobacter spp. Within 24 h of collection, following confirmation that the samples contained approximately 106 CFU g−1 campylobacter, the feces were administered to 16-day-old Ross birds (PD Hook Hatcheries, Ltd., Bampton, United Kingdom) housed in 3.50- by 2.90-m rooms. Two groups of birds (n = 20), housed separately, were exposed to 170 g feces, from either fecal sample 1 or 2, scattered over an area approximately 1 m by 0.5 m between the feed and water. Serial dilutions of each sample were made to determine Campylobacter numbers at the time of challenge. In addition, 5 g of each sample was retained and placed in a tray of litter at room temperature on an open bench and monitored for Campylobacter viability over time. Three days postchallenge, the birds were sampled by cloacal swabbing. Ten birds from each group were killed at 5 and 7 days postchallenge, and cecal colonization levels were determined as previously described (42). Where available, 20 colonies per sample were isolated for storage by freezing and subsequent PFGE typing.
RESULTS
Identification of Campylobacter in the target house and farm environment.
All 15 crop cycles reared in the target house during the study period were sampled. However, several outbreaks of avian influenza (AI) were experienced in other areas of the United Kingdom during the study period, which resulted in restricted access to poultry farms in general. During these restricted periods, the environment of the study farm could not be sampled, resulting in an incomplete sampling regimen for three of the 15 crop cycles (Table 1). When such events occurred, the status of the flock was ascertained from cecal samples taken at slaughter.
Five of the 15 crops were found to be already colonized with Campylobacter by the time of the first visit at 20 to 24 days. Three further crops were positive at the second visit; one of these was in the early stages of colonization, as indicated by low in-flock prevalence. Four further crops were positive at clearance; again, two of these crops were in the early stages of colonization. The remaining three crops were still negative at clearance and slaughter, two of which were sampled in winter during a period of enhanced biosecurity measures in response to a threat of avian influenza. These procedures included restrictions on visitors entering the farm and extra washing of vehicles and equipment.
From the 38 sampling visits, a total of 1,337 samples were taken from the farm and adjoining dairy environment and 506 from the broilers chickens. Overall 49.4% of flock samples were positive. Of the samples from the farm and adjoining dairy environment overall Campylobacter was isolated from 407 (30.4%), of which 55.9% of 136 samples from calf feces or overshoes from the surroundings of associated calf pens were positive. Campylobacter was also recovered from 55 (47.4%) of a further 116 samples taken from vehicles and equipment on arrival on the farm at depopulation events.
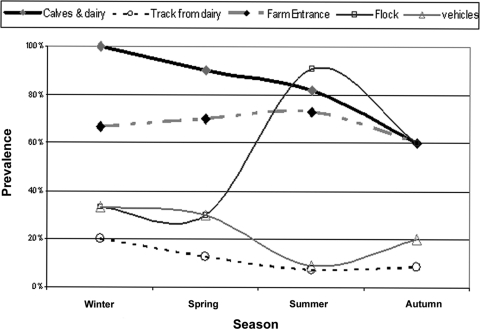
The downtime between flocks ranged from 7 to 16 days, with an average of 10.2 days (median = 10). The average downtime of the positive flocks (13.6 days) was not significantly different from that of negative flocks (14.5 days) (P = 0.468; t test). As expected, variation was found in the prevalence of positive flocks according to age and to season. Negative crops had an average age of 24 days, while positive crops were older (mean age = 31 days) (P = 0.007). Flock samples were most likely to yield campylobacters in summer (P = 0.002) (Fig. 1) than at other times of the year. In marked contrast, no corresponding seasonal increases in prevalence for any single environmental category were observed (Fig. 1); for example, samples from the calves and dairy yard were consistently positive throughout the year. Contamination of vehicles and equipment also varied between seasons but showed an inverse relationship with the summer peak of Campylobacter in the flocks. However, overall such vehicles could only be infrequently sampled. A drop in the number of Campylobacter-positive flocks occurred in August, which may have reflected the rapid drying of surfaces such as surrounding concreted areas.
FIG. 1.
Relationship between season and Campylobacter prevalence in different sample categories at the visit level.
Environmental samples, which were positive while the concomitant flock remained negative, included the house surroundings and track; main entrance; anteroom floors and doors; puddles; effluent tank; catchers' vehicles, crates, and modules; and feces from adjacent calves and the dairy area (Table 2). Samples from water from the mains, drinker lines, and cups, as well as dead bird storage, insect, and air samples, were all negative before the respective flock was colonized.
TABLE 2.
Numbers of samples taken, Campylobacter status, and matches to flock isolates tested for the various samples at all visits and at visits before the concomitant flock was positive
| Source category | All visits |
Visits at which flock tested negativea |
Visits at which flock was positive |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. positive/total collected | % positive | No. of flock matching typesb | % of flock matching types | No. positive/total collected | % positive | No. of flock matching typesb | % of flock matching types | No. positive/total collected | % positive | No. of flock matching typesb | % of flock matching types | |
| House surroundings, including vents and track | 164/642 | 25.5 | 58/88 | 65.9 | 30/295 | 10.2 | 1/21 | 4.8 | 134/347 | 38.6 | 57/67 | 85.1 |
| Track | 20/74 | 27.0 | 6/14 | 42.9 | 3/32 | 9.4 | 0/3 | 0 | 17/42 | 40.5 | 6/11 | 54.5 |
| Main entrance and wheel wash | 66/152 | 43.4 | 21/33 | 63.6 | 19/68 | 27.9 | 1/7 | 14.3 | 47/84 | 56 | 20/25 | 80 |
| Anteroom floor and doors | 38/152 | 25.0 | 15/22 | 68.2 | 4/68 | 5.9 | 1/4 | 25.0 | 34/84 | 40.1 | 11/18 | 61.1 |
| Farm vehicles and equipment | 11/62 | 17.7 | 4/5 | 80 | 0/24 | 0 | NAc | NA | 11/38 | 28.9 | 4/5 | 80 |
| Catchers' vehicles, crates, and modules | 55/116 | 47.4 | 3/19 | 15.8 | 7/23 | 30.4 | 0/2 | 0 | 48/93 | 51.6 | 3/17 | 17.6 |
| Puddles | 18/57 | 31.6 | 6/11 | 54.5 | 8/27 | 29.6 | 1/3 | 33.3 | 10/30 | 33.3 | 5/8 | 62.5 |
| Effluent tank | 3/18 | 16.7 | 1/1 | 100 | 1/9 | 11.1 | 1/1 | 100 | 2/9 | 22.2 | NTd | NT |
| Water from mains | 1/33 | 3.0 | 1/1 | 100 | 0/11 | 0 | NA | NA | 1/22 | 0.05 | 1/1 | 100 |
| Drinker lines and cups | 11/31 | 35.5 | 2/2 | 100 | 0/9 | 0 | NA | NA | 11/22 | 50 | 2/2 | 100 |
| Insectse | 0/4 | 0 | NA | NA | 0/3 | 0 | NA | NA | 0/1 | 0 | NA | NA |
| Other animalsf | 0/4 | 0 | NA | NA | 0/1 | 0 | NA | NA | 0/3 | 0 | NA | NA |
| Dead bird bags | 3/4 | 75.0 | 1/1 | 100 | 0/1 | 0 | NA | NA | 3/3 | 100 | 1/1 | 100 |
| Flock | 239/431 | 55.5 | ||||||||||
| Additional cecag | 11/75 | 14.7 | ||||||||||
| Calves/dairy yard | 76/136 | 55.9 | 21/43 | 48.8 | 35/63 | 55.6 | 1/20 | 5.0 | 41/73 | 55.6 | 20/23 | 87 |
| Air | 16/42 | 38.1 | 11/12 | 91.7 | 0/6 | 0 | 16/36 | 44.4 | 11/12 | 91.7 | ||
Includes flocks 12, 13, and 15, which were not detectably colonized by postclearance slaughter.
Excludes negative flocks.
NA, not applicable.
NT, no isolates typed.
Black beetles in litter close to doors and flies in houses.
Feces from wild animals on site, including rabbits and wild birds.
Taken due to AI restrictions.
Campylobacter environmental contamination and the colonization status of the target broiler flocks were analyzed at both the visit and the sample levels. For two of the visits (crop 4, visit 1; crop 12, visit 2), no positive samples were identified from the environmental sampling undertaken. From the remaining 36 visits, positive environmental samples were identified during 20 visits (57.1% [20/36]) when the flocks were colonized compared with 16 (42.9%) when the flocks were still negative (P = 0.843). There was no environmental location that was consistently positive while the flock was negative, indicative of a potential persistent source. For every environmental sample analyzed at the visit level (house surrounds, calves/dairy, farm entrance, anteroom/doors, farm equipment, puddles, catchers/equipment, dead bird storage, and water from the mains and drinkers), contamination increased when the flock was positive. However, the track to the dairy farm was the only source significantly associated with flock positivity at the visit level; on 66.7% (20/36) of the visits, this source was positive when the flock was infected, compared to 33.3% (10/36) when the flock was negative (P = 0.035; analyzed by univariable logistic regression with the flock as a random effect).
A similar profile of contamination was detected when data were analyzed at the sample level (Table 2). Only 17.1% (104/608) of environmental samples were Campylobacter positive, while the concomitant flock was uncolonized (17 visits). This environmental contamination increased to 42.4% (358/845) after flock positivity was detected (a total of 21 visits). As well as the track, increased contamination was most notable (P < 0.05) in the samples from the house surrounds, the main entrance and wheel wash area, the poultry house anteroom and doors, and farmers' vehicles and equipment. In addition, drinker lines, air, water from the mains, in-house insects, and dead bird bags only became positive after flock positivity (Table 2).
Epidemiology of Campylobacter genotypes from the poultry and dairy farms.
Forty-three SmaI PFGE genotypes were identified from the 820 flock and environmental isolates. Of the 12 positive flocks, 9 yielded only a single genotype, while the remaining 3 (crops 8, 9, and 2) were colonized by 2, 3, and 6 different genotypes, respectively. However, a marked strain succession through the crop cycle was only identified in crop 9, during which PFGE genotype 12 was succeeded by PFGE types 29 and 114.
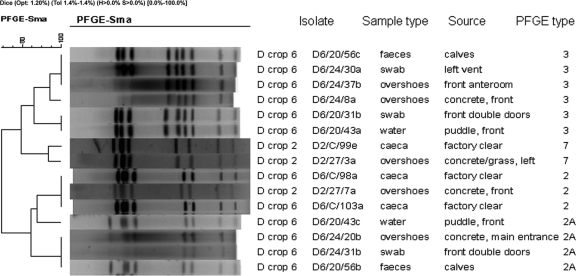
Two of the five flocks positive at the first visit (crops 3 and 10) had flock-type matching isolates from samples taken from the house surroundings and adjoining dairy area and crop 11 only from the latter site during the same visit, suggesting links between strains colonizing the cattle and broilers. The environment was Campylobacter positive prior to detectable colonization of the flocks during six crop cycles, although only two crops (crop 2 and crop 6) demonstrated Campylobacter genotypes in the farm environment that were subsequently detected in the concomitant target flock. In crop 2, isolates from the house surrounds at day 27 were identical to the predominant flock genotype, PFGE 7/flaA SVR 32/ST 48, recovered from the ceca at clearance (Fig. 2). Similarly, birds in crop 6 were colonized at clearance with a strain of genotype PFGE 2/flaA SVR 42/ST 61. This strain, or one highly related to it (PFGE 2A/flaA SVR 42/ST 61), was detected in calf feces and puddles and at the main entrance prior to flock colonization (Fig. 2). However, in the same crop, another calf-associated strain contaminating the surrounds and the poultry house anteroom was not detected in the flock (Fig. 2). Genotyping confirmed the widespread contamination of the farm by flock-colonizing strains. The routes and vehicles for such dissemination are unclear, but all campylobacters recovered from air and drinker samples matched or were closely related to strains already colonizing the respective flocks; this included strains recovered from ambient air 20 m from the nearest poultry house.
FIG. 2.
Dendrogram derived from SmaI PFGE patterns showing matches to flock genotypes detected in environmental samples from crops 2 and 6 prior to detectable colonization. The band position tolerance was set at 1.4%, and clustering was performed using UPGMA. The scale indicates the percentage of similarity as determined with the Pearson coefficient. Isolate references comprise crop number, day of sampling, and standardized sample number.
Environmental persistence or recycling of Campylobacter genotypes.
Six genotypes persisted and/or recurred during at least two crop cycles (Table 3). The most commonly observed persistent PFGE genotype, 7, was identified in 199 out of 768 isolates (25.9%) from six crop cycles, but was found only in the flock of three of these cycles. Selected isolates typed by multiple methods were all of the same genotype (PFGE 7/flaA SVR 32/ST 48). This strain was first identified in a sample from the house surrounds during crop 2, prior to the strain being detected in flock samples and spreading to a number of environmental sites, including in-house water, air samples, vents, guttering, and concreted areas around the houses and puddles. However, the strain then remained undetected until crop 9, when it was next identified in calf feces and a puddle near the track used by dairy staff. Thereafter, this strain contaminated the calf and dairy areas, general surrounds, and flocks of the two subsequent crop cycles. The strain then persisted in the dairy area and farm surroundings through to crop 15, but was not recovered from the flocks of crops 12 to 15. Thus, despite persistence in the immediate environment, there appeared to be restricted transmission of this strain in the broiler chickens.
TABLE 3.
Recurring PFGE genotypes identified in different crop cycles on the broiler farm and adjoining dairy
| Source category | Result for PFGE type: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2/2a |
3 |
4 |
7 |
12 |
13 |
|||||||
| No. of crops (crop no.) | No. of samples | No. of crops (crop no.) | No. of samples | No. of crops (crop no.) | No. of samples | No. of crops (crop no.) | No. of samples | No. of crops (crop no.) | No. of samples | No. of crops (crop no.) | No. of samples | |
| House surrounds, track | 4 (2, 3, 6, 7) | 12 | 2 (5, 6) | 9 | 4 (1, 8, 11, 12) | 5 | 5 (2, 10, 11, 12, 14, 15) | 69 | 2 (8, 9) | 39 | 2 (7, 8) | 5 |
| Calves and dairy | 4 (3, 6, 7, 8) | 7 | 3 (3, 6, 15) | 19 | 3 (3, 8, 11) | 3 | 6 (9, 10, 11, 12, 14, 15) | 42 | 2 (8, 9) | 7 | 4 (6, 7, 8, 9) | 9 |
| Main entrance, wheel-wash | 3 (1, 3, 6) | 8 | 2 (6, 15) | 6 | 1 (8) | 1 | 3 (2, 10, 12, 15) | 17 | 1 (9) | 11 | 0 | 0 |
| Anteroom floor and doors | 2 (1, 6) | 2 | 1 (6) | 8 | 0 | 0 | 2 (10, 11) | 5 | 1 (9) | 13 | 0 | 0 |
| Equipment | 1 (6) | 1 | 0 | 0 | 0 | 0 | 1 (2) | 1 | 1 (9) | 11 | 0 | 0 |
| Catchers | 2 (6, 9) | 3 | 1 (6) | 1 | 0 | 0 | 2 (9, 14) | 7 | 1 (9) | 1 | 0 | 0 |
| Puddles | 2 (1, 6) | 3 | 1 (6) | 4 | 1 (1) | 1 | 3 (2, 9, 15) | 9 | 1 (9) | 9 | 0 | 0 |
| Main water source | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Drinker lines | 0 | 0 | 0 | 0 | 0 | 0 | 2 (2, 10) | 5 | 1 (9) | 3 | 0 | 0 |
| Effluent tank, trough | 1 (6) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Insects | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (9) | 1 | 0 | 0 |
| Flock | 2 (3, 6) | 20 | 1 (2) | 5 | 1 (2) | 4 | 3 (2, 10, 11) | 52 | 1 (9) | 14 | 1 (9) | 1 |
Several other strains also demonstrated persistence in the environment but apparent restricted infectivity in broilers. Strains of PFGE genotype 2 and its subtype, 2A (Fig. 2), were identified in four crop cycles (Table 3). The strain was first recovered from surrounds and adjacent dairy animals during crop 1 and at the front of the target house during crop 2. During crop 3, this strain was again isolated from farm surrounds and the dairy environment, as well as colonizing the target flock. However, this genotype was only identified once more, in the environment and flock of crop 6. Isolates of genotype PFGE 13/flaA SVR 41/ST 270 were recovered from 3.9% of all samples, particularly in the calf and dairy areas during crops 7, 8, and 9 (Table 3). However, this strain was only recovered in the flock of crop 9. Similarly, isolates of PFGE type 12 were associated with the house surrounds during crop 8 but only colonized birds of crop 9 (Table 3).
Interactions between Campylobacter strains in broilers and cattle.
Throughout the study, several strains intermittently occurred in both broiler flocks and cattle samples, indicating a dynamic interaction between the two reservoirs. Several of these have been mentioned above, in particular PFGE 7/flaA SVR 32/ST 48. In addition, strains of genotype PFGE 3/flaA SVR 41/ST 21 were first identified in the target flock feces during crop 2 (Table 3) and then later in the calf and dairy yard during crops 3 and 4. Similarly, strains of genotype PFGE 4/flaA SVR 52, first identified in the house surrounds of crop 1, were recovered from the broiler chickens of crop 2 and then from cattle samples of crops 3, 8, and 11. These results suggested that some strains, particularly those carried by cattle and recoverable from the poultry environment, are ineffective chicken colonizers. To investigate whether this was a reflection of host adaptation the colonization potential of strain PFGE 7/flaA SVR 32/ST 48 was experimentally investigated for chicken colonization potential.
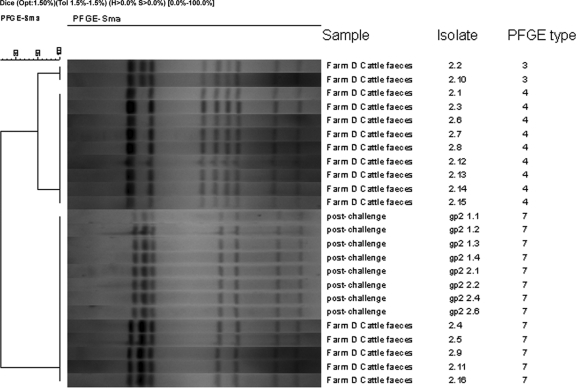
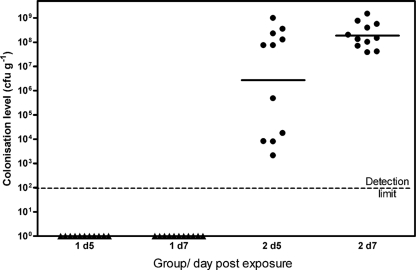
Two groups of Ross broiler chickens (groups 1 and 2) were exposed to naturally occurring Campylobacter-contaminated cattle feces from sample 1 or 2. Preliminary studies showed that these samples contained C. jejuni at levels of 1.1 × 106 and 2.0 × 106 CFU g−1, respectively, and PFGE analysis identified up to three distinct strains, with types 7 and 4 present in approximately equal proportions (Fig. 3). Cloacal swab samples taken 3 days postexposure indicated that 0/20 and 5/20 birds from groups 1 and 2, respectively, were shedding campylobacters (Fig. 3). By 5 and 7 days postexposure, all the chickens in group 2, but none in group 1, were colonized (Fig. 4). Both fecal samples demonstrated a similar but small drop in Campylobacter recovery over the first 24 h of storage. However, although no campylobacters were recovered from fecal sample 1 stored for 4 days, >105 CFU g−1 campylobacters remained detectable in fecal sample 2 after the same period. Thus, the poor survival in fecal sample 1 probably explains why the birds exposed to this sample remained Campylobacter negative. The reason for the failure of the organisms to survive in this fecal sample is unclear.
FIG. 3.
Dendrogram derived from SmaI PFGE patterns of C. jejuni strains recovered from cattle feces samples and from the ceca of experimentally exposed broiler chickens.
FIG. 4.
Colonization of two independently housed groups of broiler chickens at 5 and 7 days following exposure to naturally occurring C. jejuni-contaminated calf feces samples at 16 days of age.
Only organisms of PFGE type 7 were recovered from the experimentally exposed birds. By additional typing, the strain was confirmed as genotype PFGE 7/flaA SVR 32, identical to the strain predominating in the cattle feces sampled throughout the study period. The failure of the strain belonging to PFGE type 4 to colonize the birds from the same fecal exposure remains unknown.
It is impossible to determine the dose received by each bird in such exposure experiments. Nevertheless, estimates indicate that the dose would be high and possibly as high as 107 CFU per bird. In a second experiment, Ross birds (n = 20) were challenged again with the same weight of cattle feces containing PFGE type 7, but at a level of only 200 CFU g−1. Despite the 1,000-fold-lower challenge level, by 2 days postexposure, 5/10 birds were colonized at levels of between 1 × 103 and 7 × 108 CFU g−1 cecal contents.
DISCUSSION
In order to ensure the best chance of identifying the sources of Campylobacter causing broiler flock colonization, extensive, structured sampling coupled to best practice culture and typing methodologies was undertaken on one poultry farm and its environment during 15 crop cycles. There was considerable variation in the Campylobacter status of the flocks at the various sampling times. Five flocks were positive at the first sampling, while three flocks remained negative through to slaughter. However, there were no obvious changes in flock management or biosecurity over the period, which might account for these differences.
As expected from previous studies, campylobacters were recoverable from various locations around the farm even while the flock was Campylobacter negative (17, 20). However, once a flock became positive, the degree of environmental contamination around the farm increased substantially despite the employment of standard biosecurity measures. Positive aerosol samples were even detectable 20 m from the house. This is a clear indicator that standard biosecurity measures are incapable of confining Campylobacter within the poultry house. By careful analysis of the routes of transmission out of a positive house, some indication of the routes of ingress might be obtained.
By taking a longitudinal sampling approach, it was anticipated that the most likely environmental sources of flock infection might be identified. However, five flocks were already positive on the first visit (between 20 and 24 days of age), when no preceding environmental isolates for that crop were available. Nevertheless, in those seven flocks which became positive by the second or third sampling, some strains cultured from the environment were genotypically identical to isolates subsequently recovered from concomitant flock samples. Most frequently, such strains were recovered from cattle on the adjacent farm and the main poultry farm entrance and driveway. Many of these strains persisted in the environment, intermittently recoverable at various sites, and occasionally infected the broiler chickens. Similar observations on the intermittent recurrence of some Campylobacter clones have recently been reported in other studies (20, 26, 43).
Overall the longitudinal observations suggested that cattle housed in the yard adjoining the broiler chicken farm may have constituted a reservoir (i.e., a site of amplification) for certain Campylobacter strains. Contamination from the yard could have been spread onto the broiler chicken farm by dairy farm vehicles using the track to the side of the target house. However, some of the recurring strains first appeared in this study of broiler flocks sampled during visits when calves were either not present in adjacent pens or were not detectably infected, suggesting that transmission from broiler chickens to the calves also occurred. Previous studies (5, 31, 43) have also indicated the cross-infection of broiler chickens and cattle by the same Campylobacter strains.
Cattle are well-recognized reservoirs of C. jejuni and C. coli and fecally contaminate the environment (38). The presence of cattle on or near the poultry farm statistically increases the likelihood of positive flocks (11). PFGE 7/flaA SVR 32/ST 48, PFGE 2 (2A)/flaA SVR 42/ST 61, and PFGE 3/flaA SVR 41/ST 21 strains circulating in the dairy and broiler chicken farm environments from our study matched MLST sequence types reported to be dominant in dairy cattle and associated environments in recent studies based in the United Kingdom (13, 21, 33), supporting a role for cattle as a reservoir for flock-colonizing campylobacters.
Interestingly, some strains (for example, PFGE types 3 or 7) recoverable from the cattle and the poultry house surroundings failed to colonize or only occasionally colonized the broiler flocks. There are many potential reasons for this observation. Such strains may be poor environmental survivors in cattle feces, and the experimental evidence from preliminary storage studies with type 7 indicates that there may be some evidence for this under certain circumstances. An alternative explanation might be that these genotypes were host adapted to cattle and that their potential to colonize chickens was compromised.
Some host specificity of certain C. jejuni genotypes has already been identified by population-based studies using serotyping (29) and MLST (13, 23, 34), suggesting that some strains have a preference for the bovine intestinal environment. In order to determine whether the strains found in the calves which showed an apparently restricted colonization ability for the broiler chickens in later crop cycles had become host restricted, experimental challenge studies were undertaken using 17-day-old Ross broiler chickens. These studies showed that the strain of PFGE type 7, but not that of type 4, naturally occurring in cattle feces could effectively colonize chickens, even at relatively low exposure levels. These findings confirmed that cattle feces were a potential vehicle for Campylobacter transmission and suggest that host specificity is not the cause of the observed strain-restricted colonization.
Toward the end of the study period, some changes in farm management practices that were highlighted by analysis of the questionnaire and observations recorded during individual farm visits were implemented. In particular, there was an increased frequency of removal of calf fecal material and of washing down of the calf pens, together with a reduction of dairy farm traffic along the common track. Such enhanced biosecurity may have contributed to the reduced transmission of Campylobacter from the cattle to the broilers and thus to the strain restrictions observed in colonization.
In conclusion, this detailed longitudinal study of one farm over 15 crop cycles has clearly demonstrated the widespread contamination of the poultry environment with campylobacters. This environmental contamination is markedly enhanced once the flock becomes Campylobacter positive, indicating a continuous and extensive leakage of Campylobacter-containing material out from the poultry houses. The presence of certain Campylobacter strains in the external environment of the poultry farm prior to the detection of identical strains in the broilers provides confirmatory evidence of the horizontal transmission of these bacteria into the flock. The intermittent recovery of some Campylobacter strains in and around the farm environment appears to reflect the presence of local reservoirs: as represented in the case of this study farm by an adjacent dairy farm and calf-rearing pens. The longitudinal nature of the study enabled confirmation that bovine fecal Campylobacter strains initially recovered from the dairy yard can subsequently colonize the poultry. Surprisingly, this transmission route was intermittently effective as a source of flock infection. The possibility of host adaptation of the bovine strains in fecal material was experimentally investigated and shown not to be a factor. However, the introduction of some simple biosecurity measures, including vehicle movement restrictions between dairy farm and broiler houses, such as use of the track and the removal of the calves to an alternative site on the dairy farm to prevent bovine fecal material from reaching the poultry farm, may have been effective.
Acknowledgments
We gratefully acknowledge the assistance given by farmers, managers, and technical staff of the poultry companies concerned. Fabrizio Lemma, Meenaxi Sharma, Dawn Harrison, Vicky Tucker, and Robert Atterbury are thanked for provision of microbiological expertise. Fran Colles of the Peter Medawar Building for Pathogen Research and the Department of Zoology, University of Oxford, is thanked for assistance with MLST typing.
This study was part of a project funded by the United Kingdom Department for Environment, Food and Rural Affairs (OZ0610) and made use of the Campylobacter Fla database (http://pubmlst.org/campylobacter/flaA/) hosted at the University of Oxford, funded by DEFRA grant OZ0615.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Adkin, A., E. Hartnett, L. Jordan, D. Newell, and H. Davison. 2006. Use of a systematic review to assist the development of Campylobacter control strategies in broilers. J. Appl. Microbiol. 100:306-315. [DOI] [PubMed] [Google Scholar]
- 2.Allen, V. M., H. Weaver, A. M. Ridley, J. A. Harris, M. Sharma, J. Emery, N. Sparks, M. Lewis, and S. Edge. 2008. Sources and spread of thermophilic Campylobacter spp. during partial depopulation of broiler chicken flocks. J. Food Prot. 71:264-270. [DOI] [PubMed] [Google Scholar]
- 3.Best, E. L., E. J. Powell, C. Swift, K. A. Grant, and J. A. Frost. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 229:237-241. [DOI] [PubMed] [Google Scholar]
- 4.Bouwknegt, M., A. W. van de Giessen, W. D. Dam-Deisz, A. H. Havelaar, N. J. Nagelkerke, and A. M. Henken. 2004. Risk factors for the presence of Campylobacter spp. in Dutch broiler flocks. Prev. Vet. Med. 62:35-49. [DOI] [PubMed] [Google Scholar]
- 5.Bull, S. A., V. M. Allen, G. Domingue, F. Jorgensen, J. A. Frost, R. Ure, R. Whyte, D. Tinker, J. E. Corry, J. Gillard-King, and T. J. Humphrey. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, S. A., A. Thomas, T. Humphrey, J. Ellis-Iversen, A. J. Cook, R. Lovell, and F. Jorgensen. 2008. Flock health indicators and Campylobacter spp. in commercial housed broilers reared in Great Britain. Appl. Environ. Microbiol. 74:5408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callicott, K. A., V. Friethriksdottir, J. Reiersen, R. Lowman, J. R. Bisaillon, E. Gunnarsson, E. Berndtson, K. L. Hiett, D. S. Needleman, and N. J. Stern. 2006. Lack of evidence for vertical transmission of Campylobacter spp. in chickens. Appl. Environ. Microbiol. 72:5794-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis-Iversen, J., A. J. Cook, R. P. Smith, G. C. Pritchard, and M. Nielen. 2009. Temporal patterns and risk factors for Escherichia coli O157 and Campylobacter spp., in young cattle. J. Food Prot. 72:490-496. [DOI] [PubMed] [Google Scholar]
- 11.Ellis-Iversen, J., F. Jorgensen, S. Bull, L. Powell, A. J. Cook, and T. J. Humphrey. 2009. Risk factors for Campylobacter colonisation during rearing of broiler flocks in Great Britain. Prev. Vet. Med. 89:178-184. [DOI] [PubMed] [Google Scholar]
- 12.Evans, S. J., and A. R. Sayers. 2000. A longitudinal study of campylobacter infection of broiler flocks in Great Britain. Prev. Vet. Med. 46:209-223. [DOI] [PubMed] [Google Scholar]
- 13.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, E., H. Barnhart, D. W. Dreesen, N. J. Stern, and J. L. Corn. 1997. Epidemiological study of Campylobacter spp. in broilers: source, time of colonization, and prevalence. Avian Dis. 41:890-898. [PubMed] [Google Scholar]
- 15.Guerin, M. T., W. Martin, J. Reiersen, O. Berke, S. A. McEwen, J. R. Bisaillon, and R. Lowman. 2007. House-level risk factors associated with the colonization of broiler flocks with Campylobacter spp. in Iceland, 2001-2004. BMC Vet. Res. 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hald, B., H. Skovgard, D. D. Bang, K. Pedersen, J. Dybdahl, J. B. Jespersen, and M. Madsen. 2004. Flies and Campylobacter infection of broiler flocks. Emerg. Infect. Dis. 10:1490-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansson, I., I. Vagsholm, L. Svensson, and E. Olsson Engvall. 2007. Correlations between Campylobacter spp. prevalence in the environment and broiler flocks. J. Appl. Microbiol. 103:640-649. [DOI] [PubMed] [Google Scholar]
- 18.Havelaar, A. H., M. J. Mangen, A. A. de Koeijer, M. J. Bogaardt, E. G. Evers, W. F. Jacobs-Reitsma, W. van Pelt, J. A. Wagenaar, G. A. de Wit, H. van der Zee, and M. J. Nauta. 2007. Effectiveness and efficiency of controlling Campylobacter on broiler chicken meat. Risk. Anal. 27:831-844. [DOI] [PubMed] [Google Scholar]
- 19.Humphrey, T., M. Mason, and K. Martin. 1995. The isolation of Campylobacter jejuni from contaminated surfaces and its survival in diluents. Int. J. Food Microbiol. 26:295-303. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen, G., H. Kruse, and M. Hofshagen. 2006. Genetic diversity and description of transmission routes for Campylobacter on broiler farms by amplified-fragment length polymorphism. J. Appl. Microbiol. 101:1130-1139. [DOI] [PubMed] [Google Scholar]
- 21.Kwan, P. S., A. Birtles, F. J. Bolton, N. P. French, S. E. Robinson, L. S. Newbold, M. Upton, and A. J. Fox. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyngstad, T. M., M. E. Jonsson, M. Hofshagen, and B. T. Heier. 2008. Risk factors associated with the presence of Campylobacter species in Norwegian broiler flocks. Poult. Sci. 87:1987-1994. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy, N. D., F. M. Colles, K. E. Dingle, M. C. Bagnall, G. Manning, M. C. Maiden, and D. Falush. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg. Infect. Dis. 13:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDowell, S. W., F. D. Menzies, S. H. McBride, A. N. Oza, J. P. McKenna, A. W. Gordon, and S. D. Neill. 2008. Campylobacter spp. in conventional broiler flocks in Northern Ireland: epidemiology and risk factors. Prev. Vet. Med. 84:261-276. [DOI] [PubMed] [Google Scholar]
- 25.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messens, W., L. Herman, L. De Zutter, and M. Heyndrickx. 2009. Multiple typing for the epidemiological study of contamination of broilers with thermotolerant Campylobacter. Vet. Microbiol. 138:120-131. [DOI] [PubMed] [Google Scholar]
- 27.Milnes, A. S., I. Stewart, F. A. Clifton-Hadley, R. H. Davies, D. G. Newell, A. R. Sayers, T. Cheasty, C. Cassar, A. Ridley, A. J. Cook, S. J. Evans, C. J. Teale, R. P. Smith, A. McNally, M. Toszeghy, R. Futter, A. Kay, and G. A. Paiba. 2008. Intestinal carriage of verocytotoxigenic Escherichia coli O157, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica, in cattle, sheep and pigs at slaughter in Great Britain during 2003. Epidemiol. Infect. 136:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 30.Ogden, I. D., M. MacRae, M. Johnston, N. J. Strachan, A. J. Cody, K. E. Dingle, and D. G. Newell. 2007. Use of multilocus sequence typing to investigate the association between the presence of Campylobacter spp. in broiler drinking water and Campylobacter colonization in broilers. Appl. Environ. Microbiol. 73:5125-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley, A. M., V. M. Allen, M. Sharma, J. A. Harris, and D. G. Newell. 2008. Real-time PCR approach for detection of environmental sources of Campylobacter strains colonizing broiler flocks. Appl. Environ. Microbiol. 74:2492-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridley, A. M., M. J. Toszeghy, S. A. Cawthraw, T. M. Wassenaar, and D. G. Newell. 2008. Genetic instability is associated with changes in the colonization potential of Campylobacter jejuni in the avian intestine. J. Appl. Microbiol. 105:95-104. [DOI] [PubMed] [Google Scholar]
- 33.Rotariu, O., J. F. Dallas, I. D. Ogden, M. Macrae, S. K. Sheppard, M. Maiden, F. J. Gormley, K. J. Forbes, and N. J. Strachan. 2009. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across NE and SW Scotland. Appl. Environ. Microbiol. 75:6275-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard, S. K., J. F. Dallas, M. MacRae, N. D. McCarthy, E. L. Sproston, F. J. Gormley, N. J. Strachan, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheppard, S. K., J. F. Dallas, N. J. Strachan, M. MacRae, N. D. McCarthy, D. J. Wilson, F. J. Gormley, D. Falush, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shreeve, J. E., M. Toszeghy, M. Pattison, and D. G. Newell. 2000. Sequential spread of Campylobacter infection in a multipen broiler house. Avian Dis. 44:983-988. [PubMed] [Google Scholar]
- 37.Stafford, R. J., P. J. Schluter, A. J. Wilson, M. D. Kirk, G. Hall, and L. Unicomb. 2008. Population-attributable risk estimates for risk factors associated with Campylobacter infection, Australia. Emerg. Infect. Dis. 14:895-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94(Suppl.):104S-113S. [DOI] [PubMed] [Google Scholar]
- 39.Stern, N. J., P. Fedorka-Cray, J. S. Bailey, N. A. Cox, S. E. Craven, K. L. Hiett, M. T. Musgrove, S. Ladely, D. Cosby, and G. C. Mead. 2001. Distribution of Campylobacter spp. in selected U.S. poultry production and processing operations. J. Food Prot. 64:1705-1710. [DOI] [PubMed] [Google Scholar]
- 40.Tam, C. C., C. D. Higgins, K. R. Neal, L. C. Rodrigues, S. E. Millership, and S. J. O'Brien. 2009. Chicken consumption and use of acid-suppressing medications as risk factors for Campylobacter enteritis, England. Emerg. Infect. Dis. 15:1402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 43.Zweifel, C., K. D. Scheu, M. Keel, F. Renggli, and R. Stephan. 2008. Occurrence and genotypes of Campylobacter in broiler flocks, other farm animals, and the environment during several rearing periods on selected poultry farms. Int. J. Food Microbiol. 125:182-187. [DOI] [PubMed] [Google Scholar]