Abstract
The intestinal microbe Escherichia coli is subject to fecal deposition in secondary habitats, where it persists transiently, allowing for the opportunity to colonize new hosts. Selection in the secondary habitat can be postulated, but its impact on the genomic diversity of E. coli is unknown. Environmental selective pressure on extrahost E. coli can be revealed by landscape genetic analysis, which examines the influences of dispersal processes, landscape features, and the environment on the spatiotemporal distribution of genes in natural populations. We conducted multilocus sequence analysis of 353 E. coli isolates from soil and fecal samples obtained in a recreational meadow to examine the ecological processes controlling their distributions. Soil isolates, as a group, were not genetically distinct from fecal isolates, with only 0.8% of genetic variation and no fixed mutations attributed to the isolate source. Analysis of the landscape genetic structure of E. coli populations showed a patchy spatial structure consistent with patterns of fecal deposition. Controlling for the spatial pattern made it possible to detect environmental gradients of pH, moisture, and organic matter corresponding to the genetic structure of E. coli in soil. Ecological distinctions among E. coli subpopulations (i.e., E. coli reference collection [ECOR] groups) contributed to variation in subpopulation distributions. Therefore, while fecal deposition is the major predictor of E. coli distributions on the field scale, selection imposed by the soil environment has a significant impact on E. coli population structure and potentially amplifies the occasional introduction of stress-tolerant strains to new host individuals by transmission through water or food.
Escherichia coli bacteria are widespread commensal and pathogenic members of the vertebrate gut microbiota and are considered to be an indicator of fecal pollution in water. The fecal-oral route of transmission often requires transient passage in secondary (i.e., extrahost) habitats, where E. coli must survive environmental stressors to colonize new hosts. Common secondary habitats into which E. coli is transmitted include surface and groundwaters, soils, plant surfaces, and a variety of domestic and agricultural environments. However, soil is a particularly interesting secondary habitat, because its chemical and physical heterogeneity on small spatial scales may provide a mechanism for generating and maintaining biodiversity within microbial species, including extrahost E. coli. Fecal deposition of E. coli into soil represents an intermediate step in a host-soil-water cycle that is one mechanism by which E. coli may colonize new hosts (8). E. coli abundance declines over months in soil, but persistent strains can be mobilized in overland or groundwater flow, leading to redeposition in a new soil environment or entry into surface waters and community water supplies (2, 20, 44).
Extrahost persistence implies that E. coli strains in secondary habitats are subject to environmental stressors following deposition. Environmental selection may impact the genetic diversity of host-adapted E. coli populations by driving evolution of traits that favor persistence in secondary habitats in combination with those promoting fitness in the gut. Indeed, half of the total E. coli population might reside in secondary habitats (35), but the role of the environment in structuring populations of E. coli, whether naturalized or host adapted, has not been examined. Recent studies have concluded that E. coli may establish stable, replicating (i.e., naturalized) populations in secondary habitats, resulting in genetic distinction from the original host-adapted population (6, 7, 42). Data supporting the naturalized E. coli hypothesis suggest that environmental populations might interfere with estimates of fecal pollution in waterways, because they would falsely resemble recent fecal contamination (13).
Landscape genetics is a field of study that uses population genetics, spatial statistics, and landscape ecology to understand the processes structuring a population across environments while accounting for independent geographic, landscape, and temporal patterns (23, 37). These methods provide a framework to test whether changes in the distribution of extrahost E. coli strains are due to selective pressures imposed in a heterogeneous secondary habitat such as soil. If environmental (i.e., edaphic) variation selects for persistent E. coli genotypes in soil, then the landscape genetic distribution in soil will change in response to edaphic gradients. The contribution of fecal deposition to E. coli spatial patterns must be examined in addition to edaphic variables, because deposition is the process that controls E. coli introduction into soil (14, 21). Therefore, landscape genetic analysis of E. coli distributions can help to clarify how deposition in soil changes extrahost populations (7, 14, 39). For example, if E. coli populations in soil were found to be structured along a pH gradient, then soil pH might be a useful predictor for mapping fecal pollution risk or potential environmental reservoirs of fecal bacteria.
We conducted a spatially and temporally explicit genetic analysis of isolates from the topsoil of a recreational meadow to quantify the roles of fecal deposition and environmental selection in constraining the extrahost distribution of E. coli strains. We chose to examine an area of recreational land use with a modest fecal input, because this approach was expected to be a strong initial test of environmental selective pressure on E. coli. The goals of this study were (i) to determine the extent of genetic isolation between our soil isolates and isolates represented in a global database of E. coli gene sequences, (ii) to examine the role of relationship between the spatial distributions of fecal deposition events and E. coli isolates, and (iii) to examine the role of temporal, landscape, and environmental (i.e., edaphic) factors in the genetic structure of E. coli populations in soil.
MATERIALS AND METHODS
Soil sampling procedure.
Because the spatial scales of genetic and environmental variation were unknown, random soil sample coordinates were generated in GRASS GIS 6.2.3 (15) to provide a wide range of spatial relationships for each of three sampling dates. Three topsoil cores measuring 3 cm2 by 4 cm deep were collected at each site and sieved through a sterile 2-mm mesh to remove rocks and larger organic matter. Soil pH, gravimetric soil moisture (%M, wt/wt), and percent organic matter (%OM, wt/wt; estimated by using loss on ignition) were determined according to standard methods (5). Soil series polygon, elevation, slope, aspect, and orthoimagery data were gathered from the Cornell University Geospatial Information Repository (http://cugir.mannlib.cornell.edu).
To isolate E. coli from soils, 8-g portions of sieved soil from the three combined soil cores obtained at each site were suspended in 80 ml of broth containing EC medium with 4-methylumbelliferyl-β-d-glucuronide (EC-MUG). Suspensions were divided among 384 subsamples and incubated at 37°C. Isolates from EC-MUG agar subcultures were screened with biochemical tests for glutamate decarboxylase and beta-glucuronidase activity (31, 33). Positive results for these two tests accurately identified E. coli, as confirmed by later multilocus sequence analysis (MLSA).
Fecal survey.
Parallel east-west transects were laid on the field site at 10-m intervals. Personnel documented the abundance, animal origin, and location (±1 m) of fecal deposits. The spatial distribution of fecal deposits was analyzed using the univariate modified Ripley's K statistic as implemented in the R package splancs (http://www.r-project.org) (32). Statistical significance was determined by using 1,000 simulations of fecal events under complete spatial randomness (CSR).
Geostatistical analysis.
To aid interpretation of ecological data, a geostatistical technique for the interpolation of spatially structured data, universal kriging, was used to predict the spatial distribution of soil variables as implemented in the R package gstat. Theoretical variogram models were fit to experimental variograms of soil characteristics by using a reweighted least-squares approach (22). Because soil data means and variances were not statistically stationary between soil series, variography and kriging were performed within soil series, and these results were joined at their boundaries. A Gaussian variographic model produced the best fit to the experimental variogram of %OM in Hudson silt loam soil, but exponential variographic models were best in all other cases. Comparison of interpolated data to data for soil samples withheld from the interpolation revealed that predicted values coarsely represented the spatial pattern (n = 7 pairs; P > 0.05 in paired t tests between interpolated and withheld values).
MLSA.
Genomic DNA was isolated from E. coli by alkaline lysis of biomass in 50 mM NaOH at 95°C. Genomic fingerprints were generated for each isolate using repetitive sequence-based PCR (rep-PCR) (29). Two representatives of each rep-PCR fingerprint from each soil sample were subjected to sequencing of the aspC, clpX, fadD, icdA, lysP, mdh, and uidA genes by Sanger cycle sequencing at the Cornell University Life Sciences Core Laboratory (42). Evaluation of sequence read quality and assembly of forward and reverse reads were performed using Perl scripts which iterated runs of phred and CAP3, respectively (12, 17). Where sequence read quality had a probability of error of >0.005 (Q score < 23), sequences were edited manually. Assembled sequences of each MLSA locus were aligned and trimmed to standard base positions matching the E. coli K-12 sequence type from the STEC Center website (http://www.shigatox.net).
Population structure analysis.
Population genetic parameters from MLSA data were estimated using DnaSP (for polymorphism within collections [π] and nucleotide divergence between collections x and y [Dx vs y]) and ClonalFrame (10, 34). ClonalFrame was used to reconstruct the pattern of vertical inheritance in the presence of modest levels of recombination between E. coli strains. ClonalFrame was run for 2.5 × 105 burn-in iterations followed by 2.5 × 105 post-burn-in iterations. Convergence of parameter estimates between duplicated runs was confirmed with Gelman-Rubin statistics, and 5 × 105 total iterations were always sufficient to produce Gelman-Rubin statistics of <1.1 for all parameters (41). The clonal phylogeny of E. coli was reconstructed from the majority consensus of 501 neighbor-joining trees that were sampled every 500 iterations after the burn-in period and was displayed using a consensus splits network in SplitsTree v.4 (18). This dendrogram was used to assign isolates to E. coli subpopulations.
Spatial maps of E. coli genotype variation were generated using the first two eigenvectors resulting from principal coordinate analysis of ClonalFrame distance matrices as implemented in the R package ade4. The spatial distribution of isolate scores on the eigenvectors was mapped using an inverse squared distance weighting function in the R package gstat.
Ecological analysis.
All methods described herein were performed in R and, unless otherwise noted, in the vegan package (22). Only unconstrained analyses were performed (i.e., unexplained variation was never discarded).
To test for genetic isolation of soil E. coli away from the global E. coli gene pool, all available sequence sets containing sequences of the seven MLSA genes (see “MLSA” above) from the GenBank nucleotide database and the STEC reference collection were combined with clade ET-1 sequence types from Walk et al. (42), and the resulting collection was referred to as the global E. coli data set. Analysis of distance (ANODIS) was performed on the ClonalFrame distance matrix with an ANODIS model consisting of fixed terms for subpopulation membership, isolate collection membership (global versus field or soil versus fecal), and the interaction between those terms (24). The significance of F statistics from ANODIS was evaluated by using 999 Monte Carlo permutations of the ClonalFrame distance matrix.
Mantel correlograms were used to examine the spatial structure of E. coli subpopulations in soil (22). The range of distances between soil samples was divided into windows measuring 5 m across by using a spatial connectivity matrix. The significance of autocorrelation values within spatial lags was evaluated with 999 Monte Carlo permutations of the genetic distance matrix. The overall significance of autocorrelation functions was evaluated using the Holm method of multiple-testing correction.
Long-distance trends in the data were analyzed by computing the correlation between the ClonalFrame distance matrix and matrices of absolute (i.e., Manhattan) edaphic, landscape, and temporal distances while correcting for the effect of Euclidean geographic distance using partial Mantel tests (22). Environmental and landscape variables included in partial Mantel tests and variance partitioning were chosen using a stepwise selection technique based on the Akaike information criterion (AIC) as implemented in the stepAIC function of the MASS package in R. Stepwise model selection reduces the complexity of ecological analyses by using automated procedures to determine the combinations of measured factors that best explain the landscape genetic structure. Where both temporal and geographic distances had significant effects, the temporal effect was removed by detrending prior to tests. The significance of Mantel tests was evaluated with 106 Monte Carlo permutations of the ClonalFrame distance matrix.
Variance partitioning of the landscape genetic structure of soil E. coli isolates across spatial scales was performed (26). Briefly, ClonalFrame distance matrices were transformed into eigenvector matrices (i.e., artificial variables explaining the variation in genetic relationships) by using principal coordinate analysis, and all resulting eigenvectors were used in the variance partitioning analysis. Since partial Mantel tests had already been used to analyze effects on the largest spatial scale, nonspatial variables were detrended to remove the long-range spatial gradients from the data. The resulting residuals were used in the variance partitioning analysis to test the effects of nonspatial variables on spatial scales smaller than the whole field site. The Euclidean geographic distance between soil samples was decomposed to an eigenvector matrix by using the principal components on a neighbor matrix (PCNM) technique (11). Prior to variance partitioning, the combination of PCNM eigenvectors that best explained the genetic structure of soil E. coli isolates was determined using an orthogonal AIC procedure in the spacemakeR package, because the sampling scheme was spatially irregular (11). The significance of variance partitioning results was evaluated by analysis of variance (ANOVA) with 999 Monte Carlo permutations.
Accession numbers.
The MLSA sequences and corresponding environmental data were deposited in GenBank with accession numbers HM219874 to HM222344.
RESULTS
E. coli isolate collection.
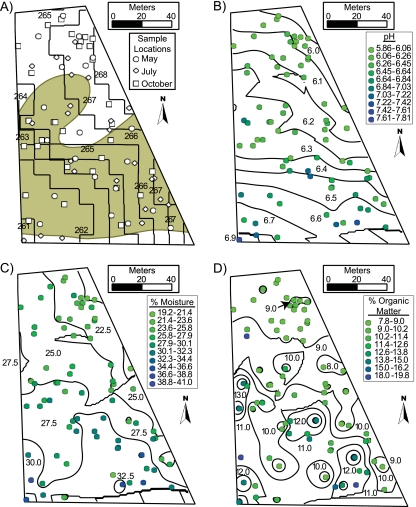
Topsoil cores from a recreational meadow in the Mitchell Street Natural Area (42°26′5.295″N, 76°28′16.97″W) of the Cornell Plantations were sampled in May, July, and October of 2008. Two soil series were present in the field: Hudson silt loam (HsB) and the related, albeit less water-permeable, Rhinebeck silt loam (RkB). The field was dominated by two hay grass species: Festuca elatior and Phleum pratense. A topographic depression in the southwest led to a short, steep gradient in soil moisture, and soil moisture decreased by approximately 1.5% per month over the three sample dates (Fig. 1 A). Otherwise, the site was characterized by increasing northeast-to-southwest gradients in pH, soil moisture (%M), and total organic matter (%OM) (Fig. 1B to D).
FIG. 1.
Contour plots of soil variables. (A) Contour plot of elevation. Black lines represent elevation contours, and black text indicates the contour values (10-m areal resolution). Points represent soil sample locations in May, July, and October 2008. The Rhinebeck silt loam area is shaded tan, and the Hudson silt loam area is unshaded. (B, C, and D) Contour plots of soil pH, %M, and %OM, respectively. Black contours represent the trend surface output from universal kriging accounting for a gradient in the data mean. Black text indicates the contour values. Colored points represent the measured value for each soil sample.
Attempts to isolate E. coli from topsoil yielded 394 isolates from 49 of 78 soil samples and 77 more isolates from six fecal deposit samples originating from deer (n = 2 samples), a small rodent, rabbits (n = 2 samples), and a dog. After genomic fingerprinting with rep-PCR to limit genotype redundancy within samples, 297 soil isolates and 56 fecal isolates were analyzed by MLSA. Nonsynonymous mutations were not observed in the data set, suggesting that the seven MLSA genes were not the direct targets of natural selection in the field site but might exhibit signals of selective pressures acting on the whole genome.
Results from a series of statistical tests to determine the spatial, temporal, landscape, and environmental structures imposed on E. coli populations in soil are presented here. These tests search for linear relationships between genetic distances and site characteristics. Modest recombination during the evolution of these isolates could conceal the pattern of vertical descent generated by the accumulation of mutations. Therefore, ClonalFrame was used to estimate genetic distances between isolates while correcting for the historic impact of recombination between E. coli lineages. The clonal phylogeny of field E. coli isolates consisted of 5 major clades, and analysis of field isolate relationships to E. coli reference collection (ECOR) strains allowed the majority of isolates to be assigned to existing ECOR groups (see Table S1 in the supplemental material). ECOR B1 constituted 40% of soil genotypes and could be subdivided into three clades, designated B1A, B1B, and ET-1. Eighteen isolates occupied divergent branches in the network and were excluded from genetic structure analyses due to their rarity. It is tempting to speculate that these long-branch isolates may represent cryptic lineages of the genus Escherichia (43), but further tests of this hypothesis are needed.
Lack of genetic isolation.
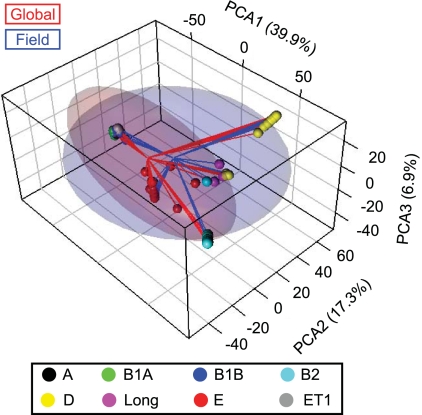
If E. coli in soils displayed genetic isolation from fecal and clinical isolates, then soil isolates might be on the path to speciation away from the global E. coli population. An ANODIS model was applied to test for evidence of DNA sequence divergence between the collection of field isolates and all comparison data available in the GenBank and STEC reference sequence databases for pathogenic, commensal, and environmental strains from a variety of hosts, clinical specimens, and habitats, designated the global E. coli collection. This test was designed to detect significant genetic differences between collections of organisms based on ClonalFrame estimates of genetic distance. The model contained terms for ECOR subpopulation membership, isolate collection membership, and their interaction. There was a small but statistically significant average divergence, with 1.9% of the total variance in genetic distance explained by the difference between global and field isolate collections and 2.4% of the variance in genetic distance explained by isolate collection within ECOR subpopulations (Fig. 2). The average nucleotide divergence between collections (Dx vs y) was similar to nucleotide polymorphism within collections (π; π for the field collection [πField] = 0.0160; πGlobal = 0.0155; DField vs Global = 0.0162). However, no fixed mutations between the global and field sequences were observed. Therefore, only small amounts of genetic differentiation exist between the E. coli population in the field site and a global sample of E. coli.
FIG. 2.
Principal coordinate analysis (PCA) of average genetic distance estimates from 501 ClonalFrame dendrograms. Sphere colors indicate subpopulation membership of isolates. Purple spheres depict isolates from long branches. Lines connect isolate positions to sequence collection centroids. Translucent ellipses represent 67% confidence interval clouds around the genetic distribution of sequence collections. Field E. coli isolates (n = 353) consisted of soil and fecal isolates from this study. Global E. coli isolates (n = 438) consisted of sequence types of diverse clinical, food, and environmental specimens from the GenBank and shigatox.net databases.
If soil isolates from this field site showed less divergence from fecal isolates than from the global E. coli sample, that would indicate that soil E. coli strains are not genetically distinct from fecal E. coli strains. The genetic divergence between field isolates from soil and fecal sources was statistically significant but explained only 0.2% of the total variance in genetic distance, with another 0.8% explained by soil versus fecal isolates within subpopulations. The soil and fecal isolates had no fixed mutations between them, but π was smaller for fecal isolates than for soil isolates, due probably to the small number of sampled fecal deposits (πSoil = 0.0168; πFecal = 0.0113; DFecal vs Soil = 0.0152). Therefore, field E. coli isolates represented a geographic subset of the global gene pool, and soil isolates have not substantially diverged from fecal isolates in the same field.
ANODIS also revealed that genetic divergence between E. coli subpopulations explained 63% of the total variation in genetic distance. This result suggested that even subtle shifts of subpopulation representation in soil samples could confound the analysis of ecological gradients by producing incorrect estimates of correlations between environmental and genetic variations. Therefore, ecological analyses within subpopulations were conducted. ClonalFrame analysis of MLSA data within subpopulations indicated that ECOR B2, D, and E were mainly clonal (see Table S1 in the supplemental material). The B1A, B1B, and ET-1 clades had a relatively high recombination rate, with 6 to 8 times more nucleotide changes due to homologous recombination than due to mutation, but small π values within these clades makes recombination rate estimates less precise (see Table S1 in the supplemental material).
Spatial structure of E. coli.
If fecal deposition was the main driver of E. coli distributions in soil, the spatial pattern of fecal deposition events and that of E. coli genotypes should coincide. The spatial structure of sampled data is analyzed by using autocorrelation coefficients which describe spatial gradients, patches (i.e., local aggregates), or boundaries (21). Mantel correlograms were generated for each E. coli subpopulation to examine patterns of spatial autocorrelation among isolate genotypes. The presence of autocorrelation peaks over increasing distance indicated that genotype similarity within subpopulations was structured in spatial patches that varied in size from <5 m to 10 m (Table 1; see also Fig. S1 in the supplemental material). Subpopulations B1A, B1B, B2, and E exhibited interpatch distances of 20 to 35 m (Table 1; see also Fig. S1 in the supplemental material), while interpatch distances for ECOR D were 40 to 45 m and those for clade ET-1 were 65 to 70 m.
TABLE 1.
Summary of E. coli spatial patterns and large-scale sources of genotype variationa
| Population or point pattern | Patch size (m) | Interpatch distance (m) | Spatial gradientb | Landscape gradientb | Edaphic gradientb | Temporal gradientb |
|---|---|---|---|---|---|---|
| B1A | 5 to 10 | 20 to 30 | 0.178* | 0.223** (ES) | 0.187** (PMO) | 0.163* |
| B1B | <5c | 20 to 30 | 0.295*** | NS | NS | 0.094* |
| B2 | <5c | 20 to 30 | 0.141** | NS | NS | 0.384*** |
| D | <5c | 40 to 45 | 0.117** | 0.261*** (S) | 0.231*** (PO) | 0.086** |
| E | <5c | 30 to 35 | 0.152* | 0.149* (E) | NS | 0.110* |
| ET1 | <5c | 65 to 70 | NS | NS | NS | 0.146* |
| Soil isolates | <5c | NS | NS | 0.065*** (S) | 0.149*** (O) | 0.095*** |
| Fecal deposition | 3 to 9 | 19 to 30 | NA | NA | NA | NA |
Summary of spatial autocorrelation patterns and trends in genetic structure on the scale of the entire field site. ND, not determined; NS, not significant.
Partial Mantel's R values are reported with P values of <0.05 (*), <0.01 (**), and <0.001 (***). Variables included in the edaphic and landscape matrices are listed in parentheses. P, pH; M, %M; O, %OM; E, elevation; S, slope.
Significant positive autocorrelation was not observed into the 10-m distance lag, preventing estimation of the smallest patch size.
A survey of mammalian fecal deposits was conducted to describe the spatial pattern of fecal deposition in the field. The survey revealed that the dominant source was deer (57 of 80 deposits), followed by dog (10 of 80), and rabbit (5 of 80). A single small rodent deposit was counted, but the contribution of small rodents to fecal deposition is certainly underestimated due to the difficulty in cataloging small fecal deposits. Origin could not be assigned for seven deposits. The modified Ripley's K statistic (Lhat) was used to analyze the spatial distribution of fecal deposition events in relation to simulations under spatial randomness. Negative Lhat values indicated that fecal deposition was clustered in spatial patches with variable patch sizes between 3 and 9 m and interpatch distances of 19 to 30 m (Table 1; see also Fig. S2 in the supplemental material). Therefore, patches of related E. coli genotypes had a spatial scale similar to that of fecal patches. Related genotypes and fecal events were both located at 20- to 30-m distances more often than expected under spatial randomness for four of six subpopulations, indicating that host deposition of E. coli strains is controlling much of the soil E. coli spatial pattern (see Fig. S3 in the supplemental material).
Nonspatial effects on E. coli structure.
If fecal deposition were the sole driver of landscape genetic structure, then E. coli isolate genotypes would exhibit no dependence on edaphic variation. Edaphic and landscape variables that correlated with genotype variation were selected using a stepwise model selection technique within subpopulations. Partial Mantel tests revealed significant correlations between genetic distance within subpopulations and nonspatial variables on the scale of the entire field site (Table 1). However, the majority of E. coli genotypes were structured in spatial patches with variable patch sizes and variable interpatch distances, so a linear gradient did not represent all of the spatial variation in genotypes (see Fig. S1 in the supplemental material). Geographic variation on the scales of patches and interpatch distances was analyzed using the PCNM technique. The PCNM analysis identified from 9 to 15 orthogonal spatial variables within subpopulations, and a model selection procedure identified 2 to 8 spatial variables that explained the genetic relationships within subpopulations.
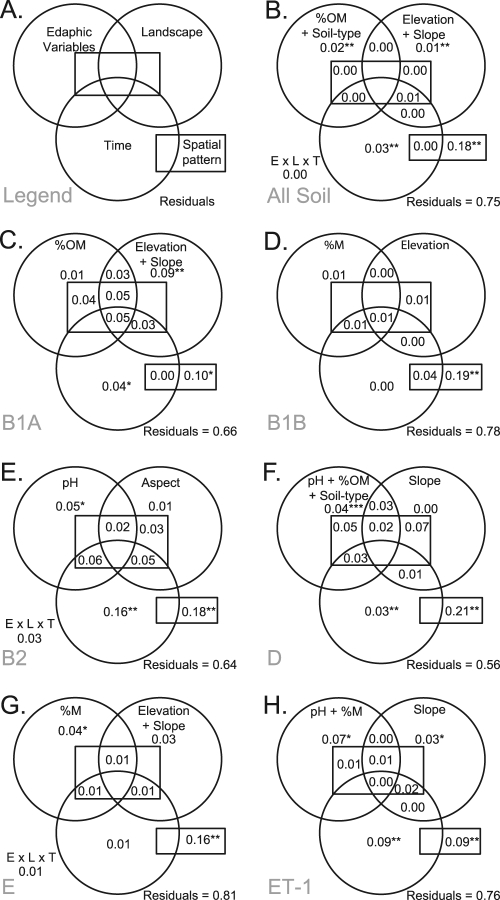
Landscape genetic structure within subpopulations was analyzed across spatial scales by variance partitioning on ordinations after detrending of the data to remove the large-scale gradients represented in Table 1. Variance partitioning identified %OM, terrain slope, elevation, time, and PCNM spatial variables as factors explaining significant variation in the landscape genetic structure of all soil E. coli strains in this field site (Fig. 3 B). When variance partitioning was applied within subpopulations, spatial variables explained significant amounts of landscape genetic structures, but less so in clades ET-1 and B1A (Fig. 3). Spatial variation was the largest component of the landscape genetic structure of E. coli populations, reinforcing the role of patchy fecal deposition in distributing E. coli. Clade B1B exhibited only a spatial pattern in genotypes, with large patches that lacked correspondence to measured nonspatial variables (see the B1B eigenvector 1 and 2 maps in Fig. S3 in the supplemental material).
FIG. 3.
Venn diagrams from variance partitioning of subpopulation genetic structures between edaphic, landscape, temporal, and spatial variables. Variables included in partitions are listed in each panel. The central rectangle represents interactions between the three nonspatial variance partitions and the spatial partition. Nonspatial interactions among edaphic, landscape, and temporal variables (E × L × T) are indicated near the lower left corners of diagrams. Proportions of variance explained by each partition are displayed, with the significance of F tests annotated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. (A) Legend; (B) all soil E. coli isolates; (C) clade B1A; (D) clade B1B; (E) ECOR B2; (F) ECOR D; (G) ECOR E; and (H) clade ET-1.
The combinations of landscape, temporal, and edaphic variables that best explained the landscape structure of E. coli strains differed among subpopulations (Fig. 3). Landscape variables had a significant relationship to some subpopulation distributions, and these effects are likely explained by host behaviors. For example, muddy lower elevations or steep slopes in this field may deter foraging by some animal hosts more than others, influencing the deposition of E. coli into soil. However, further work is needed to clarify the cause of this genetic correlation with landscape features. For example, clade B1A displayed very low genetic diversity in the north half of the field, while different B1A genotypes were isolated in the south, resulting in a relationship between site elevation and genetic structure (see the B1A eigenvector 2 map in Fig. S3 in the supplemental material). Sample date (i.e., time) had a significant relationship to landscape genetic structure in four subpopulations, with the largest impact on ECOR B2 distribution (Fig. 3E). The number of ECOR B2 isolates in soil decreased over time (number in May [nMay] = 28; nJul = 16; and nOct = 4), and the average genetic divergence between ECOR B2 isolates increased over time (DMay vs Jul. = 0.0067; DMay vs Oct. = 0.0071; and DJul. vs Oct. = 0.0047).
The soil environment predicted small but significant amounts of the landscape genetic structure for soil isolates, refuting the model that fecal deposition alone explains the distribution of E. coli in soil. Soil pH explained significant genetic variation in ECOR B2, ECOR D, and clade ET-1, soil %M explained the structures of ECOR E and clade ET-1, and soil %OM and soil type partially explained the structure of ECOR D (Fig. 3). Edaphic variables explained the distributions of clade ET-1 and ECOR E only on small spatial scales, whereas clade B1A genotypes displayed edaphic variation only on the scale of the whole field (Fig. 3 and Table 1). This result suggests that ET-1 and ECOR E distributions may be affected by short gradients in soil characteristics but that B1A genotypes may tolerate wider ranges of edaphic variation. In clade B1A, ECOR B2, and ECOR D, spatial and spatiotemporal patterns in edaphic variables also explained substantial amounts of the genetic distribution.
DISCUSSION
Density-dependent decline of E. coli abundance in soil and months-long persistence at 102 to 103 CFU per g soil have been reported in numerous publications (20, 40). In contrast, storm events and grazing may mobilize E. coli from soil toward new hosts over substantially shorter time intervals (14, 25). While transmission of E. coli through extrahost habitats is not the most efficient means to reach new hosts, the deposition of E. coli into soil provides a means for environmental selection to enrich locally adapted genotypes that may contribute to the genomic diversity of the E. coli species. It is possible that some E. coli genotypes are adapted for extrahost persistence, and these strains may act as a reservoir of stress tolerance genes in the global population. Neither the role of the environment in enriching locally adapted genotypes nor the contribution of these genotypes to the diversity of the E. coli species has been examined. The present work attempts to test for adaptation of E. coli strains to local soil environments as the first step in understanding the role that secondary habitats may play in generating and maintaining genomic diversity in the E. coli species.
MLSA examines genetic variation in housekeeping genes and provides only a coarse measure of the impact of environmental selection on genomes, with the majority of variation in MLSA genes attributable to random genetic drift punctuated by demographic events (38, 46). The presented MLSA scheme was chosen as a means of detecting environmental selection, because local soil populations are small, declining, and limited in dispersal, suggesting that selection for persistent strains should be detectable at the whole-genome level. Analysis of other genetic loci that are under direct selection by edaphic variables would likely yield stronger relationships between environment and gene sequence than were observed here. However, the direct genetic targets of environmental selection in soil were difficult or impossible to postulate based on the data available prior to this study. The results of this study provide a path toward the determination of genetic loci important for environmental persistence through continued genomic and phenotypic analyses of collections of locally adapted strains from geographic subsets of the E. coli species.
The present analysis of E. coli distributions in soil supports a model of a fecal deposition-driven spatial pattern overlaid by abiotic sorting of strains. The observed spatial patterns indicate that fecal deposition is the dominant field-scale process distributing E. coli. Correlation of E. coli genetic structure with landscape variables that likely affect host behavior supports this interpretation. The high proportion of ECOR B1 isolates in both the fecal and the soil isolate collections (40% in the soil collection and 38% in the fecal collection), combined with the low abundance of ECOR A (2% of all field isolates), is in good agreement with a recent meta-analysis of commensal E. coli population structure which concluded that ECOR B1 is the predominant subpopulation in wild and domestic animals while ECOR A and B2 are the predominant subpopulations in humans (38).
If the soil environment were indiscriminately lethal across E. coli genotypes, spatial and landscape effects should have been the only significant predictors of genetic relationships. Rather, edaphic and temporal patterns explained some of the genetic structure both within and across E. coli subpopulations, indicating that the soil environment is selectively sorting E. coli strains. Moreover, the observed impact of edaphic variables on genetic structure is probably a conservative estimate, because (i) the MLSA genes were not the direct targets of selection and (ii) unmeasured yet spatially structured edaphic variation might appear as pure spatial variation in both partial Mantel tests and variance partitioning. Even small impacts of secondary habitats on genotype distributions support the hypothesis that the soil environment affects the opportunity for E. coli genotypes to reach new hosts in the host-soil-water cycle. However, further testing is required to determine whether the soil environment selects for phenotypes beneficial for colonizing new hosts. In contrast, persistent genotypes may be subject to evolutionary trade offs demanding decreased fitness in the host gut in exchange for environmental persistence.
Changes in E. coli population structure following fecal deposition have been reported previously (16, 39, 45), but the present work links those changes to patterns of fecal deposition coupled with environmental selection. The utility of E. coli as a fecal indicator bacterium (FIB) in pollution surveillance is dependent on the predictability of the population changes imposed by secondary habitats. While the usefulness of E. coli as a FIB is a topic of active debate (4, 39), persistence of E. coli in soil may enable the application of landscape genetics to develop GIS models of soil reservoirs and transport paths in the environment from data about environmental isolates (9). Such methods might obviate source attribution, instead yielding geographic predictions of fecal pollution risk to water and food supplies and improving the ability of regulators to target landscapes for monitoring. Since E. coli ecology in secondary habitats is heavily impacted by land and waste management practices, the landscape genetics of E. coli should be considered across land uses (36).
It was important to define the extent of genetic isolation between soil isolates from this field and the global E. coli gene pool, because evidence of genetic isolation would suggest that soil isolates cannot recombine with other E. coli strains, including pathogens. Several studies have reported the genetic differentiation of E. coli populations in coastal and riverine soils versus fecal isolates, leading the investigators to conclude that naturalized E. coli populations may be confounding the analysis of fecal pollution in the water supply (6, 19). Two studies found only limited evidence for the genetic isolation of E. coli in soil and beach sands (4, 42) when genetic distinctions between ECOR subpopulations were taken into account (38). ANODIS indicated that the population structure of E. coli is a modern genetic continuum across geography and environment within mostly clonal ECOR groups and that isolates from the soils in this field site constitute a genetic subset of the E. coli species rather than a divergent lineage on the path to speciation. However, if the soil E. coli isolates had been examined as a single group, without division into subpopulations, the fecal and soil E. coli isolates might appear to be genetically differentiated, because ECOR E isolates were a larger proportion of the fecal than of the soil collection (40% of fecal versus 10% of soil isolates). This change in proportion of ECOR E generated a substantial shift in the fecal isolate collection away from the soil isolate collection, increasing the apparent genetic divergence between those groups 5-fold.
While the majority of E. coli subpopulations were structured on spatial scales that corresponded to the spatial pattern of fecal deposition, the genotypes in clades ET-1 and ECOR D were dispersed on considerably larger spatial scales. In particular, the low nucleotide diversity of clade ET-1 in 18 soil samples suggests that the ecological process dispersing ET-1 is operating on a spatial scale larger than those measured in this study. Walk and colleagues observed clade ET-1 to be abundant across six beach sites, and this wide distribution was taken as evidence for the naturalized state of clade ET-1 (42). While a broad geographic distribution does not absolutely exclude unknown host-driven processes, the present study supports the hypothesis that ET-1 is a naturalized subpopulation that may have colonized environments with diverse chemical and physical properties.
The ability to distinguish between ECOR subpopulations in this study was central to observing the landscape ecology of E. coli in soil. Ecological distinctions among the E. coli subpopulations were observed, with differing combinations of edaphic and landscape variables explaining their genetic structure. While the present work is the first quantitative analysis of edaphic impacts on E. coli landscape genetic structure, numerous studies have examined the effect of soil characteristics on E. coli survival. In particular, %M has often been hypothesized as a determinant of E. coli survival rates in secondary habitats (14). While %M partially explained the genetic structure of ECOR E and clade ET-1, the genetic structure of other subpopulations was explained by a variety of factors. Soil pH has also been hypothesized to be an important environmental variable for E. coli survival in soil (14), and our results suggest that E. coli genotypes display variation in persistence at different soil pH values. Moreover, the gradient in soil pH should induce variation in the bioavailability of toxic heavy metals in the study site, potentially compounding selective pressures on E. coli in soil.
No attempt was made to distinguish pathogens from nonpathogens in this study. In the environment, ecological differences between E. coli pathogens and nonpathogens have been observed to be small (1, 3, 14), but concerns have been raised about the utility of nonpathogenic FIB strains in estimating fecal pollution risk, mainly resulting from reports of naturalized E. coli (39). In this context, the strong temporal trend in ECOR B2 genotypes is interesting, because this subpopulation contains the greatest proportion of virulence factors (38). We interpret the temporal structure of ECOR B2 strains to indicate that turnover of this subpopulation due to death and deposition is fast relative to other E. coli subpopulations, and this may have implications for the ability of pathogens to persist outside the host relative to commensal strains. However, pathogens exist in all E. coli subpopulations, so direct analysis of pathogen ecology in comparison with nonpathogen ecology is warranted.
Ecological analysis of microbial communities and populations is often hindered by two strongly interdependent assumptions: (i) that species under study are ecologically homogeneous units and (ii) that environmental selection is the sole force in microbial biogeography. At issue is the choice of both the genetic and geographic scales for analysis of any ecological process (28). Population genetic analysis permitted the division of E. coli isolates into discrete subpopulations and partially circumvented noise due to conflicting ecological processes acting on these subpopulations. When variance partitioning was applied to Fusarium and to Burkholderia cepacia complex genotypes, edaphic variation explained 2 and 1.1% of genotype variation, respectively, with larger amounts of variation attributed to spatial variation in edaphic conditions (30, 47). A similar amount of genotypic variation was explained by edaphic variation within subpopulations of E. coli. Small-scale spatial processes were to be expected in this study, because E. coli populations were examined after deposition in a secondary habitat. However, even native soil microbes have displayed limited spatial ranges on field, landscape, and regional scales (27, 30, 47). It is increasingly clear that population genetic structure and the spatial distribution of microbes cannot be ignored if nonspatial ecological processes are to be elucidated.
Supplementary Material
Acknowledgments
This work was supported by USDA-Hatch Act Federal Formula Funds award no. NYC-125438 to D.H.B.
We gratefully acknowledge the provision of ET-1 sequence data by S. T. Walk. The STEC Center at Michigan State University provided ECOR strains for the subpopulation assignment of our isolates. The Cornell University Life Sciences Core Laboratories Center (CLC) performed the nucleotide sequencing. The members of the Buckley laboratory assisted with the fecal survey. Teresa Bergholz, J. Chris Gaby, and M. Todd Walter provided useful comments during the development of this article.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, G. L., S. J. Kenney, P. D. Millner, L. R. Beuchat, and P. L. Williams. 2006. Shedding of foodborne pathogens by Caenorhabditis elegans in compost-amended and unamended soil. Food Microbiol. 23:146-153. [DOI] [PubMed] [Google Scholar]
- 2.Avery, S. M., A. Moore, and M. L. Hutchison. 2004. Fate of Escherichia coli originating from livestock faeces deposited directly onto pasture. Lett. Appl. Microbiol. 38:355-359. [DOI] [PubMed] [Google Scholar]
- 3.Barker, J., T. J. Humphrey, and M. W. R. Brown. 1999. Survival of Escherichia coli O157 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, F. P., F. Abram, F. A. Chinalia, K. G. Richards, and V. O'Flaherty. 2010. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl. Environ. Microbiol. 76:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt, R. (ed.). 2004. Soil survey investigations report, no. 42. Soil survey laboratory methods manual. USDA Natural Resources Conservation Service, Lincoln, NE.
- 6.Byappanahalli, M. N., L. W. Richard, D. A. Shivelya, F. John, S. Ishii, and M. J. Sadowsky. 2007. Population structure of cladophora-borne Escherichia coli in nearshore water of Lake Michigan. Water Res. 41:3649-3654. [DOI] [PubMed] [Google Scholar]
- 7.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 8.Collins, R., S. Elliott, and R. Adams. 2005. Overland flow delivery of faecal bacteria to a headwater pastoral stream. J. Appl. Microbiol. 99:126-132. [DOI] [PubMed] [Google Scholar]
- 9.Cushman, S. A., K. S. McKelvey, J. Hayden, and M. K. Schwartz. 2006. Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am. Nat. 168:486-499. [DOI] [PubMed] [Google Scholar]
- 10.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dray, S., P. Legendre, and P. R. Peres-Neto. 2006. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol. Modell. 196:483-493. [Google Scholar]
- 12.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Field, K. G., and M. Samadpour. 2007. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 41:3517-3538. [DOI] [PubMed] [Google Scholar]
- 14.Fremaux, B., C. Prigent-Combaret, and C. Vernozy-Rozand. 2008. Long-term survival of Shiga toxin-producing Escherichia coli in cattle effluents and environment: an updated review. Vet. Microbiol. 132:1-18. [DOI] [PubMed] [Google Scholar]
- 15.Geospatial Data Team. 2010. Geographic Resources Analysis Support System (GRASS) GIS software. Open Source Geospatial Foundation, Vancouver, Canada.
- 16.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 17.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 19.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam, M., M. P. Doyle, S. C. Phatak, P. Millner, and X. P. Jiang. 2005. Survival of Escherichia coli O157:H7 in soil and on carrots and onions grown in fields treated with contaminated manure composts or irrigation water. Food Microbiol. 22:63-70. [Google Scholar]
- 21.Legendre, P., and M.-J. Fortin. 1989. Spatial pattern and ecological analysis. Vegetatio 80:107-138. [Google Scholar]
- 22.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed., vol. 20. Elsevier, Amsterdam, Netherlands.
- 23.Manel, S., S. Joost, B. K. Epperson, R. Holderegger, A. Storfer, M. S. Rosenberg, K. T. Scribner, A. Bonin, and M. J. Fortin. 2010. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol. Ecol. 19:3760-3772. [DOI] [PubMed] [Google Scholar]
- 24.McArdle, B. H., and M. J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297. [Google Scholar]
- 25.Oliver, D. M., L. Heathwaite, P. M. Haygarth, and C. D. Clegg. 2005. Transfer of Escherichia coli to water from drained and undrained grassland after grazing. J. Environ. Qual. 34:918-925. [DOI] [PubMed] [Google Scholar]
- 26.Peres-Neto, P. R., P. Legendre, S. Dray, and D. Borcard. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87:2614-2625. [DOI] [PubMed] [Google Scholar]
- 27.Philippot, L., D. Bru, N. P. Saby, J. Cuhel, D. Arrouays, M. Simek, and S. Hallin. 2009. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 11:3096-3104. [DOI] [PubMed] [Google Scholar]
- 28.Prosser, J. I., B. J. Bohannan, T. P. Curtis, R. J. Ellis, M. K. Firestone, R. P. Freckleton, J. L. Green, L. E. Green, K. Killham, J. J. Lennon, A. M. Osborn, M. Solan, C. J. van der Gast, and J. P. Young. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384-392. [DOI] [PubMed] [Google Scholar]
- 29.Rademaker, J. L. W., F. W. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 3.4.3:1-3.4.3:27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 30.Ramette, A., and J. M. Tiedje. 2007. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. U. S. A. 104:2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice, E. W., C. H. Johnson, M. E. Dunnigan, and D. J. Reasoner. 1993. Rapid glutamate decarboxylase assay for detection of Escherichia coli. Appl. Environ. Microbiol. 59:4347-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ripley, B. D. 1976. The second-order analysis of stationary point processes. J. Appl. Probab. 13:255-266. [Google Scholar]
- 33.Robison, B. J. 1984. Evaluation of a fluorogenic assay for detection of Escherichia coli in foods. Appl. Environ. Microbiol. 48:285-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozas, J. 2009. DNA sequence polymorphism analysis using DnaSP. Methods Mol. Biol. 537:337-350. [DOI] [PubMed] [Google Scholar]
- 35.Savageau, M. A. 1983. Escherichia coli habitats, cell-types, and molecular mechanisms of gene-control. Am. Nat. 122:732-744. [Google Scholar]
- 36.Semenov, A. V., E. Franz, L. van Overbeek, A. J. Termorshuizen, and A. H. van Bruggen. 2008. Estimating the stability of Escherichia coli O157:H7 survival in manure-amended soils with different management histories. Environ. Microbiol. 10:1450-1459. [DOI] [PubMed] [Google Scholar]
- 37.Storfer, A., M. A. Murphy, J. S. Evans, C. S. Goldberg, S. Robinson, S. F. Spear, R. Dezzani, E. Delmelle, L. Vierling, and L. P. Waits. 2007. Putting the “landscape” in landscape genetics. Heredity 98:128-142. [DOI] [PubMed] [Google Scholar]
- 38.Tenaillon, O., D. Skurnik, B. Picard, and E. Denamur. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207-217. [DOI] [PubMed] [Google Scholar]
- 39.Texier, S., C. Prigent-Combaret, M. H. Gourdon, M. A. Poirier, P. Faivre, J. M. Dorioz, J. Poulenard, L. Jocteur-Monrozier, Y. Moenne-Loccoz, and D. Trevisan. 2008. Persistence of culturable Escherichia coli fecal contaminants in dairy alpine grassland soils. J. Environ. Qual. 37:2299-2310. [DOI] [PubMed] [Google Scholar]
- 40.van Elsas, J. D., P. Hill, A. Chronakova, M. Grekova, Y. Topalova, D. Elhottova, and V. Kristufek. 2007. Survival of genetically marked Escherichia coli O157:H7 in soil as affected by soil microbial community shifts. ISME J. 1:204-214. [DOI] [PubMed] [Google Scholar]
- 41.Vos, M., and X. Didelot. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199-208. [DOI] [PubMed] [Google Scholar]
- 42.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 43.Walk, S. T., E. W. Alm, D. M. Gordon, J. L. Ram, G. A. Toranzos, J. M. Tiedje, and T. S. Whittam. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh, C. J., and J. Kunapo. 2009. The importance of upland flow paths in determining urban effects on stream ecosystems. J. North Am. Benthol. Soc. 28:977-990. [Google Scholar]
- 45.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Van Leeuwenhoek J. Microbiol. 55:23-32. [DOI] [PubMed] [Google Scholar]
- 46.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yergeau, E., K. Labour, C. Hamel, V. Vujanovic, A. Nakano-Hylander, R. Jeannotte, and M. St.-Arnaud. 2010. Patterns of Fusarium community structure and abundance in relation to spatial, abiotic and biotic factors in soil. FEMS Microbiol. Ecol. 71:34-42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.